In silico investigation of DNA minor groove binding bibenzimidazoles in the context of UVA phototherapy†
Raymond C.
Beh
 ab,
Eleni
Pitsillou
ac,
Julia J.
Liang
ab,
Eleni
Pitsillou
ac,
Julia J.
Liang
 ac,
Andrew
Hung
c and
Tom C.
Karagiannis
*ab
ac,
Andrew
Hung
c and
Tom C.
Karagiannis
*ab
aEpigenomic Medicine, Department of Diabetes, Central Clinical School, Monash University, Melbourne, VIC 3004, Australia. E-mail: tom.karagiannis@monash.edu; Tel: +61 400 857 906
bDepartment of Clinical Pathology, The University of Melbourne, Parkville, VIC 3052, Australia
cSchool of Science, College of Science, Engineering & Health, RMIT University, VIC 3001, Australia
First published on 6th December 2021
Abstract
The versatility of DNA minor groove binding bibenzimidazoles extends to applications in cancer therapy, beyond their typical use as DNA stains. In the context of UVA phototherapy, a series of halogenated analogues designated ortho-, meta-, and para-iodoHoechst have been investigated. Phototoxicity involves dehalogenation of the ligands following exposure to UVA light, resulting in the formation of a carbon-centred radical. While the cytotoxic mechanisms have been well established, the nature and severity of DNA damage induced by the ortho-, meta-, and para-iodoHoechst isomers requires clarification. Our aims were to measure and compare the binding constants of iodoHoechst analogues, and to determine the proximity of the carbon-centred radicals formed following photodehalogenation to the C1′, C4′, and C5′ DNA carbons. We performed molecular docking studies, as well as classical molecular dynamics simulations to investigate the interactions of Hoechst ligands with DNA including a well-defined B-DNA dodecamer containing the high affinity AATT minor groove binding site. Docking highlighted the binding of Hoechst analogues to AATT regions in oligonucleotides, nucleosomes, and origami DNA helical bundles. Further, MD simulations demonstrated the stability of Hoechst ligands in the AATT-containing minor groove over microsecond trajectories. Our findings reiterate that the efficiency of dehalogenation per se, rather than the proximity of the carbon-centred radicals to the DNA backbone, is responsible for the extreme phototoxicity of the ortho- isomer compared to the meta- and para-iodoHoechst isomers. More generally, our analyses are in line with the potential utility of ortho-iodoHoechst in DNA-targeted phototherapy, particularly if combined with a cell-specific delivery system.
Introduction
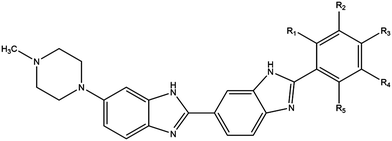
Phototherapy involves the use of ultraviolet light (UV) to treat skin lesions in particular cutaneous T-cell lymphoma (CTCL), psoriasis, and vitiligo.1–3 While UVB (280–315 nm) is used alone (especially in narrowband UVB 311–312 nm), UVA therapy is combined with a photosensitiser and can be referred to as photochemotherapy.4–6 Initial attempts involved incorporation of a halogenated DNA nucleoside analogue into the DNA followed by UVA-irradiation to induce DNA damage; bromouracil is a prototypical example.7–11 The most commonly used modality involves the combination of UVA with psoralens known as psoralen and UVA therapy (PUVA).12–15 The plant-derived phototoxic psoralens can be administered either orally or topically and this is followed by UVA-irradiation to induce photolesions; fluences in the order of kJ m−2 are required for the phototoxic effects.12–15 We have developed experimental DNA minor groove binding ligands which are significantly more potent requiring fluences in the order of J m−2 for effectiveness.16,17The commercially available DNA minor groove binding bibenzimidazole analogues, Hoechst 33258, and Hoechst 33342, were developed in the early 1970s by the Hoechst chemical company as potential antihelminths.18 Given their intrinsic fluorescent properties, they are currently widely used as vital DNA stains, taking advantage of the increase in fluorescence yield upon binding to DNA. Hoechst 33258 and 33342 are planar drug molecules consisting of two consecutive bibenzimidazole rings, a positively charged N-methylpiperazine group and a substituted aromatic ring; they differ only in the substitution on the terminal ring – Hoechst 33258 contains a phenolic moiety whereas Hoechst 33342 contains a methoxy group (Fig. 1). The structural motif produces an arc conformation of the planar drug molecules, which allows them to fit into the minor groove of the DNA in regions where there are predominantly adenine (A) and thymine (T) base pairs.19
By substituting chemical moieties on the terminal phenolic ring, the versatility of Hoechst-based DNA minor groove binding ligands has been extended to numerous other applications. Synthetic analogues have been developed as potential radiation protectors, exemplified by methylproamine.20,21 Radioiodinated analogues have potential utility in nuclear medicine as highlighted by DNA-targeted Auger radiotherapy (123I, 125I) and positron emission tomography (124I, PET).22–27 By halogenating Hoechst ligands with stable iodine atoms (127I), utilisation in DNA-targeted phototherapy using UVA light (∼350 nm) has been considered (Fig. 1).16,28
In the context of phototherapy, a series of iodinated analogues have been investigated as potential UVA photosensitisers.29 Directly iodinated Hoechst 33258, referred to as iodoHoechst 33258, has been shown to be a potent UVA sensitiser in cell-free systems (DNA) and cell culture experiments.17,30,31 Subsequently, three iodoHoechst isomers – with the only difference being the position of the iodine atom on the phenyl ring – designated ortho- (also referred to as UVASens), meta-, and para-iodoHoechst have been synthesised.29,32 Phototoxicity involves dehalogenation of the ligands following exposure to UVA light, resulting in the formation of a carbon-centred radical on the DNA causing the formation of DNA lesions and cell-death.17,30,31 Clonogenic survival assays indicate that the potency of phototoxicity for the iodinated ligands is ortho- ≫ meta-, para-iodoHoechst > iodoHoechst 33258.29
While the in vitro mechanisms have been well established, the nature and severity of DNA damage induced by the different isomers requires clarification. Here, we performed a series of in silico experiments to highlight the binding of Hoechst DNA minor groove binding bibenzimidazoles to the various forms of DNA (B-, A-, and Z-DNA), nucleosome core particles (containing the core histones), and to elongated strands of origami DNA. Our main aims were to compare the binding constants of the three iodoHoechst (ortho-, meta-, and para) isomers to DNA, and importantly to measure the proximity of the carbon-centred radicals formed following dehalogenation to the C1′, C4′, and C5′ carbons, to evaluate lesions on the DNA. Along with docking studies, we performed classical molecular dynamics simulations on a microsecond timescale to gain new atomistic insights between the three isomers, with respect to binding stability and DNA damaging propensity.
Materials and methods
Molecular docking and blind docking of Hoechst and analogues to different genomic structures
The crystal structures of A-DNA (PDB ID: 1QPH), B-DNA (PDB ID: 1DNH, 129D, 5DAM, and 448D), and Z-DNA (PDB ID: 4OCB) were obtained from the RCSB Protein Data Bank.33–39 Crystal structures of the human nucleosome core particle (PDB ID: 2CV5), human telomeric nucleosome (PDB ID: 6KE9), nucleosome containing methylated SAT2R DNA (PDB ID: 5CPJ), and nucleosome containing acetylated histones (PDB ID: 4YS3) were also obtained from the RCSB PDB.40–43 Additionally, the cryo-EM structure of a 16-helix bundle DNA origami object was utilised (PDB ID: 7ARQ).44 The co-crystallised ligands and water molecules were removed from the structures in preparation for molecular docking. The chemical structures of Hoechst 33258, Hoechst 33342, iodoHoechst 33258, meta-iodoHoechst, para-iodoHoechst, and phenylHoechst were drawn using ChemDraw 19.0 (PerkinElmer, MA, USA). The three-dimensional structures of the ligands were obtained using Chem3D 19.0 (PerkinElmer, MA, USA). The structure of ortho-iodoHoechst was obtained from the National Center for Biotechnology Information PubChem.45The DNA and nucleosome structures were imported into AutoDockTools-1.5.6 and were prepared as macromolecules.46 The compounds were also prepared as ligands in AutoDockTools-1.5.6.46 The receptor grids were generated using the coordinates of the co-crystallised ligands present in the B-DNA crystal structures. The grids were 20 × 20 × 20 Å in size. Blind docking was also performed on the A-DNA, B-DNA, Z-DNA, and nucleosome structures. The receptor grid was generated around the entire crystal structure. An exhaustiveness of 2048 was used and the number of modes was set to 20.
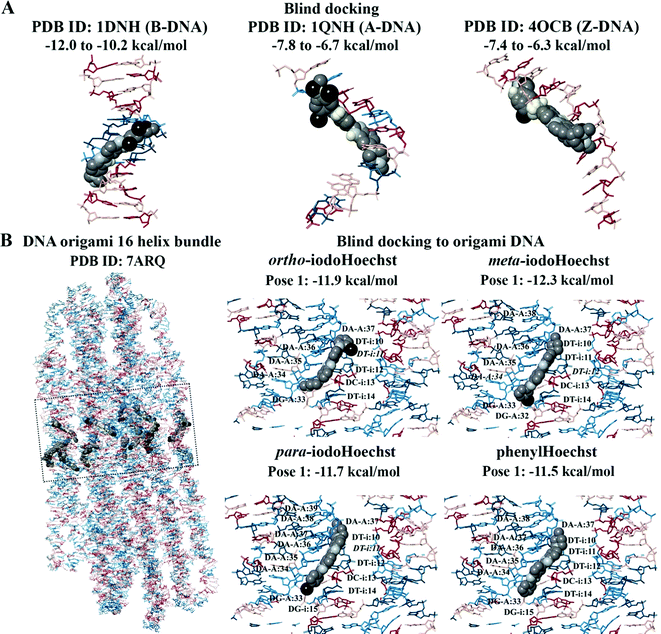
For the 16-helix bundle DNA origami object, blind docking was performed using an exhaustiveness of 8 and the grid was 50 × 150 × 50 Å in size (centre x: 403.3, y: 413.6, z: 418.1). Iterative docking was performed with ortho-iodoHoechst, where co-ordinates of the docked ligand were retained in subsequent docking calculations. Docking was performed (exhaustiveness = 8) in this manner for a total of 64 iterations, with the final ligand-nucleosome complex (PDB ID: 2CV5) containing 64 ligands. AutoDock Vina was used to perform the docking calculations and the jobs were run on the Spartan High Performance Computing (HPC) system.47,48 The output files were saved using PyMOL and the results were visualised in Maestro.49,50
Molecular dynamics (MD) simulation procedures
Molecular dynamics (MD) simulations were carried out using GROMACS 2018.251,52 using the AMBER parmbsc1 force field for DNA53 and TIP3P water.54 The initial structure for DNA was obtained from the crystal structure 1DNH.34 The docked ligands as described above served as starting structures, with their topologies generated using ACPYPE.55 A total of six systems were studied: ligand-free DNA, and DNA bound with Hoechst 33258, ortho-iodoHoechst, meta-iodoHoechst, para-iodoHoechst, and phenylHoechst. Systems were solvated in a triclinic box under period boundary conditions with a minimum distance 1.0 nm between any DNA atom to the box edge. The solvated system was neutralised and salted with 0.15 M NaCl. Energy minimisation was performed using the steepest-descent gradient method. NVT equilibration was performed for 100 ps at a temperature of 310 K with a modified Berendsen thermostat,56 followed by 100 ps under the NPT ensemble for 100 ps using the Parrinello–Rahman barostat57 to maintain pressure at 1.0 bar. Bond lengths constrained using the LINCS algorithm,58 long-range electrostatic forces were calculated using particle-mesh Ewald scheme (PME)59 (grid spacing 0.16 nm). Cut-off ratios of 1.2 nm were used for Coulomb and van der Waals potentials for calculation of short-range nonbonded interactions. Simulations performed with a time-step of 2 fs for 1 μs in triplicate for each system with different starting velocities.Analysis was performed on the 1 μs production runs using gmx rms, gms rmsf, and gmx distance within GROMACS. The molecular mechanics Poisson–Boltzmann surface area (MM-PBSA) method was employed for the calculation of binding free energy using g_mmpbsa.60 MM-PBSA calculations were performed on the final 100 ns segment of triplicate trajectories. Energy contributions from electrostatic, van der Waals, and polar solvation terms were calculated using the adaptive Poisson–Boltzmann Solver (APBS).61 Grid spacing was set to 0.05 nm, and values of 80 and 2 were used for the solvent and solute dielectric constants, respectively. Solvent-accessible surface area (SASA) was used to approximate the non-polar energy contribution, with the probe radius was set to 0.14 nm. Entropic energy terms were excluded from the calculations. Trajectories were visualised using VMD 1.9.3.62
Results and discussion
The binding of Hoechst-based bibenzimidazoles to consecutive AATT base pairs in the minor groove of DNA has been well-investigated. The sequence selectivity was first identified using 125I-labelled Hoechst 33258, prepared by direct iodination of Hoechst 33258 by electrophilic aromatic substitution, and plasmid DNA.63,64 This landmark study was then confirmed with Hoechst 33258 using DNA footprinting assays.65,66 These studies validated the high affinity binding site in A–T rich regions in the minor groove. The initial crystal structure (PDB ID: 8BNA) using the DNA dodecamer CGCGAATTCGCG indicated, in contrast to the footprinting experiments, binding of Hoechst 33258 to ATTC instead of AATT.67 The subsequent X-ray crystal structure (PDB ID: 1DNH) of the same complex was in agreement with the earlier experimental findings.34 The crystal structure indicated that binding of Hoechst 33258 covers the four AATT base pairs. Here, we investigated the binding characteristics of Hoechst binding ligands to different DNA structures.Molecular docking of Hoechst ligands to DNA
In the crystal structure (PDB ID: 1DNH), the co-crystallised Hoechst 33258 ligand is surrounded by the bases DA-B:17 to DG-B:22 and DG-A:4 to DG-A:10 (within 5 Å).34 The co-crystallised Hoechst 33258 ligand was predicted to bind with an affinity of −11.7 kcal mol−1 and the RMSD was found to be 2.02 Å (Fig. 2A). When the Hoechst ligands were docked to the B-DNA crystal structure, they were found to be surrounded by the AATT base pairs and the binding affinities can be seen in Fig. 2B. Hoechst 33258, iodoHoechst 33258, ortho-iodoHoechst, and phenylHoechst were predicted to form hydrogen bonds with DT-B:20, in the AT binding site.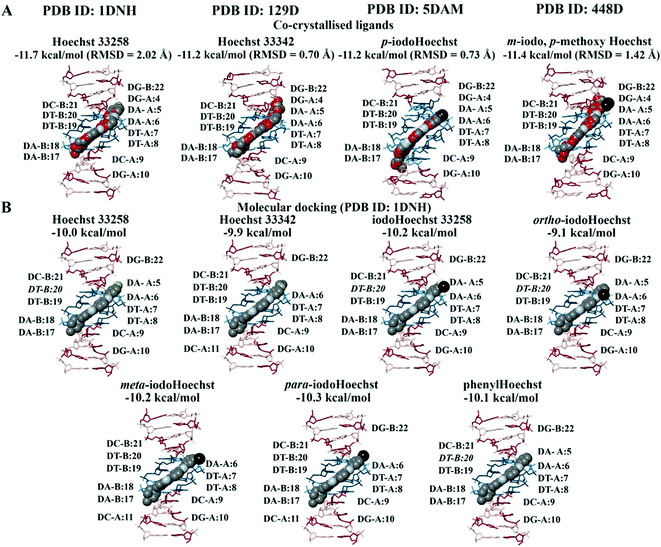 | ||
| Fig. 2 Molecular docking of the Hoechst ligands to the minor groove in the B-DNA. The orientation and position of the co-crystallised Hoechst analogues (A) in the high affinity AATT site are depicted. The binding affinities and RMSD values of the Hoechst ligands are provided (PDB ID: 1DNH, 129D, 5DAM, and 448D). The docking results for Hoechst 33258, Hoechst 33342, iodoHoechst 33258, ortho-, meta, and para-iodoHoechst can be seen (B). The bases A, T, C, and G are coloured cyan, dark teal, dark red, and pink, respectively. Iodine atoms are coloured black, hydrogen atoms are coloured light yellow, nitrogen atoms are coloured silver, and oxygen atoms are coloured tan. For the co-crystallised ligands, the carbon atoms are coloured red. | ||
The Hoechst analogues were also docked to several additional crystal structures of B-DNA that were available on the RCSB PDB for comparison.37–39 This included the structure of d(CGCAATTCGCG) in complex with Hoechst 33342 (PDB ID: 129D), d(CGCAAATTTGCG) in complex with p-iodoHoechst (PDB ID: 5DAM), and d(CGCGAATTCGCG) in complex with m-iodo, p-methoxy Hoechst (PDB ID: 448D). In comparison to the co-crystallised ligands, our calculated RMSD values were 0.70 Å for Hoechst 33342, 0.73 Å for p-iodoHoechst, and 1.42 Å for m-iodo, p-methoxy Hoechst (Fig. 2A). Like the co-crystallised ligands, the molecular docking results showed that the Hoechst analogues were binding to the AATT site in the B-DNA and were forming hydrogen bonds with the relevant base pairs. The binding affinities are provided in Fig. S1 and S2 (ESI†). Overall, these findings are in accordance with those previously published.68–70
Blind docking was performed on the crystal structures of A-DNA, B-DNA, and Z-DNA (Fig. 3A). The results revealed that Hoechst 33258, Hoechst 33342, iodoHoechst 33258, meta-iodoHoechst, ortho-iodoHoechst, para-iodoHoechst, and phenylHoechst were positioned in the minor groove of the B-DNA oligonucleotide. The binding affinities of the ligands were stronger for B-DNA (−12.0 to −10.2 kcal mol−1) compared to A-DNA (−7.8 to −6.7 kcal mol−1) and Z-DNA (−7.4 to −6.3 kcal mol−1). Furthermore, blind docking to a portion of an origami DNA cryo-EM structure indicated that the top-ranking pose of each ligand bound, in the minor groove, to a section of DNA that was comprised of AT base pairs, consistent with our expectations (Fig. 3B).
Blind and iterative docking to human nucleosome core particles
We investigated a generic human nucleosome core particle (PDB ID: 2CV5), a telomeric nucleosome (PDB ID: 6KE9), and a nucleosome containing acetylated histones (PDB ID: 4YS3), for which the binding characteristics of the Hoechst ligands was similar (Fig. 4A). For the human nucleosome core particle and nucleosome with acetylated histones, the blind docking results demonstrated that the Hoechst analogues were preferentially binding to the DNA and that there were no poses positioned in the histone complex. There were 21 ligand poses that were binding to the AATT site in the nucleosomal DNA (PDB ID: 2CV5) and this region consisted of the bases DA-72 to DT-75 on chain I and DA-218 to DT-221 on chain J. The binding affinities of the ligand poses that were predicted to bind to this site ranged from −12.5 to −10.8 kcal mol−1. Moreover, 53 ligand poses were positioned in a region of the DNA that was comprised of the bases DG-J:185 to DC-J:195 and DA-I:99 to DA-I:111.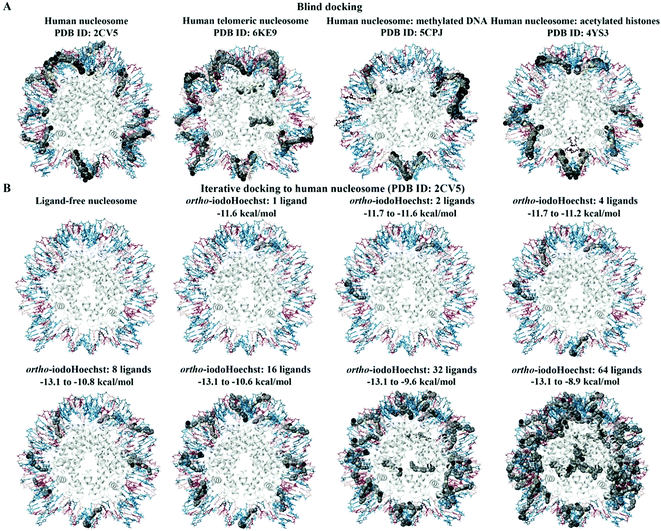 | ||
| Fig. 4 Blind and iterative docking to human nucleosomes. The blind docking results for the human nucleosome (PDB ID: 2CV5), human telomeric nucleosome (PDB ID: 6KE9), human nucleosome with methylated DNA (PDB ID: 5CPJ), and human nucleosome with acetylated histones (PDB ID: 4YS3) are shown (A). The methylated DNA and acetylated histones are coloured brown. ortho-IodoHoechst was docked to the generic nucleosome core particle (PDB ID: 2CV5) for a total of 64 consecutive iterations (B). The results for 1, 2, 4, 8, 16, 32, and 64 ligands can be seen. The bases A, T, C, and G are coloured cyan, dark teal, dark red, and pink, respectively. Iodine atoms are coloured black, hydrogen atoms are coloured light yellow, nitrogen atoms are coloured silver, and oxygen atoms are coloured tan. The histone chains are coloured grey. | ||
The major difference in binding properties was observed in the nucleosome containing methylated DNA (Fig. 4A). The blind docking results for the human nucleosome with methylated DNA (PDB ID: 5CPJ) revealed that the ligands were preferentially binding to a site that consisted of the bases DG-123 to DG-133 on chain I and DG-15 to DG-25 on chain J. There were 103 ligand poses within this region of DNA. Similarly, several ligands were predicted to bind to the bases DC-67 to DC-81 on chain J and DA-68 to DG-80 on chain I. Although most of the ligands were binding to the DNA, there were some poses that were predicted to bind to the histones for the human nucleosome with methylated DNA and human telomeric nucleosome, with significantly lower affinity compared to the AT binding site.
The human nucleosome core particle (PDB ID: 2CV5) and ortho-iodoHoechst were used for docking for a total of 64 consecutive iterations (Fig. 4B). The first 20 ligands preferentially bound to the nucleosomal DNA, with the majority in the AT-rich region. The 21st ligand and only a total of 11 ligands were bound exclusively to histones with a lower affinity than the high-affinity AT binding site on DNA. Essentially, binding to histones only occurred once the high affinity binding sites were saturated, again reiterating the preference of the DNA minor groove binding ligand for AT regions in the DNA (Table S1, ESI†).
Binding and stability of Hoechst ligands to DNA dodecamer
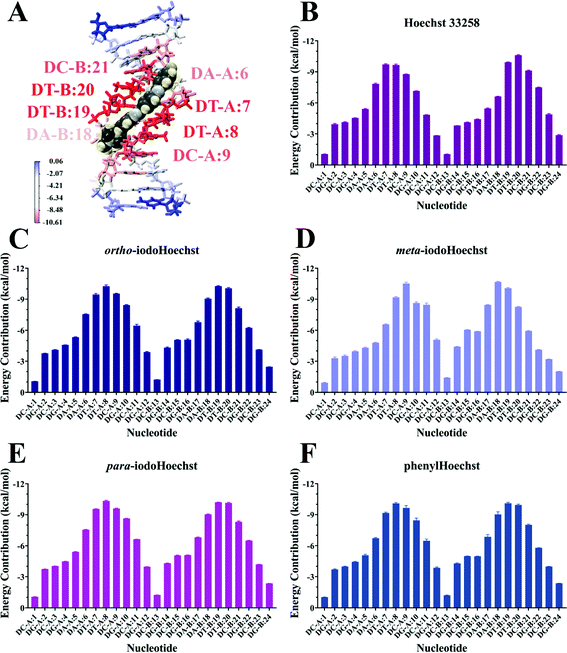
MD simulations demonstrate the stability of Hoechst ligands in the minor groove of the DNA dodecamer over a microsecond, demonstrated by the stability of both the DNA and ligands in all systems (Movies S1–S6, ESI†). RMSD remained largely the same across the entire course of the trajectory, with an average of 0.18 nm for DNA all systems (Fig. S3, ESI†). Ligand RMSD was also stable throughout the trajectory: 0.05 nm for Hoechst 33258, 0.08 nm for ortho- and meta-iodoHoechst, 0.06 nm for para-iodoHoechst, and 0.10 nm for phenylHoechst (Fig. S3, ESI†). Compared to ligand-free DNA, RMSF analysis indicated a modest suppression of ∼0.02 nm in fluctuation of nucleotides in the AATT region of the dodecamer upon binding of Hoechst ligands (Fig. S3, ESI†).MM-PBSA analysis indicates that ΔGbinding for all Hoechst ligands were similar, with electrostatic forces largely driving binding to DNA (Table 1). The accuracy of MM-PBSA has previously been discussed, and it is known that MM-PBSA can overestimate binding free energies.71–73 Nevertheless, calculated binding free energies of Hoechst ligands exhibit relative binding energies consistent with docking results (Fig. 2). Per-nucleotide binding energies indicate stronger energy contributions from AT bases in the dodecamer, with nucleotides contributing approximately −10 kcal mol−1 to binding of the ligands (Fig. 5). Our overall aim was to evaluate the binding of a series of halogenated iodoHoechst ligands, including the ortho-, meta-, and para-iodoHoechst isomers, to clarify their potential mechanism of action in the context of DNA-targeted phototherapy. Previously, using extensive cell culture experiments, we have highlighted the efficient dehalogenation of the ortho isomer (D37% = 49 ± 2 J m−2) compared to the meta (D37% = 251 ± 32 J m−2) and para isomers (D37% = 327 ± 29 J m−2).29 Further, the inefficient repair of DNA lesions induced by dehalogenation of the ortho isomer compared to the meta and para isomers has been highlighted, using γH2AX as a molecular marker.74
| Hoechst 33258 | ortho-IodoHoechst | meta-IodoHoechst | para-IodoHoechst | phenylHoechst | |
|---|---|---|---|---|---|
| ΔEvdW | −62.8 ± 0.1 | −65.2 ± 0.0 | −65.6 ± 0.2 | −66.8 ± 0.0 | −63.4 ± 0.2 |
| ΔEelec | −293.6 ± 0.1 | −300.8 ± 0.3 | −283.3 ± 1.2 | −300.7 ± 0.2 | −299.2 ± 2.1 |
| ΔGpolar | 88.3 ± 0.3 | 93.6 ± 0.1 | 87.4 ± 0.8 | 93.4 ± 0.2 | 93.7 ± 2.2 |
| ΔGnonpolar | −4.7 ± 0.0 | −4.9 ± 0.0 | −4.8 ± 0.0 | −5.0 ± 0.0 | −4.7 ± 0.1 |
| ΔG bind | −272.9 ± 0.3 | −277.2 ± 0.2 | −266.3 ± 0.6 | −279.1 ± 0.4 | −273.6 ± 0.3 |
A key question that remains is whether the different cross-sections for dehalogenation are responsible for the extreme photopotency of the ortho isomer compared to meta- and para-iodoHoechst. Previous studies have tentatively assigned photocleavage to the C1′ carbon for ortho-iodoHoechst and C5′ carbon for meta- and para-iodoHoechst.16,31 According to DNA minor groove binding ligand![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
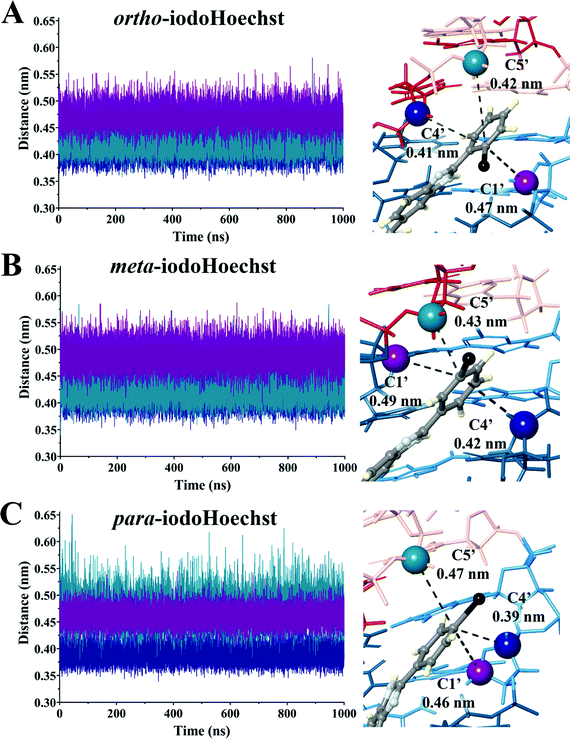
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DNA ratios, complex patterns of photolesions could be anticipated.75 Here, we focused on the high affinity Hoechst ligand binding sites which would most accurately reflect the situation in vitro and in vivo given the very low concentration required to induce DNA damage and cell death.29 We investigated the proximity of the carbon-centred radical formed following dehalogenation of each ligand to deoxyribose backbone carbons to estimate varying targets for photoextraction of hydrogen atoms (Table 2 and Fig. S4–S6, ESI†). These were calculated as an average between triplicate simulations over the entire trajectory for ortho-, meta-, and para-iodoHoechst (Fig. 6). Distances fluctuated between approximately 0.35 to 0.60 nm for all systems, except for the distance between the para-iodoHoechst carbon-centred radical and C5′ of the deoxyribose backbone which exhibited a slightly higher range in distances. Despite this, the average distance between the examined backbone carbons remained less than 0.50 nm, and the phototoxicity of the ligands cannot be attributed to the distance of the carbon-centred radical to the deoxyribose carbon.
DNA ratios, complex patterns of photolesions could be anticipated.75 Here, we focused on the high affinity Hoechst ligand binding sites which would most accurately reflect the situation in vitro and in vivo given the very low concentration required to induce DNA damage and cell death.29 We investigated the proximity of the carbon-centred radical formed following dehalogenation of each ligand to deoxyribose backbone carbons to estimate varying targets for photoextraction of hydrogen atoms (Table 2 and Fig. S4–S6, ESI†). These were calculated as an average between triplicate simulations over the entire trajectory for ortho-, meta-, and para-iodoHoechst (Fig. 6). Distances fluctuated between approximately 0.35 to 0.60 nm for all systems, except for the distance between the para-iodoHoechst carbon-centred radical and C5′ of the deoxyribose backbone which exhibited a slightly higher range in distances. Despite this, the average distance between the examined backbone carbons remained less than 0.50 nm, and the phototoxicity of the ligands cannot be attributed to the distance of the carbon-centred radical to the deoxyribose carbon.
| Ligand | DehalogenationaD37% (J m−2) | Dehalogenation events for cell-death #a (N50) | Carbon-centred radical distance from deoxyribose carbon (nm) | ||
|---|---|---|---|---|---|
| C1′ | C4′ | C5′ | |||
| a From Karagiannis et al. (2006).24 Dehalogenation of 37% assuming a Poisson distribution (D37%), and the number of dehalogenation events required to kill the average cell for each ligand (N50). | |||||
| ortho-IodoHoechst | 49 ± 2 | 1.23 ± 0.04 × 104 | 0.47 | 0.41 | 0.42 |
| meta-IodoHoechst | 251 ± 32 | 3.92 ± 0.29 × 104 | 0.49 | 0.42 | 0.43 |
| para-IodoHoechst | 327 ± 29 | 11.6 ± 0.90 × 104 | 0.46 | 0.39 | 0.47 |
Conclusions
Overall our results are consistent with the known high affinity binding of DNA minor groove binding bibenzimidazoles in consecutive AT regions. Binding of ligands to oligonucleotides, nucleosomes, and origami DNA reiterates the preference of the ligands for the AT-rich minor groove compared to alternative binding sites and proteins. MD simulations demonstrate the extreme stability of the ligands in the minor groove over the course of all-atom microsecond trajectories. With respect to DNA-targeted phototherapy, our findings indicate that the efficiency of dehalogenation per se, rather than the proximity of the carbon-centred radicals to the DNA backbone, is responsible for the extreme phototoxicity of the ortho-isomer compared to the meta- and para-iodoHoechst isomers. More generally, our analyses are in line with the potential utility of halogenated DNA minor groove binding bibenzimidazoles in DNA-targeted phototherapy, particularly if combined with a cell-specific delivery system.Author contributions
T. C. K. and A. H. conceptualized the aims and methodology, were involved in supervision, and production of the first draft of the manuscript. R. C. B., E. P. and J. J. L. performed data analysis and curated data and produced the first draft of the manuscript. All authors contributed to editing and reviewing the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We would like to acknowledge intellectual and financial support by McCord Research (Iowa, USA). JJL is supported by an Australian Government Research Training Program Scholarship. We are indebted to Alfonso Perez Escudero and the team for enabling access to supercomputing facilities. We thank the National Computing Infrastructure (NCI), and the Pawsey Supercomputing Centre in Australia (funded by the Australian Government). Further, we thank the Spartan High Performance Computing service (University of Melbourne), and the Partnership for Advanced Computing in Europe (PRACE) for awarding the access to Piz Daint, hosted at the Swiss National Supercomputing Centre (CSCS), Switzerland.References
- R. Vangipuram and S. R. Feldman, Oral Diseases, 2016, 22, 253–259 CrossRef CAS PubMed.
- S. Ahmed, H. Khan, M. Aschner, H. Mirzae, E. Küpeli Akkol and R. Capasso, Int. J. Mol. Sci., 2020, 21(16), 5622 CrossRef CAS PubMed.
- J. A. Parrish, T. B. Fitzpatrick, L. Tanenbaum and M. A. Pathak, N. Engl. J. Med., 1974, 291, 1207–1211 CrossRef CAS PubMed.
- T. Gambichler, F. Breuckmann, S. Boms, P. Altmeyer and A. Kreuter, J. Am. Acad. Dermatol., 2005, 52, 660–670 CrossRef PubMed.
- H. Hönigsmann, W. Brenner, W. Rauschmeier, K. Konrad and K. Wolff, J. Am. Acad. Dermatol., 1984, 10, 238–245 CrossRef.
- H. Hönigsmann and A. Tanew, in Dermatological Phototherapy and Photodiagnostic Methods, ed. H. Hönigsmann, C. A. Elmets and J. Krutmann, Springer Berlin Heidelberg, Berlin, Heidelberg, 2009, pp. 135–149 Search PubMed.
- J. Zimbrick, J. Ward and L. Myers, Int. J. Radiat. Biol. Relat. Stud. Phys., Chem. Med., 1969, 16, 505–523 CrossRef CAS PubMed.
- J. D. Zimbrick, J. F. Ward and L. S. Myers, Jr., Int. J. Radiat. Biol. Relat. Stud. Phys., Chem. Med., 1969, 16, 525–534 CrossRef CAS PubMed.
- F. Hutchinson, Q. Rev. Biophys., 1973, 6, 201–246 CrossRef CAS PubMed.
- F. Hutchinson and W. Köhnlein, Progress in molecular and subcellular biology, Springer, 1980, pp. 1–42 Search PubMed.
- C. Limoli and J. Ward, Radiat. Res., 1993, 134, 160–169 CrossRef CAS PubMed.
- M. Grundmann-Kollmann, R. Ludwig, T. M. Zollner, F. Ochsendorf, D. Thaci, W.-H. Boehncke, J. Krutmann, R. Kaufmann and M. Podda, J. Am. Acad. Dermatol., 2004, 50, 734–739 CrossRef PubMed.
- P. Vieyra-Garcia, R. Fink-Puches, S. Porkert, R. Lang, S. Pöchlauer, G. Ratzinger, A. Tanew, S. Selhofer, S. Paul-Gunther, A. Hofer, A. Gruber-Wackernagel, F. Legat, V. Patra, F. Quehenberger, L. Cerroni, R. Clark and P. Wolf, JAMA Dermatol., 2019, 155, 538–547 CrossRef PubMed.
- Z. Liu, Y. Lu, M. Lebwohl and H. Wei, Free Radical Biol. Med., 1999, 27, 127–133 CrossRef CAS PubMed.
- D. Papadopoulo and D. Averbeck, Mutat. Res., Fundam. Mol. Mech. Mutagen., 1985, 151, 281–291 CrossRef CAS PubMed.
- R. F. Martin, D. P. Kelly, M. Roberts, P. Nel, J. Tursi, L. Denison, M. Rose, M. Reum and M. Pardee, Int. J. Radiat. Biol., 1994, 66, 517–521 CrossRef CAS PubMed.
- R. F. Martin, D. P. Kelly, M. Roberts, A. Green, L. Denison, M. Rose, M. Reum and M. Pardee, Int. J. Radiat. Oncol., Biol., Phys., 1994, 29, 549–553 CrossRef CAS.
- H. Loewe and J. Urbanietz, Arzneim. Forsch., 1974, 24, 1927–1933 CAS.
- M. A. Carrondo, M. Coll, J. Aymami, A. H. Wang, G. A. van der Marel, J. H. van Boom and A. Rich, Biochemistry, 1989, 28, 7849–7859 CrossRef CAS PubMed.
- C. N. Sprung, R. S. Vasireddy, T. C. Karagiannis, S. J. Loveridge, R. F. Martin and M. J. McKay, Mutat. Res., Fundam. Mol. Mech. Mutagen., 2010, 692, 49–52 CrossRef CAS PubMed.
- R. F. Martin, S. Broadhurst, M. E. Reum, C. J. Squire, G. R. Clark, P. N. Lobachevsky, J. M. White, C. Clark, D. Sy, M. Spotheim-Maurizot and D. P. Kelly, Cancer Res., 2004, 64, 1067 CrossRef CAS PubMed.
- M. A. Walicka, Y. Ding, A. M. Roy, R. S. Harapanhalli, S. J. Adelstein and A. I. Kassis, Int. J. Radiat. Biol., 1999, 75, 1579–1587 CrossRef CAS PubMed.
- P. Lobachevsky, G. R. Clark, P. D. Pytel, B. Leung, C. Skene, L. Andrau, J. M. White, T. Karagiannis, C. Cullinane, B. Q. Lee, A. Stuchbery, T. Kibedi, R. J. Hicks and R. F. Martin, Int. J. Radiat. Biol., 2016, 92, 686–697 CrossRef CAS PubMed.
- P. N. L. R. F. M. Tom and C. Karagiannis, Acta Oncol., 2000, 39, 681–685 CrossRef PubMed.
- V. Murray and R. F. Martin, J. Mol. Biol., 1988, 203, 63–73 CrossRef CAS PubMed.
- V. Murray and R. F. Martin, J. Mol. Biol., 1988, 201, 437–442 CrossRef CAS PubMed.
- R. S. Harapanhalli, L. W. McLaughlin, R. W. Howell, D. V. Rao, S. J. Adelstein and A. I. Kassis, J. Med. Chem., 1996, 39, 4804–4809 CrossRef CAS PubMed.
- T. C. Karagiannis, P. N. Lobachevsky, B. K. Y. Leung, J. M. White and R. F. Martin, Cancer Res., 2006, 66, 10548 CrossRef CAS PubMed.
- T. C. Karagiannis, P. N. Lobachevsky and R. F. Martin, J. Photochem. Photobiol., B, 2006, 83, 195–204 CrossRef CAS PubMed.
- R. F. Martin, V. Murray, G. D'Cunha, M. Pardee, E. Kampouris, A. Haigh, D. P. Kelly and G. S. Hodgson, Int. J. Radiat. Biol., 1990, 57, 939–946 CrossRef CAS PubMed.
- V. Murray and R. F. Martin, Nucleic Acids Res., 1994, 22, 506–513 CrossRef CAS PubMed.
- D. P. Kelly, S. A. Bateman, R. J. Hook, R. F. Martin, M. E. Reum, M. Rose and A. R. D. Whittaker, Aust. J. Chem., 1994, 47, 1751–1769 CrossRef CAS.
- H. M. Berman, J. Westbrook, Z. Feng, G. Gilliland, T. N. Bhat, H. Weissig, I. N. Shindyalov and P. E. Bourne, Nucleic Acids Res., 2000, 28, 235–242 CrossRef CAS PubMed.
- M. K. Teng, N. Usman, C. A. Frederick and A. H. Wang, Nucleic Acids Res., 1988, 16, 2671–2690 CrossRef CAS PubMed.
- Z. Luo, M. Dauter and Z. Dauter, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2014, 70, 1790–1800 CrossRef CAS PubMed.
- H. Raaijmakers, D. Suck and C. Mayer, Crystal Structure Analysis of the A-DNA Dodecamer GACCACGTGGTC, 1999, Unpublished.
- M. Sriram, G. A. van der Marel, H. L. Roelen, J. H. van Boom and A. H. Wang, EMBO J., 1992, 11, 225–232 CrossRef CAS PubMed.
- G. R. Clark, Crystal Structure of p-iodoHoechst bound to d(CGCAAATTTGCG), Int. J. Radiat. Biol., 2016 Search PubMed.
- C. J. Squire, L. J. Baker, G. R. Clark, R. F. Martin and J. White, Nucleic Acids Res., 2000, 28, 1252–1258 CrossRef CAS PubMed.
- Y. Tsunaka, N. Kajimura, S.-I. Tate and K. Morikawa, Nucleic. Acids Res., 2005, 33, 3424–3434 CrossRef CAS PubMed.
- A. Soman, C. W. Liew, H. L. Teo, N. V. Berezhnoy, V. Olieric, N. Korolev, D. Rhodes and L. Nordenskiöld, Nucleic Acids Res., 2020, 48, 5383–5396 CrossRef CAS PubMed.
- A. Osakabe, F. Adachi, Y. Arimura, K. Maehara, Y. Ohkawa and H. Kurumizaka, Open Biol., 2015, 5, 150128 CrossRef PubMed.
- N. Chatterjee, J. A. North, M. L. Dechassa, M. Manohar, R. Prasad, K. Luger, J. J. Ottesen, M. G. Poirier and B. Bartholomew, Mol. Cell. Biol., 2015, 35, 4083–4092 CrossRef CAS PubMed.
- M. Kube, F. Kohler, E. Feigl, B. Nagel-Yüksel, E. M. Willner, J. J. Funke, T. Gerling, P. Stömmer, M. N. Honemann, T. G. Martin, S. H. W. Scheres and H. Dietz, Nat. Commun., 2020, 11, 6229 CrossRef CAS PubMed.
- S. Kim, J. Chen, T. Cheng, A. Gindulyte, J. He, S. He, Q. Li, B. A. Shoemaker, P. A. Thiessen, B. Yu, L. Zaslavsky, J. Zhang and E. E. Bolton, Nucleic Acids Res., 2021, 49, D1388–D1395 CrossRef CAS PubMed.
- G. M. Morris, R. Huey, W. Lindstrom, M. F. Sanner, R. K. Belew, D. S. Goodsell and A. J. Olson, J. Comput. Chem., 2009, 30, 2785–2791 CrossRef CAS PubMed.
- L. Lafayette, G. Sauter, L. Vu and B. Meade, OpenStack Summit, Barcelona, 2016.
- O. Trott and A. J. Olson, J. Comput. Chem., 2010, 31, 455–461 CAS.
- Schrödinger, The PyMOL Molecular Graphics System, Version 1.7.4.5 Edu edn.
- Schrödinger, New York, NY, Release 2021-3 edn., 2021.
- H. J. C. Berendsen, D. van der Spoel and R. van Drunen, Comput. Phys. Commun., 1995, 91, 43–56 CrossRef CAS.
- M. J. Abraham, T. Murtola, R. Schulz, S. Páll, J. C. Smith, B. Hess and E. Lindahl, SoftwareX, 2015, 1-2, 19–25 CrossRef.
- I. Ivani, P. D. Dans, A. Noy, A. Pérez, I. Faustino, A. Hospital, J. Walther, P. Andrio, R. Goñi, A. Balaceanu, G. Portella, F. Battistini, J. L. Gelpí, C. González, M. Vendruscolo, C. A. Laughton, S. A. Harris, D. A. Case and M. Orozco, Nat. Methods, 2016, 13, 55–58 CrossRef CAS PubMed.
- D. J. Price and C. L. Brooks, 3rd, J. Chem. Phys., 2004, 121, 10096–10103 CrossRef CAS PubMed.
- A. W. Sousa da Silva and W. F. Vranken, BMC Research Notes, 2012, 5, 367 CrossRef PubMed.
- H. J. C. Berendsen, J. P. M. Postma, W. F. V. Gunsteren, A. DiNola and J. R. Haak, J. Chem. Phys., 1984, 81, 3684–3690 CrossRef CAS.
- M. Parrinello and A. Rahman, Phys. Rev. Lett., 1980, 45, 1196–1199 CrossRef CAS.
- B. Hess, H. Bekker, H. J. Berendsen and J. G. Fraaije, J. Comput. Chem., 1997, 18, 1463–1472 CrossRef CAS.
- T. Darden, D. York and L. Pedersen, J. Chem. Phys., 1993, 98, 10089–10092 CrossRef CAS.
- R. Kumari, R. Kumar and A. Lynn, J. Chem. Inf. Model., 2014, 54, 1951–1962 CrossRef CAS PubMed.
- K. Robert, A. B. Nathan and J. A. McCammon, Computat. Sci. Discovery, 2012, 5, 015005 CrossRef PubMed.
- W. Humphrey, A. Dalke and K. Schulten, J. Mol. Graphics, 1996, 14(33–38), 27–38 Search PubMed.
- R. F. Martin and N. Holmes, Nature, 1983, 302, 452–454 CrossRef CAS PubMed.
- R. F. Martin and M. Pardee, Int. J. Appl. Radiat. Isotopes, 1985, 36, 745–747 CrossRef CAS.
- R. P. Hertzberg and P. B. Dervan, Biochemistry, 1984, 23, 3934–3945 CrossRef CAS PubMed.
- K. D. Harshman and P. B. Dervan, Nucleic Acids Res., 1985, 13, 4825–4835 CrossRef CAS PubMed.
- P. E. Pjura, K. Grzeskowiak and R. E. Dickerson, J. Mol. Biol., 1987, 197, 257–271 CrossRef CAS PubMed.
- X.-X. Zhang, S. L. Brantley, S. A. Corcelli and A. Tokmakoff, Commun. Biol., 2020, 3, 525 CrossRef CAS PubMed.
- U. Issar, T. Kumari and R. Kakkar, J. Comput. Sci., 2015, 10, 166–177 CrossRef.
- F. G. Loontiens, P. Regenfuss, A. Zechel, L. Dumortier and R. M. Clegg, Biochemistry, 1990, 29, 9029–9039 CrossRef CAS PubMed.
- S. Genheden and U. Ryde, Expert Opin. Drug Discovery, 2015, 10, 449–461 CrossRef CAS PubMed.
- H. Sun, Y. Li, S. Tian, L. Xu and T. Hou, Phys. Chem. Chem. Phys., 2014, 16, 16719–16729 RSC.
- T. Hou, J. Wang, Y. Li and W. Wang, J. Chem. Inf. Model., 2011, 51, 69–82 CrossRef CAS PubMed.
- B. Briggs, K. Ververis, A. L. Rodd, L. J. L. Foong, F. M. D. Silva and T. C. Karagiannis, J. Photochem. Photobiol., B, 2011, 103, 145–152 CrossRef CAS PubMed.
- A. Adhikary, V. Buschmann, C. Müller and M. Sauer, Nucleic Acids Res., 2003, 31, 2178–2186 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1cp04841d |
| This journal is © the Owner Societies 2022 |