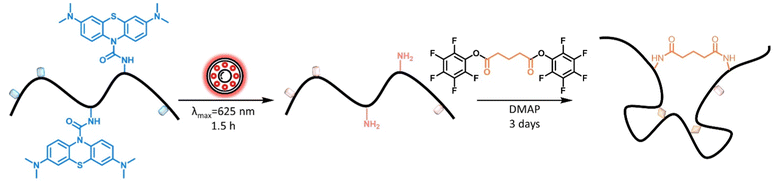
Red light induced folding of single polymer chains†
Ishrath
Mohamed Irshadeen
 ab,
Vinh X.
Truong
ab,
Vinh X.
Truong
 *abd,
Hendrik
Frisch
*abd,
Hendrik
Frisch
 *ab and
Christopher
Barner-Kowollik
*ab and
Christopher
Barner-Kowollik
 *abc
*abc
aSchool of Chemistry and Physics, Queensland University of Technology (QUT), Brisbane 2 George Street, QLD 4000, Australia. E-mail: christopher.barnerkowollik@qut.edu.au; h.frisch@qut.edu.au
bCentre for Materials Science, Queensland University of Technology (QUT), Brisbane 2 George Street, QLD 4000, Australia
cInstitute of Nanotechnology, Karlsruhe Institute of Technology (KIT), Hermann-von-Helmholtz-Platz 1, 76344 Eggenstein-Leopoldshafen, Germany
dInstitute of Materials Research and Engineering (IMRE), Agency for Science, Technology and Research (A*STAR), 2 Fusionopolis Way, Singapore 138 634, Singapore. E-mail: vinh_truong@imre.a-star.edu.sg
First published on 27th October 2022
Abstract
We pioneer the photochemical generation of single chain nanoparticles (SCNPs) at the to-date mildest reported wavelength of 625 nm by exploiting the photochemical uncaging of methylene blue protected amines. The protected amines are tethered to polymers prepared via reversible addition–fragmentation chain transfer (RAFT) polymerisation, and subsequently undergo intrachain crosslinking by amide formation.
Single chain nanoparticles (SCNPs) take inspiration from the control over sequence and 3D architecture observed in their natural analogues, e.g. enzymes and proteins.1–4 Early SCNP systems consisted of homopolymers with pendant functionalities randomly dispersed along the chain, which were subsequently utilised to form crosslinks—either by reacting with each other or via the aid of an added crosslinker—thus leading to a macromolecule with a more compact structure. While the field is still far from achieving the fine control over sequence and folding seen in polypeptides and proteins, SCNPs have evolved significantly over the last decade to include reversibly folded structures,5,6 multiple sequentially controlled folding moieties,7–11 embedded catalysts at their core12–14 or featuring fluorescent properties.6,15,16
Photoreactions have been extensively utilised for the controlled folding of single polymer chains in recent years, due to the powerful spatiotemporal control afforded by light.10,17–20 A significant disadvantage of photoreactions is that many of them require high energy UV light activation, making them non-viable to be used with biological tissues or for applications that require large penetration depths. Some reactions, including the [2+2] photocycloadditions of styrylpyrene and pyrene chalcone,21,22 have been shown to function efficiently in the visible range. Our team recently reported the pyridinepyrene unit, a new chromophore undergoing [2+2] photocycloaddition with orange light (λmax = 590 nm) at low pH within single polymer chains—perhaps the longest reported wavelength to trigger photocycloadditions.23
However, photocleavage reactions have a potentially even wider range of trigger wavelengths, from the UV light triggered ortho-nitrobenzyl and photoenol reaction24 to Near Infrared (NIR) triggered Photoactivatable Protecting Groups (PPGs).25,26 PPGs have been widely employed in light-induced ligations by caging and subsequent photo-uncaging of a reactive function e.g., thiol or amine for rapid thermal ligation.27,28 While many cycloadditions have been constrained to higher energy wavelengths, photo-induced ligations using PPGs have been successfully performed with long wavelength visible (green and red) light to date.25,26,29–35 However, to the best of our knowledge, none of these visible-light induced reactions have been employed in single chain nanoparticle folding. Herein, we demonstrate the folding of SCNPs induced by red light (λmax = 625 nm), which is the longest wavelength utilised to date.
We caged 3-aminopropyl methacrylate with methylene blue, affording a protected monomer (MB-MMA) that is capable of undergoing reversible deactivation radical polymerisation (RDRP). Here, the protection of the amine prevents its interference with the RAFT agent (Scheme 1).36 The methylene blue solution has an intense blue colour, while its reduced form, the leuco-methylene blue that was used as the caged moiety, is colourless;37–39 thus, the caged amine monomer solution in acetonitrile is nearly colourless—its UV-Vis absorbance only displays a small absorption band at 650 nm (Fig. 1).
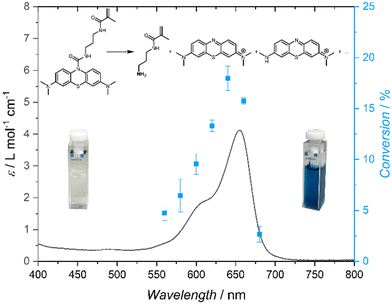 | ||
| Fig. 1 Action plot of MB-MMA in acetonitrile-d3 at a concentration of 1.1 mmol L−1, irradiated with 630 μmol of photons at each wavelength. | ||
While UV-Vis spectra were commonly used to determine the photoreactivity of chromophores, our group has discovered that the reactivity maxima do not necessarily overlap with the absorption maxima, and the two are often offset by tens of nanometres.24 An action plot, wherein a tuneable laser is employed to produce monochromatic light at a range of selected wavelengths, enables the determination of the actual wavelength-dependent reactivity of the chromophore. When recording an action plot, the number of photons is identical at all wavelengths along with concentrations, solvent, and temperature. Thus, we initially probed the wavelength-dependent reactivity of MB-MMA in acetonitrile across a range of wavelengths from 560 to 700 nm. All samples featured a concentration of 1.1 mmol L−1 in deuterated acetonitrile and were irradiated with 630 μmol of photons at each wavelength. The conversion of the photocleavage reaction was recorded via1H NMR spectroscopy against an internal standard of 1,3,5-trimethoxybenzene dissolved at an equimolar concentration with MB-MMA.
The photochemical characteristics of MB-MMA were determined by UV-vis absorption spectroscopy and an action plot analysis,24 which – jointly – show the wavelength resolved reactivity of the MB-MMA from 580 nm up to 700 nm (Fig. 1). The action plot demonstrates that the photocleavage of the monomer was most efficient at 640 nm, yet can be initiated by light up to 680 nm. While most action plots of photocycloadditions display a pronounced bathochromic shift of the peak reactivity compared to the λmax of the absorption spectrum,24 this phenomenon has so far not been observed for photocleavable protecting groups.40,41
MB-MMA was subsequently irradiated with red light from an LED (λmax = 625 nm) for 1.5 h and analysed by hyphenated liquid chromatography-mass spectrometry (LC-MS). Here, the methylene group was observed to successfully cleave, generating a free amine (ESI,† Fig. S17) for the subsequent coupling reaction. The solution turned from colourless to deep blue post-irradiation (Fig. 1), which suggests the presence of free methylene blue after photocleavage.
To exploit the amine groups released upon low energy visible light irradiation of the caged amine, an external crosslinker capable of efficiently binding amine groups on both ends to fold the polymer is required. Thus, we synthesised a glutaric acid based crosslinker that was capped on both ends with pentafluorophenol ester (ESI,†M2, Fig. S13). Pentafluorophenol (PFP) is an efficient leaving group that is stable for long-term storage and efficiently reacts with amines to form amide adducts.42 To determine the efficacy of the crosslinker, we tested its reaction with butylamine, catalysed by DMAP. The product was subsequently analysed via1H NMR spectroscopy (ESI,† Fig. S16). The reaction was found to successfully establish amide bonds at both ends of the crosslinker, deeming it suitable for use with unprotected MB-MMA.
We subsequently copolymerised the functional monomer (MB-MMA) with methyl methacrylate (MMA) via RAFT polymerisation. The polymer was isolated via precipitation and characterised via NMR spectroscopy and size exclusion chromatography (SEC) (P1Mn = 19.6 kg mol−1|Đ = 1.2; ESI,† Fig. S8).
The polymer was estimated, by integration of 1H NMR spectrum, to have 13 methylene blue units per polymer chain on average (9.8 wt%). In a one-pot reaction, we dissolved polymer P1 (0.25 mg mL−1), M2, and DMAP catalyst in ACN, and irradiated the mixture for 1.5 h with a red light LED (λmax = 625 nm) under stirring in the dark for 3 days at ambient conditions, before precipitating the polymer in diethyl ether and characterization (P1′′).
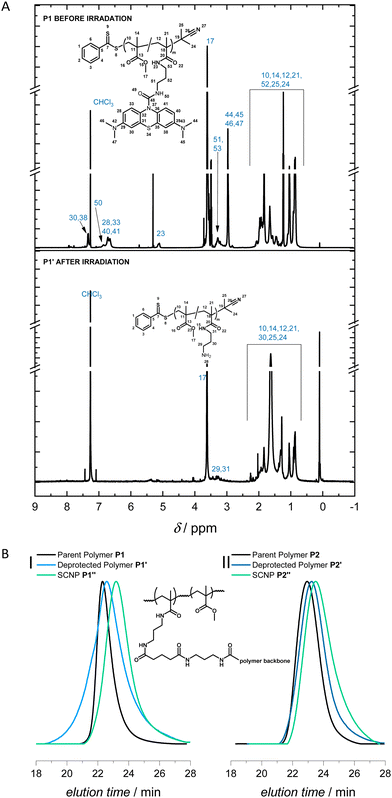
The SEC of P1′′ shows a clear shift towards longer elution times compared to the parent polymer P1, which suggests the polymer compacted successfully after the photodeprotection, resulting in the formation of SCNPs, via the external crosslinker (Fig. 2B(I)). Comparison of the peak molecular weights, Mp, showed an apparent compaction of 15.5% and 29.4% for P1′ (photodeprotected polymer) and P1′′ (SCNP), respectively, relative to the parent polymer P1. P1′′ displayed an apparent compaction of 16.5% relative to P1′. The successful photodeprotection was further substantiated via1H NMR spectroscopy of the purified deprotected polymer compared to the parent linear polymer. The deprotected polymer lacked the aromatic resonances (δ = 6.5–7.5 ppm) and the resonance at δ = 2.92 ppm from the methyl groups of MB-MMA observed in the parent polymer (Fig. 2A).
To ensure the robustness and repeatability of our one-pot visible light SCNP folding system, we synthesised a second polymer P2 (Mn = 11.7 kg mol−1|Đ = 1.3) featuring close to 6 MB-MMA units on average per polymer (ESI,† Fig. S12). The comparison of the SEC traces of P2, P2′, and P2′′ indicate increasing elution times after the photodeprotection and the subsequent folding, respectively (Fig. 2B(II)). Comparison of the Mp values showed an apparent compaction of 5.1% and 27.6% for P2′ and P2′′, respectively, compared to the parent polymer P2 (ESI,† Table S4). P2′′ features an apparent compaction of 27.7% relative to P2′.
Longer wavelengths allow for deeper penetration, especially through biological tissues. In particular, light within the biological window of λ = 650 to 1350 nm43,44 has been established to have the best penetration into live tissue. Thus, we sought to establish the effectiveness of our MB-folding system with the interception of a barrier between the light source and the chromophore sample.
Initially, a single layer of 80 gsm bright white paper was wrapped around the sample vial, which contained P1 (0.25 mg mL−1, in ACN) and was irradiated with a red light LED (λmax = 625 nm) for 6 h before comparing the SEC trace of the resultant polymer with the parent polymer (ESI,† Fig. S19). The reaction was successful as the red light was able to penetrate through the paper to reach the sample (ESI,† Fig. S18) and the SCNP formed had an apparent compaction of 38.9% relative to P1 (ESI,† Table S5). A longer irradiation time was used to compensate for the scattering of photons as the light passed through a barrier. To replicate the penetration of light through biological tissue, we subsequently used a single slice of chicken breast (1.1 mm thick) (ESI,† Fig. S20) in place of the paper and repeated the irradiation experiment. Analysis of the SEC traces showed that the folding was successful and the resultant SCNP had an apparent compaction of 38.5% compared to P1, and 27.2% compared to P1′ based on the Mp values (ESI,† Table S6), thus further substantiating the claim that red light penetrates through biological tissues.
In summary, we introduce a methylene blue based chromophore, which acts as a photolabile protecting group for primary amines. Upon incorporation into a copolymer, red light induced the photodeprotection of pendant amines, which readily underwent intramacromolecular crosslinking in the presence of bivalent active esters to form SCNPs. Seizing the penetration depth of such low-energy light, we further demonstrate that the photoinduced folding occurs successfully through a paper and a biological tissue barrier. The action plot shows that the monomer is capable of undergoing photocleavage triggered by light up to λ = 680 nm, which is advantageous for future biological applications, for example in photodynamic therapy and light-mediated conjugation of bioactive components deep within the tissue.27,45
C. B.-K. acknowledges funding from the Australian Research Council (ARC) in the form of a Laureate Fellowship (FL170100014) enabling his photochemical research program as well as continued key support from the Queensland University of Technology (QUT). I. M. I. gratefully acknowledges QUT for a PhD Research Scholarship. H. F. acknowledges support by the ARC in the form of a DECRA Fellowship. The Central Analytical Research Facility (CARF) at QUT is gratefully acknowledged for access to analytical instrumentation.
Conflicts of interest
There are no conflicts to declare.References
- E. J. Stollar and D. P. Smith, Essays Biochem., 2020, 64(4), 649–680 CrossRef CAS PubMed.
- M. Bajaj and T. Blundell, Annu. Rev. Biophys. Bioeng., 1984, 13(1), 453–492 CrossRef CAS PubMed.
- C. K. Lyon, A. Prasher, A. M. Hanlon, B. T. Tuten, C. A. Tooley, P. G. Frank and E. B. Berda, Polym. Chem., 2015, 6(2), 181–197 RSC.
- A. M. Hanlon, C. K. Lyon and E. B. Berda, Macromolecules, 2016, 49(1), 2–14 CrossRef CAS.
- J. Willenbacher, B. V. K. J. Schmidt, D. Schulze-Suenninghausen, O. Altintas, B. Luy, G. Delaittre and C. Barner-Kowollik, Chem. Commun., 2014, 50(53), 7056–7059 RSC.
- B. T. Tuten, D. Chao, C. K. Lyon and E. B. Berda, Polym. Chem., 2012, 3(11), 3068–3071 RSC.
- T. S. Fischer, D. Schulze-Sünninghausen, B. Luy, O. Altintas and C. Barner-Kowollik, Angew. Chem., Int. Ed., 2016, 55(37), 11276–11280 CrossRef CAS PubMed.
- O. Altintas, P. Krolla-Sidenstein, H. Gliemann and C. Barner-Kowollik, Macromolecules, 2014, 47(17), 5877–5888 CrossRef CAS.
- J. Rubio-Cervilla, H. Frisch, C. Barner-Kowollik and J. A. Pomposo, Macromol. Rapid Commun., 2019, 40(1), 1800491 CrossRef PubMed.
- H. Frisch, D. Kodura, F. R. Bloesser, L. Michalek and C. Barner-Kowollik, Macromol. Rapid Commun., 2020, 41(1), 1900414 CrossRef CAS PubMed.
- T. K. Claus, J. Zhang, L. Martin, M. Hartlieb, H. Mutlu, S. Perrier, G. Delaittre and C. Barner-Kowollik, Macromol. Rapid Commun., 2017, 38(16), 1700264 CrossRef PubMed.
- H. Rothfuss, N. D. Knöfel, P. W. Roesky and C. Barner-Kowollik, J. Am. Chem. Soc., 2018, 140(18), 5875–5881 CrossRef CAS PubMed.
- I. Berkovich, V. Kobernik, S. Guidone and N. G. Lemcoff, Metal Containing Single-Chain Nanoparticles, Single-Chain Polymer Nanoparticles, 2017, pp. 217–257 Search PubMed.
- E. Verde-Sesto, A. Blázquez-Martín and J. A. Pomposo, Polymers, 2019, 11(11), 1903 CrossRef CAS PubMed.
- C. Heiler, J. T. Offenloch, E. Blasco and C. Barner-Kowollik, ACS Macro Lett., 2017, 6(1), 56–61 CrossRef CAS PubMed.
- F. R. Bloesser, S. L. Walden, I. M. Irshadeen, L. C. Chambers and C. Barner-Kowollik, Chem. Commun., 2021, 57(42), 5203–5206 RSC.
- H. Frisch, F. R. Bloesser and C. Barner-Kowollik, Angew. Chem., Int. Ed., 2019, 58(11), 3604–3609 CrossRef CAS PubMed.
- H. Frisch, J. P. Menzel, F. R. Bloesser, D. E. Marschner, K. Mundsinger and C. Barner-Kowollik, J. Am. Chem. Soc., 2018, 140(30), 9551–9557 CrossRef CAS PubMed.
- H. Frisch, B. T. Tuten and C. Barner-Kowollik, Isr. J. Chem., 2020, 60(1–2), 86–99 CrossRef CAS.
- W. Fan, X. Tong, G. Li and Y. Zhao, Polym. Chem., 2017, 8(22), 3523–3529 RSC.
- I. M. Irshadeen, K. De Bruycker, A. S. Micallef, S. L. Walden, H. Frisch and C. Barner-Kowollik, Polym. Chem., 2021, 12(34), 4903–4909 RSC.
- D. E. Marschner, H. Frisch, J. T. Offenloch, B. T. Tuten, C. R. Becer, A. Walther, A. S. Goldmann, P. Tzvetkova and C. Barner-Kowollik, Macromolecules, 2018, 51(10), 3802–3807 CrossRef CAS.
- D. Kodura, L. L. Rodrigues, S. L. Walden, A. S. Goldmann, H. Frisch and C. Barner-Kowollik, J. Am. Chem. Soc., 2022, 144(14), 6343–6348 CrossRef CAS PubMed.
- I. M. Irshadeen, S. L. Walden, M. Wegener, V. X. Truong, H. Frisch, J. P. Blinco and C. Barner-Kowollik, J. Am. Chem. Soc., 2021, 143(50), 21113–21126 CrossRef CAS PubMed.
- A. Y. Vorobev and A. E. Moskalensky, Comput. Struct. Biotechnol. J., 2020, 18, 27–34 CrossRef CAS PubMed.
- R. Weinstain, T. Slanina, D. Kand and P. Klán, Chem. Rev., 2020, 120(24), 13135–13272 CrossRef CAS PubMed.
- V. X. Truong, ChemPhotoChem, 2020, 4(8), 564–570 CrossRef CAS.
- S. Jia and E. M. Sletten, ACS Chem. Biol., 2021 DOI:10.1021/acschembio.1c00518.
- V. X. Truong, K. M. Tsang, F. Ercole and J. S. Forsythe, Chem. Mater., 2017, 29(8), 3678–3685 CrossRef CAS.
- V. X. Truong and C. Barner-Kowollik, ACS Macro Lett., 2021, 10(1), 78–83 CrossRef CAS PubMed.
- R. R. Nani, A. P. Gorka, T. Nagaya, T. Yamamoto, J. Ivanic, H. Kobayashi and M. J. Schnermann, ACS Cent. Sci., 2017, 3(4), 329–337 CrossRef CAS PubMed.
- M. Li, A. P. Dove and V. X. Truong, Angew. Chem., Int. Ed., 2020, 59(6), 2284–2288 CrossRef CAS PubMed.
- A. Poryvai, M. Galkin, V. Shvadchak and T. Slanina, Angew. Chem., Int. Ed., 2022, 61(34), e202205855 CAS.
- H. Janeková, M. Russo, U. Ziegler and P. Štacko, Angew. Chem., Int. Ed., 2022, 61(33), e202204391 CrossRef PubMed.
- K. Kalayci, H. Frisch, C. Barner-Kowollik and V. X. Truong, Chem. Commun., 2022, 58(44), 6397–6400 RSC.
- G. Moad, E. Rizzardo and S. H. Thang, Aust. J. Chem., 2005, 58(6), 379–410 CrossRef CAS.
- S.-K. Lee and A. Mills, Chem. Commun., 2003, 2366–2367 RSC.
- H. M. Dao, C.-H. Whang, V. K. Shankar, Y.-H. Wang, I. A. Khan, L. A. Walker, I. Husain, S. I. Khan, S. N. Murthy and S. Jo, Chem. Commun., 2020, 56(11), 1673–1676 RSC.
- I. Khan, K. Saeed, I. Zekker, B. Zhang, A. H. Hendi, A. Ahmad, S. Ahmad, N. Zada, H. Ahmad, L. A. Shah, T. Shah and I. Khan, Water, 2022, 14(2), 242 CrossRef CAS.
- C. Petit, J. Bachmann, L. Michalek, Y. Catel, E. Blasco, J. P. Blinco, A.-N. Unterreiner and C. Barner-Kowollik, Chem. Commun., 2021, 57(23), 2911–2914 RSC.
- J. Bachmann, C. Petit, L. Michalek, Y. Catel, E. Blasco, J. P. Blinco, A.-N. Unterreiner and C. Barner-Kowollik, ACS Macro Lett., 2021, 10(4), 447–452 CrossRef CAS PubMed.
- M. I. Gibson, E. Fröhlich and H.-A. Klok, J. Polym. Sci., Part A: Polym. Chem., 2009, 47(17), 4332–4345 CrossRef CAS.
- S. He, J. Song, J. Qu and Z. Cheng, Chem. Soc. Rev., 2018, 47(12), 4258–4278 RSC.
- A. M. Smith, M. C. Mancini and S. Nie, Nat. Nanotechnol., 2009, 4(11), 710–711 CrossRef CAS PubMed.
- S. L. H. Higgins and K. J. Brewer, Angew. Chem., Int. Ed., 2012, 51(46), 11420–11422 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2cc05415a |
| This journal is © The Royal Society of Chemistry 2022 |