 Open Access Article
Open Access ArticleSelective NMR detection of N-methylated amines using cavitand-decorated silica nanoparticles as receptors†‡
Andrea
Cesari§
 a,
Daniele
Rosa-Gastaldo§
a,
Daniele
Rosa-Gastaldo§
 a,
Alessandro
Pedrini
a,
Alessandro
Pedrini
 b,
Federico
Rastrelli
b,
Federico
Rastrelli
 a,
Enrico
Dalcanale
a,
Enrico
Dalcanale
 b,
Roberta
Pinalli
b,
Roberta
Pinalli
 *b and
Fabrizio
Mancin
*b and
Fabrizio
Mancin
 *a
*a
aDipartimento di Scienze Chimiche, Università di Padova, via Marzolo 1, 35131 Padova, Italy. E-mail: fabrizio.mancin@unipd.it
bDipartimento di Scienze Chimiche, della Vita e della Sostenibilità Ambientale and INSTM UdR Parma - Università di Parma, Parco Area delle Scienze, 17/A, 43124, Parma, Italy. E-mail: roberta.pinalli@unipr.it
First published on 29th August 2022
Abstract
We report a strategy for the realization of NMR chemosensors based on the spontaneous self-assembly of lower rim pyridinium-functionalized tetraphopshonate cavitands on commercial silica nanoparticles. These nanohybrids enable the selective detection of physiologically relevant N-methylated amines, with a limit of detection of 31 μM, via STD-based NMR experiments, achieving for the first time fine structural selectivity in nanoparticle-assisted NMR chemosensing.
Nuclear Magnetic Resonance (NMR) spectroscopy shares with a few other techniques the advantage of identifying analytes according to their spectral fingerprint. This virtually eliminates the possibility of false positives in analytic applications and allows the identification of unknown species.1,2a,b However, in most of the cases, the number of signals obtained by analysing a mixture is overwhelming, and overlapping prevents the individuation of the single components. To overcome this issue, we perfected over the last years the “nanoparticle-assisted NMR chemosensing” technique.3 This protocol combines the recognition capabilities of nanoparticle receptors with NOE-based NMR experiments, which enable to transfer magnetization between the nanoparticles and the bound analytes. In this way, it is possible to tag and extract only the target signals form the spectrum of the mixture analysed.
Over the years, this technique has been gradually improved reaching limits of detection (LOD) in the low μM range.3 This required the design of high-performance NMR experiments4,5 and the optimization of the nanoreceptors.6,7 Beside recognizing the analyte, the fundamental role of the nanoreceptor is to enable an efficient magnetization transfer thanks to its long correlation time (τc).8 In this perspective, the 2 nm ligand-coated gold nanoparticles (AuNPs, τc ≈ 10 ns) used as first-generation self-assembled nanoreceptors were underperforming. In fact, although theory predicts that optimal NOE enhancements should be reached at τc longer than 1 ns,9 the finite association lifetime between the analyte and the receptor itself moves this threshold towards longer correlation times (about 1 μs),9 which are attained by nanoscopic objects with diameters larger than 12 nm.10 A relevant improvement was hence achieved when a second-generation nanoreceptors was conceived, based on the self-assembly of charged AuNPs on larger (15–20 nm) silica nanoparticles bearing the opposite charge.9
In the evolution of these chemosensing systems, a still lacking feature is selectivity. Indeed, AuNPs used so far as receptors could only distinguish broad classes of analytes (cations, anions).6 To address this issue we reasoned that the self-assembly of charged supramolecular hosts on the surface of silica nanoparticles would result in nanohybrid receptors large enough to allow the effective transfer of magnetization (Fig. 1a), providing a general strategy to transfer the selectivity of supramolecular hosts to NMR chemosensing.
 | ||
| Fig. 1 (a) Sensing scheme studied in this work; (b) left: chemical structure of Tiiii-Py cavitand (with included NMPEA), right: analytes used in this work. | ||
To test this hypothesis, we selected a tetraphosphonate cavitand bearing four propyl pyridinium moieties at the lower rim (Tiiii-Py, Fig. 1b).11 Tetraphosphonate cavitands are well known for their ability to selectively recognize biologically relevant N-methylated ammonium derivatives.11 Such features allowed the successful coupling of these hosts with different techniques for the detection of tumour markers12–14 and psychoactive substances.15,16 To form the nanohybrids, we selected commercial LUDOX HS-30 silica nanoparticles, which are readily available, cheap, and have an average size of 13–15 nm (Fig. S1 and S2, ESI‡). The four positive charges of the Tiiii-Py should grant its adsorption on the negatively charged surface of silica nanoparticles. The sensing performances were tested on three different phenethylamine analogues (Fig. 1b), since this family of compounds is of great relevance as neurotransmitters, drugs and even biomarkers.17
At first, we investigated the association of Tiiii-Py to LUDOX nanoparticles. Preliminary 1H-NMR experiments showed that the addition of LUDOX to a 0.5 mM Tiiii-Py solution in buffered D2O (phosphate buffer 5 mM, pD = 7.0) resulted in the progressive disappearance of the Tiiii-Py signals, reaching saturation at 400 cavitand molecules per particle (Fig. S3 and S4, ESI‡). To obtain more information on the interaction of Tiiii-Py with silica nanoparticle we used 3-methoxytyramine (3-MT, Fig. 1b) as probe molecule. Being a primary amine, 3-MT is not recognized by Tiiii-Py and indeed no changes were observed in the NMR spectrum when the two were mixed (Fig. 2a and b). On the contrary, broadening of the 3-MT signals was observed when it was added to a suspension of LUDOX (Fig. 2c). The subsequent addition of Tiiii-Py caused the re-sharpening of the 3-MT signals (Fig. 2d) to linewidths similar to those recorded in samples containing 3-MT alone (Fig. 2a).
Taken together, the above NMR experiments confirmed the absorption of Tiiii-Py on the surface of the nanoparticles to form Tiiii-Py@LUDOX nanohybrids. The grafting of small molecules to nanoparticles causes the broadening of their NMR signals because of the reduction of the tumbling rate.18 Obviously, if surface binding and dissociation are fast with respect to the NMR timescale, the extent of the observed broadening depends on the strength of the interaction, since the apparent lineshape is (approximately) averaged over the populations of the bound and unbound species. 3-MT has only one positive charge and consequently it is expected to adsorb weakly to the nanoparticles.18–20 For this reason, the broadening of its signals was moderate. On the other hand, Tiiii-Py has four positive charges that ensure a strong adsorption onto the silica surface.19,20 The consequent full adsorption of Tiiii-Py to the nanoparticles resulted in broadening of its signals beyond detection. Because of its high affinity for silica, Tiiii-Py completely displaced the absorbed 3-MT molecules from the particles when it was added to the mixture of 3-MT and LUDOX, restoring the sharpness of 3-MT signals (Fig. 2).
Interaction of polycharged species with charged nanoparticles can lead to the formation of a corona but also to nanoparticles crosslinking and aggregation.19 To investigate this point, we performed Dynamic Light Scattering (DLS) experiments. These showed that subsequent additions of Tiiii-Py (up to 100 μM) to a 1.5 μM suspension of LUDOX in 5 mM phosphate buffer (pH = 7.0) resulted in a moderate nanoparticles hydrodynamic diameter increase (from 12.8 to 17.4 nm), as expected in the case of the formation of a layer of Tiiii-Py on the surface of the nanoparticles. This diameter increase was accompanied by the formation of a small number of larger aggregates (Table S1, ESI‡). However, no relevant precipitation was observed for several hours in the conditions of the NMR sensing experiments.
Having established the formation of the Tiiii-Py@LUDOX nanohybrids, we investigated their ability to bind N-methylammonium ions. In the first experiment, 1H-NMR spectra of N-methyl phenethylamine (NMPEA, 0.5 mM in D2O buffered at pD = 7.0 with 5 mM phosphate buffer) were recorded in the presence of LUDOX, Tiiii-Py and the Tiiii-Py@LUDOX hybrids (Fig. 3a–d). As in the case of 3-MT, addition of LUDOX to a solution of NMPEA caused the broadening of all the signals of the analyte (Fig. 3d). On the other hand, the addition of the sole Tiiii-Py (Fig. 3b) caused the broadening only of the signals arising from the aliphatic residues, as well as small upfield shifts. These effects are known in the case of the formation of the Tiiii-Py/N-methylated ammonium host–guest complexes, and are ascribed to the shielding effect produced on the analyte by the inclusion in the aromatic cavity of Tiiii-Py.11 When the spectrum is recorded in the presence of the Tiiii-Py@LUDOX hybrids (Fig. 3c), the trends of broadening and chemical shift perturbations were similar to those observed in the presence of only Tiiii-Py, but less pronounced. This confirmed that silica-grafted Tiiii-Py was still capable to bind NMPEA.
Binding of NMPEA to Tiiii-Py@LUDOX was confirmed as well by a 1H-NMR titration performed on a sample containing Tiiii-Py@LUDOX (0.25 mM Tiii-Py) in D2O buffered with phosphate at pD = 7.0, providing a binding constant of (3.1 ± 0.5)·103 M−1 (Fig. S5 and S6, ESI‡), consistent with the ones previously reported for similar systems in water.11
The ability of the Tiiii-Py@LUDOX nanoconjugates to selectively bind N-methylammonium moieties, as well as the possibility to employ them in NMR chemosensing, was finally confirmed with saturation transfer difference (STD) NMR experiments on a sample containing 1 mM NMPEA and 0.25 mM Tiiii-Py or Tiiii-Py@LUDOX in buffered D2O (phosphate buffer 5 mM, pD = 7.0). This is a steady-state NOE experiment where the spin populations of the analytes interacting with the nanoparticles are altered by continuous irradiation of a selected region of the spectrum (which, in this case, includes only Tiiii-Py signals).6 As a result, in the spin diffusion limit the intensity of the analytes’ signals decreases, and subtraction from an off-resonance reference spectrum reveals only the signals of the interacting analytes. As reported in Fig. 4, no STD signal was observed when the sole Tiiii-Py was added to the NMPEA solution (Fig. 4b). This result confirmed that the small size of the Tiiii-Py·NMPEA host–guest complexes prevented an effective transfer of saturation. By the contrary, clear STD analyte signals were present (average S/N = 17 for the aromatic region) in the spectra of same sample containing the Tiiii-Py@LUDOX nanohybrids (Fig. 4d). The relevant implications of this result are many: (i) they provide another proof of the analyte binding to Tiiii-Py@LUDOX since, in the absence of interaction, no STD signal could be observed; (ii) they provide further evidence for the formation of the Tiiii-Py@LUDOX hybrids, since the cavitand alone does not produce any STD signal; (iii) they confirm that the formation of Tiiii-Py@LUDOX increases the efficiency of the saturation transfer from the cavitand to the analyte. Control experiments performed on samples containing glucose and phenylalanine as potential interferents in the presence of Tiiii-Py@LUDOX nanohybrids produced approximately zero STD response (Fig. S7, ESI‡), confirming the role of the cavitand in the selective detection of N-methylated amines. The main limit of STD-based detection is that saturation is hardly diffused across interrupted dipole–dipole coupling networks of spins and requires close analyte-receptor contacts. This can substantially affect the effectiveness of chemosensing experiments based on STD if the analyte binding sites include a limited number of hydrogen atoms and/or these belong to different spin system than the one saturated. This limitation is overcome by our recently developed HPwSTD experiment,3,4 which uses high power radiofrequency pulses to saturate both the nanohybrid spins and the slow tumbling water molecules belonging to the nanohybrid solvation shell. This ensures a larger source of saturation to be transferred to the analytes and, at the end, greater sensitivity.
To test the performances of the Tiiii-Py@LUDOX nanohybrids with the HPwSTD experiment, a mixture of three different amines, namely NMPEA, 3-MT, and 4-nitrophenetylamine (4-NO2-PEA), each at 0.25 mM concentration, was added to a suspension of Tiiii-Py@LUDOX in buffered D2O (5 mM phosphate buffer, pD = 7.0). In this case, the selectivity of the sensing system was challenged by the fact that all the analytes are phenethylamines, with the sole NMPEA being N-methylated. As expected, no HPwSTD signals from the three analytes were detected in samples devoid of the silica nanoparticles (Fig. 5(a.2)). On the other hand, broad and strong signals from all the analytes were detected in samples containing the sole silica nanoparticles (Fig. 5(a.3)). Indeed, the use of solvation water as saturation source enables also these inorganic particles as NMR nanoreceptors, but they obviously feature small selectivity. It is worth noting that the broadening of NMPEA signals appears more relevant than that of the other analytes. This likely due to the greater affinity of N-methylated ammoniums for silica with respect to primary ammonium ions.18 Still, the affinity difference is not enough to produce relevant differences in the intensities of the STD signals. Eventually, the HPwSTD spectra collected from the samples containing both Tiiii-Py and LUDOX (Fig. 5(a.4)) showed the presence of the sole signals of NMPEA, with the other analytes barely detectable.
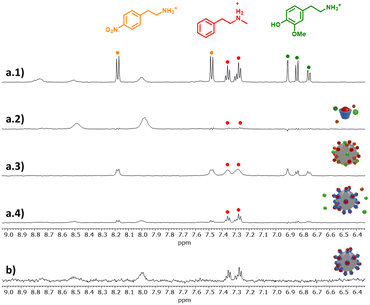 | ||
Fig. 5 (a.1) 1H-NMR subspectrum of a solution of NMPEA (red sphere), 4-NO2-PEA (orange sphere), 3-MT (green sphere) 0.25 mM each and Tiiii-Py (blue basket) 0.1 mM; (a.2) HPwSTD subspectrum of the same sample; (a.3) HPwSTD subspectrum of a solution of NMPEA, 4-NO2-PEA, 3-MT 0.25 mM each and LUDOX (grey sphere) 0.15 μM; (a.4) HPwSTD subspectrum of a solution of NMPEA, 4-NO2-PEA, 3-MT 0.25 mM each, Tiiii-Py 0.1 mM and LUDOX 0.15 μM; (b) HPwSTD subspectrum of a solution of NMPEA 50 μM, Tiiii-Py 0.1 mM and LUDOX 0.15 μM, 512 scans. (Conditions: 500 MHz, 25 °C, phosphate buffer 5 mM, pH = 7.0, H2O/D2O 90![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10). Full spectra are reported in Fig. S8, ESI.‡ 10). Full spectra are reported in Fig. S8, ESI.‡ | ||
Hence, the cavitand gives to the Tiiii-Py@LUDOX hybrids the ability to differentiate protonated N-methylated amines from protonated primary amines with very similar structure, introducing a remarkable discriminating capability. In addition, the restoration of the hyperfine structure of the signals, with respect to the sample containing only LUDOX, confirms that, notwithstanding the higher affinity for silica of N-methylated amines,18 their direct binding to the nanoparticles is prevented by the surface saturation with the cavitand.
In a final experiment, we verified the possibility to detect N-methylated amines at physiologically relevant concentrations. A sample containing 50 μM NMPEA in buffered water (5 mM phosphate buffer, pD = 7.0) was analysed with the HPwSTD protocol. Also in this case, analyte's signals were detected in the spectrum with a S/N value of 9.1 (Fig. 5(b)). Concentration dependent experiments allowed to determine a LOD value of 31 μM (Fig. S9, ESI‡). Such performance is similar or even better than that of other sensing systems based on the Tiiii-Py cavitands (Table S2, ESI‡),12–16 with the additional bonus of the virtual cancellation of any possibility of false positives due to the recording of the whole analytes’ NMR spectra.
In conclusion, we have here demonstrated as the non-covalent adsorption of the Tiiii-Py cavitand to large silica nanoparticles results in an efficient and selective NMR chemosensing systems. This strategy provides a new tool in the field of NMR spectral editing and chemosensing with nanoparticles. Application of this protocol to the detection of relevant metabolites in biological samples is under investigation.
This work was funded by the Italian Association for Cancer Research (AIRC) under the Investigators Grants scheme (IG 25003). It also benefited from the equipment and framework of the COMP-HUB Initiative, funded by the ‘Departments of Excellence’ program of the Italian Ministry for Education, University and Research (MIUR, 2018–2022).
Conflicts of interest
There are no conflicts to declare.Notes and references
- Z. Xu, C. Liu, S. Zhao, S. Chen and Y. Zhao, Chem. Rev., 2019, 119, 195–230 CrossRef CAS PubMed and references therein.
- (a) Z. Xu, S. Gu, Y. Li, J. Wu and Y. Zhao, Anal. Chem., 2022, 94, 8285–8292 CrossRef CAS PubMed; (b) C. Dong, Z. Xu, L. Wen, S. He, J. Wu, Q.-H. Deng and Y. Zhao, Anal. Chem., 2021, 93, 2968–2973 CrossRef CAS PubMed.
- F. De Biasi, F. Mancin and F. Rastrelli, Prog. Nucl. Magn. Res. Spectrosc., 2020, 117, 70–88 CrossRef CAS PubMed and references therein.
- F. De Biasi, D. Rosa-Gastaldo, X. Sun, F. Mancin and F. Rastrelli, J. Am. Chem. Soc., 2019, 141, 4870–4877 CrossRef CAS PubMed.
- F. De Biasi, B. B. Mascitti, Ē. Kupče and F. Rastrelli, J. Magn. Res., 2022, 338, 107190 CrossRef CAS PubMed.
- (a) M.-V. Salvia, F. Ramadori, S. Springhetti, M. Diez-Castellnou, B. Perrone, F. Rastrelli and F. Mancin, J. Am. Chem. Soc., 2015, 137, 886–892 CrossRef CAS PubMed; (b) L. Gabrielli, D. Rosa-Gastaldo, M. V. Salvia, S. Springhetti, F. Rastrelli and F. Mancin, Chem. Sci., 2018, 9, 4777–4784 RSC.
- X. Sun, L. Riccardi, F. De Biasi, F. Rastrelli, M. De Vivo and F. Mancin, Angew. Chem., Int. Ed., 2019, 58, 7702–7707 CrossRef CAS PubMed.
- D. Neuhaus and M. P. Williamson, Nuclear Overhauser Effect in Structural and Conformational Analysis, Wiley-VCH, 2000 Search PubMed.
- F. De Biasi, D. Rosa-Gastaldo, F. Mancin and F. Rastrelli, Chem. Commun., 2021, 57, 3002–3005 RSC.
- S.-H. Bae, H. J. Dyson and P. E. Wright, J. Am. Chem. Soc., 2009, 131, 6814–6821 CrossRef CAS PubMed.
- R. Pinalli, G. Brancatelli, A. Pedrini, D. Menozzi, D. Hernández, P. Ballester, S. Geremia and E. Dalcanale, J. Am. Chem. Soc., 2016, 138, 8569–8580 CrossRef CAS PubMed.
- M. Dionisio, J. M. Schnorr, V. K. Michaelis, R. G. Griffin, T. M. Swager and E. Dalcanale, J. Am. Chem. Soc., 2012, 134, 6540–6543 CrossRef CAS.
- G. Valenti, E. Rampazzo, E. Biavardi, E. Villani, G. Fracasso, M. Marcaccio, F. Bertani, D. Ramarli, E. Dalcanale, F. Paolucci and L. Prodi, Faraday Discuss., 2015, 185, 299–309 RSC.
- E. Biavardi, C. Tudisco, F. Maffei, A. Motta, C. Massera, G. G. Condorelli and E. Dalcanale, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 2263–2268 CrossRef CAS.
- D. Masseroni, E. Biavardi, D. Genovese, E. Rampazzo, L. Prodi and E. Dalcanale, Chem. Commun., 2015, 51, 12799–12802 RSC.
- E. Biavardi, S. Federici, C. Tudisco, D. Menozzi, C. Massera, A. Sottini, G. G. Condorelli, P. Bergese and E. Dalcanale, Angew. Chem., Int. Ed., 2014, 53, 9183–9188 CrossRef CAS PubMed.
- I. R. N. Verly, R. Leen, J. R. Meinsma, G. K. J. Hooijer, C. D. Savci-Heijink, J. van Nes, M. Broekmans, R. J. A. Wanders, A. B. P. van Kuilenburg and G. A. M. Tytgat, Eur. J. Cancer, 2019, 111, 21–29 CrossRef CAS PubMed.
- (a) B. Zhang, M. Xie, L. Bruschweiler-Li, K. Bingol and R. Brüschweiler, Anal. Chem., 2015, 87, 7211–7217 CrossRef CAS PubMed; (b) M. Xie and R. Brüschweiler, J. Phys. Chem. Lett., 2020, 11, 10401–10407 CrossRef CAS PubMed.
- T. Bian, A. Gardin, J. Gemen, L. Houben, C. Perego, B. Lee, N. Elad, Z. Chu, G. M. Pavan and R. Klajn, Nat. Chem., 2021, 13, 940–949 CrossRef CAS PubMed.
- G. Pieters, A. Cazzolaro, R. Bonomi and L. J. Prins, Chem. Commun., 2012, 48, 1916–1918 RSC.
Footnotes |
| † This paper is dedicated to Prof. Paolo Scrimin, on the occasion of its 70th birthday. |
| ‡ Electronic supplementary information (ESI) available: Experimental details, additional NMR experiments, binding isotherms. See DOI: https://doi.org/10.1039/d2cc04199e |
| § These authors equally contributed to the study. |
| This journal is © The Royal Society of Chemistry 2022 |



