 Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Glycomimetic ligands block the interaction of SARS-CoV-2 spike protein with C-type lectin co-receptors†
Sara
Pollastri‡
a,
Clara
Delaunay‡
b,
Michel
Thépaut
b,
Franck
Fieschi
*b and
Anna
Bernardi
 *a
*a
aUniversità degli Studi di Milano, Dipartimento di Chimica, via Golgi 19, Milano, Italy. E-mail: anna.bernardi@unimi.it
bUniv. Grenoble Alpes, CNRS, CEA, Institut de Biologie Structurale, 38000 Grenoble, France. E-mail: franck.fieschi@ibs.fr
First published on 5th April 2022
Abstract
The C-type lectin receptors DC-SIGN and L-SIGN bind to glycans on the SARS-CoV-2 spike glycoprotein and promote trans-infection of ACE2-expressing cells. We tested C2 triazole-modified mono- and pseudo-di-mannosides as inhibitors of DC/L-SIGN binding to a model mannosylated protein (Man-BSA) and to SARS-CoV2 spike, finding that they inhibit the interaction of both lectins with the spike glycoprotein in a Surface Plasmon Resonance (SPR) assay and are more potent than mannose by up to 36-fold (DC-SIGN) and 10-fold (L-SIGN). The molecules described here are the first known glycomimetic ligands of L-SIGN.
C-Type lectin receptors (CLRs) bind to glycan motifs expressed at the surface of pathogens using a Ca2+ ion in the binding site and respond by activating the initial steps of immune response. Multiple viruses have developed strategies to circumvent the role of CLRs in immunity activation and even to exploit CLRs to their advantage in the infection process. This was first described for HIV, which is captured by the dendritic cell CLR DC-SIGN (CD209) in genital mucosa and uses it as a carrier to reach its main cellular target, T-cells, in a process called trans-infection.1,2 DC-SIGN and its analogue L-SIGN (CD209L or DC-SIGNR) also bind to the spike glycans of SARS-CoV-2 and promote trans-infection of cells expressing ACE2, the viral entry receptor.3–5
Adhesion and entry cofactors of viruses are an important variable of the infection process and also offer additional therapeutic opportunities. In particular, competitive inhibition of virus binding to host co-receptors seems a viable strategy to develop antiviral therapies that exert low evolutionary pressure on the virus and thus do not increase the frequency of viral mutations. We have recently shown that Polyman26, a multivalent glycomimetic ligand of DC-SIGN,6,7 inhibits DC-SIGN binding to the viral spike and blocks DC-SIGN mediated trans-infection of human respiratory cells by SARS-CoV-2.3 The DC-SIGN analog L-SIGN is expressed in human lungs in type II alveolar cells and in endothelial cells and has also been characterized as an entry cofactor for SARS-CoV-2. Importantly, L-SIGN is co-expressed with ACE2 on respiratory tract cells.4,5,8 L-SIGN and DC-SIGN share 77% of their sequence and display similar activity for highly mannosylated oligosaccharides. As opposed to DC-SIGN, L-SIGN does not bind to fucosylated epitopes. The structural basis of this specificity stems from differences in the carbohydrate recognition domains of the two lectins that have been analyzed in detail.9,10 Both DC-SIGN and L-SIGN are transmembrane tetramers, featuring 4 copies of a carbohydrate recognition domain. The tetramers have similar, but not identical, topology and dynamics.11
Over the past decade we12 and others13 have reported on the development of DC-SIGN ligands as inhibitors of DC-SIGN mediate viral infections. To the best of our knowledge, however, no ligands have been reported to bind to L-SIGN. In this paper we disclose the first set of glycomimetic ligands which bind to L-SIGN in an SPR inhibition experiment, and show that they inhibit binding of both DC-SIGN and L-SIGN to immobilized SARS-CoV-2 spike protein.
All the ligands examined here (Fig. 1) contain a mannose residue modified at position 2 with a triazole moiety, deriving from our early design of triazole-modified pseudo-disaccharide scaffolds.12f All C2 triazole-mannosides 3–16 were synthesized by CuAAC reaction of the appropriate 2-azido intermediate, as shown in Scheme 1. Details of the synthesis and characterization of all new compounds are collected in the ESI.†
 | ||
| Fig. 1 Upper panel: Structure of pseudo-dimannoside based compounds; lower panel: structure of mono-mannosides. | ||
 | ||
| Scheme 1 General structure of C2-functionalized Man-based ligands and synthetic approach. Full synthetic schemes in ESI,† Schemes S1 and S2. | ||
Small oligosaccharides, from one to three units, exhibit low affinity towards CLRs, often in the mM or the high μM range and thus at the range limit of most analytical methods. For DC-SIGN, we have used over the years SPR competition tests, where glycomimetics are used to inhibit binding of the lectin to mannosylated bovine serum albumin containing on average 13 glycosylation sites displaying the Manα1–3[Manα1–6]Man trisaccharide (Man-BSA, Dextra), which is immobilized on an SPR chip. Of course, the IC50 values determined under these experimental conditions will depend on the properties of the reporter interaction (amount of ligand on the surface, concentration chosen for the lectin, KD of the reporter interaction itself). In particular, DC-SIGN was used in these assays at a concentration (20 μM) corresponding to about 4 times the apparent KD of the lectin for the surface (≈5 μM). The apparent affinity of L-SIGN for the same Man-BSA surface is over an order of magnitude lower (∼66 μM) (see comparison of the titration curves in Fig. S3, ESI†) and a concentration equal 4 times its KD for the surface is not practical, due to excessive protein consumption. Thus, on this type of Man-BSA surface, it is not possible to operate in the same relative conditions of competition for DC-SIGN and L-SIGN and the resulting IC50 are not directly comparable. Nonetheless, we initially used the Man-BSA surface to obtain a clearer comparison with previous data. Competition towards L-SIGN was thus performed at 70 μM L-SIGN, the highest concentration we could achieve, which corresponds to the KD for the surface. Hence, in this set up the IC50 values of the ligands against L-SIGN are underestimated (and therefore their activity overestimated), relative to DC-SIGN. The results of inhibition experiments with the Man-BSA chip are shown in Fig. 2. The IC50 measured for DC-SIGN can be compared to previous hits,12 while the IC50 for L-SIGN can be used to rank the compounds for L-SIGN inhibition. Although the IC50 of a given compound for DC-SIGN and L-SIGN cannot be directly compared for the reasons detailed above, the trends of the IC50 values can be compared between the two lectins and some tendencies can be appreciated.
 | ||
| Fig. 2 SPR inhibition experiments with Man-BSA chip. Performed in duplicate, using two surfaces with similar levels of Man-BSA functionalization. DC-SIGN (20 μM) or L-SIGN (70 μM) and ligands at increasing concentrations were co-injected. IC50 values (μM) are collected in Table S1 (ESI†). | ||
We started by examining the performance of a set of ligands (1–8, Fig. 1) based on the pseudo-dimannoside scaffold 1, a mimic of Manα1–2Man, that we had developed for DC-SIGN.12,14 With the possible exception of 4 and 5, they all appear to be (modestly) more active for DC-SIGN (Fig. 2, grey bars) than L-SIGN (white bars), especially considering the relative overestimation of the L-SIGN results in this type of experiment. The most active ligand in this series for both lectins is compound 7, one of the most potent monovalent DC-SIGN ligands developed so far.12f The X-ray structure of this molecule in complex with DC-SIGN revealed that its methylene amino triazole moiety engages with an ammonium-binding region in the vicinity of Phe313 and the bis-benzylamino substituents on the cyclohexane ring, which impart selectivity against Langerin, develop lipophilic interactions at the lectin surface, near the Val 351 residue (Fig. S1, ESI†).12f A similar binding mode can be assumed for L-SIGN, given the high similarity of the carbohydrate binding regions (Fig. S1, ESI†). Improvements of the affinity for DC-SIGN are also obtained with the triazolyl-pyridine moiety contained in compound 6 (fourfold increase in DC-SIGN affinity, compared to 1), which is not as effective for L-SIGN. The combination of this moiety with the amide substituents, as in 8, is detrimental for both proteins. This suggests that in DC-SIGN this ligand may adopt an alternative binding mode, where the pyridine ring competes with the amido groups for interaction with Val351. Data recently reported for similar ligands13d support this hypothesis. With the second set of compounds, the monomannosides 9–16, we sought to examine the relevance of the pseudo-mannose cyclohexane moiety on the global affinity. It is worth mentioning that for DC-SIGN the natural Manα-1,2-Man disaccharide has an IC50ca. 1 mM, similar to 1, and is more active than O-methylmannoside by a factor which, depending on the analytical method used, is estimated between three- and ten-fold. The effect is much smaller for these triazole derivatives: ligand 9 loses only about a factor of two compared to the parent pseudo-disaccharide 3 both for DC-SIGN and L-SIGN, with obvious synthetic advantages. The comparison is even favourable for the triazolyl-pyridine derivative 10 over the corresponding pseudo-disaccharide 6, further suggesting that pyridine-bearing ligands may adopt alternative binding modes in the lectins’ binding site. Additional exploration of pyridine functionalization in 11–16 reveals various derivatives that bind to DC-SIGN with an order of magnitude improvement over 1 and Manα-1,2-Man, and thus up to two orders of magnitude over mannose. None of the compounds is selective for L-SIGN, but some (e.g.13) have a clear preference for DC-SIGN.
A subset of the compounds was also tested for inhibition of the interaction of the two lectins with the spike protein of SARS-CoV-2 immobilized on an SPR chip (Fig. 3). As we have previously shown, DC-SIGN and L-SIGN display similar affinity (≈2–3 μM) towards a SARS-CoV-2 recombinant spike protein produced using an expression system well-characterized in terms of its site-specific glycosylation.3 This can be appreciated by comparing the two lectins’ titration curves on the Spike surface in Fig. S4 (ESI†). Thus, this surface allows a better analysis of L-SIGN binding inhibition and a cleaner evaluation of the relative affinity of each compound for both lectins. The results (Fig. 3) clearly support the trends already established on the Man-BSA surface, but, as expected, the IC50 values against L-SIGN display a relative increase, corresponding to a lower potency of the inhibitor. Nonetheless, these data confirm that the amino-substituted pseudo-dimannosides 3 and 7, as well as the amino-triazolyl monomannoside 9 gain one order of magnitude activity over the pseudo-disaccharide ligand 1 and mannose (D-Man, Fig. 3) and are strong candidates for the simultaneous inhibition of spike binding to DC-SIGN and L-SIGN through multivalent constructs.15 The monomannosides bearing a triazolyl-pyridine moiety, and in particular 13, display a significant selectivity and a strong affinity for DC-SIGN, together with obvious structural and synthetic simplification relative to the pseudo-disaccharides. They may be a useful and easily accessible tool to dissect the role of DC-SIGN and L-SIGN as SARS-CoV-2 co-receptors.
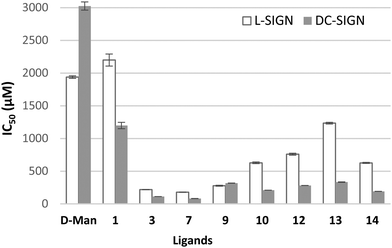 | ||
| Fig. 3 SPR inhibition experiments with spike chip. Performed in duplicate, using two surfaces with similar Spike functionalization. The lectin (20 μM) and ligands at increasing concentrations were co-injected. IC50 values (μM) are collected in Table S1 (ESI†). | ||
In conclusion, we have reported here the first group of glycomimetic ligands of L-SIGN. Their activity for L-SIGN and DC-SIGN was determined through an inhibition SPR experiment that makes use of immobilized SARS-CoV-2 spike protein as a reporter. Relative to mannose, affinity improvements by up to over one order of magnitude have been observed when using mannose residues modified at position 2 with a methylene amino triazole moiety, both as part of a pseudo-dimannoside (3–5 and 7), or as a monosaccharide (9). No selectivity has been observed so far against DC-SIGN, but, rather some of the triazolyl-pyridine derivatives, such as 13, display a moderate preference for DC-SIGN over L-SIGN.
The authors thank the CDP Glyco@Alps (ANR-15-IDEX-02) for financial support. This work used the facilities of Unitech-COSPECT at the University of Milan and the platforms of the Grenoble Instruct-ERIC centre (ISBG; UMS 3518 CNRS-CEA-UGA-EMBL) within the Grenoble Partnership for Structural Biology (PSB), supported by FRISBI (ANR-10-INBS-05-02) and GRAL, within the University Grenoble Alpes graduate school CBH-EUR-GS (ANR-17-EURE-0003).
Conflicts of interest
There are no conflicts of interest to declare.Notes and references
- T. B. H. Geijtenbeek, D. S. Kwon, R. Torensma, S. J. van Vliet, G. C. F. van Duijnhoven, J. Middel, I. L. M. H. A. Cornelissen, H. S. L. M. Nottet, V. N. KewalRamani, D. R. Littman, C. G. Figdor and Y. van Kooyk, Cell, 2000, 100, 587–597 CrossRef CAS PubMed.
- T. B. H. Geijtenbeek, R. Torensma, S. J. van Vliet, G. C. F. van Duijnhoven, G. J. Adema, Y. van Kooyk and C. G. Figdor, Cell, 2000, 100, 575–585 CrossRef CAS PubMed.
- M. Thépaut, J. Luczkowiak, C. Vivès, N. Labiod, I. Bally, F. Lasala, Y. Grimoire, D. Fenel, S. Sattin, N. Thielens, G. Schoehn, A. Bernardi, R. Delgado and F. Fieschi, PLoS Pathog., 2021, 17, e1009576 CrossRef PubMed.
- (a) R. Amraei, W. Yin, M. A. Napoleon, E. L. Suder, J. Berrigan, Q. Zhao, J. Olejnik, K. B. Chandler, C. Xia, J. Feldman, B. M. Hauser, T. M. Caradonna, A. G. Schmidt, S. Gummuluru, E. Mühlberger, V. Chitalia, C. E. Costello and N. Rahimi, ACS Cent. Sci., 2021, 7, 1156–1165 CrossRef CAS PubMed; (b) F. A. Lempp, L. B. Soriaga, M. Montiel-Ruiz, F. Benigni, J. Noack, Y.-J. Park, S. Bianchi, A. C. Walls, J. E. Bowen, J. Zhou, H. Kaiser, A. Joshi, M. Agostini, M. Meury, E. Dellota, S. Jaconi, E. Cameroni, J. Martinez-Picado, J. Vergara-Alert, N. Izquierdo-Useros, H. W. Virgin, A. Lanzavecchia, D. Veesler, L. A. Purcell, A. Telenti and D. Corti, Nature, 2021, 598, 342–347 CrossRef CAS PubMed.
- Q. Lu, J. Liu, S. Zhao, M. F. Gomez Castro, M. Laurent-Rolle, J. Dong, X. Ran, P. Damani-Yokota, H. Tang, T. Karakousi, J. Son, M. E. Kaczmarek, Z. Zhang, S. T. Yeung, B. T. McCune, R. E. Chen, F. Tang, X. Ren, X. Chen, J. C. C. Hsu, M. Teplova, B. Huang, H. Deng, Z. Long, T. Mudianto, S. Jin, P. Lin, J. Du, R. Zang, T. T. Su, A. Herrera, M. Zhou, R. Yan, J. Cui, J. Zhu, Q. Zhou, T. Wang, J. Ma, S. B. Koralov, Z. Zhang, I. Aifantis, L. N. Segal, M. S. Diamond, K. M. Khanna, K. A. Stapleford, P. Cresswell, Y. Liu, S. Ding, Q. Xie and J. Wang, Immunity, 2021, 54, 1304–1319.e9 CrossRef CAS PubMed.
- S. Ordanini, N. Varga, V. Porkolab, M. Thépaut, L. Belvisi, A. Bertaglia, A. Palmioli, A. Berzi, D. Trabattoni, M. Clerici, F. Fieschi and A. Bernardi, Chem. Commun., 2015, 51, 3816 RSC.
- A. Berzi, S. Ordanini, B. Joosten, D. Trabattoni, A. Cambi, A. Bernardi and M. Clerici, Sci. Rep., 2016, 6, 35373 CrossRef CAS PubMed.
- V. S. F. Chan, K. Y. K. Chan, Y. Chen, L. L. M. Poon, A. N. Y. Cheung, B. Zheng, K.-H. Chan, W. Mak, H. Y. S. Ngan, X. Xu, G. Screaton, P. K. H. Tam, J. M. Austyn, L.-C. Chan, S.-P. Yip, M. Peiris, U.-S. Khoo and C.-L. S. Lin, Nat. Genet., 2006, 38, 38 CrossRef CAS PubMed.
- H. Feinberg, D. A. Mitchell, K. Drickamer and W. I. Weis, Science, 2001, 294, 2163–2166 CrossRef CAS PubMed.
- Y. Guo, H. Feinberg, E. Conroy, D. A. Mitchell, R. Alvarez, O. Blixt, M. E. Taylor, W. I. Weis and K. Drickamer, Nat. Struct. Mol. Biol., 2004, 11, 591–598 CrossRef CAS PubMed.
- Y. Guo, I. Nehlmeier, E. Poole, C. Sakonsinsiri, N. Hondow, A. Brown, Q. Li, S. Li, J. Whitworth, Z. Li, A. Yu, R. Brydson, W. B. Turnbull, S. Pöhlmann and D. Zhou, J. Am. Chem. Soc., 2017, 139, 11833–11844 CrossRef CAS PubMed.
- (a) J. J. Reina, S. Sattin, D. Invernizzi, S. Mari, L. Martínez-Prats, G. Tabarani, F. Fieschi, R. Delgado, P. M. Nieto, J. Rojo and A. Bernardi, ChemMedChem, 2007, 2, 1030–1036 CrossRef CAS PubMed; (b) G. Timpano, G. Tabarani, M. Anderluh, D. Invernizzi, F. Vasile, D. Potenza, P. M. Nieto, J. Rojo, F. Fieschi and A. Bernardi, ChemBioChem, 2008, 9, 1921–1930 CrossRef CAS PubMed; (c) S. Sattin, A. Daghetti, M. Thépaut, A. Berzi, M. Sánchez-Navarro, G. Tabarani, J. Rojo, F. Fieschi, M. Clerici and A. Bernardi, ACS Chem. Biol., 2010, 5, 301–312 CrossRef CAS PubMed; (d) N. Varga, I. Sutkeviciute, C. Guzzi, J. McGeagh, I. Petit-Haertlein, S. Gugliotta, J. Weiser, J. Angulo, F. Fieschi and A. Bernardi, Chem. – Eur. J., 2013, 19, 4786–4797 CrossRef CAS PubMed; (e) V. Porkolab, E. Chabrol, N. Varga, S. Ordanini, I. Sutkevičiūtė, M. Thépaut, M. J. García-Jiménez, E. Girard, P. M. Nieto, A. Bernardi and F. Fieschi, ACS Chem. Biol., 2018, 13, 600–608 CrossRef CAS PubMed; (f) L. Medve, S. Achilli, J. Guzman-Caldentey, M. Thépaut, L. Senaldi, A. le Roy, S. Sattin, C. Ebel, C. Vivès, S. Martin-Santamaria, A. Bernardi and F. Fieschi, Chem. – Eur. J., 2019, 25, 14659–14668 CrossRef CAS PubMed.
- (a) M. J. Borrok and L. L. Kiessling, J. Am. Chem. Soc., 2007, 129, 12780–12785 CrossRef CAS PubMed; (b) S. L. Mangold, L. R. Prost and L. L. Kiessling, Chem. Sci., 2012, 3, 772–777 RSC; (c) J. Aretz, H. Baukmann, E. Shanina, J. Hanske, R. Wawrzinek, V. A. Zapol'skii, P. H. Seeberger, D. E. Kaufmann and C. Rademacher, Angew. Chem., Int. Ed., 2017, 56, 7292–7296 CrossRef CAS PubMed; (d) J. Cramer, A. Lakkaichi, B. Aliu, R. P. Jakob, S. Klein, I. Cattaneo, X. Jiang, S. Rabbani, O. Schwardt, G. Zimmer, M. Ciancaglini, T. Abreu Mota, T. Maier and B. Ernst, J. Am. Chem. Soc., 2021, 143, 17465–17478 CrossRef CAS PubMed; (e) R. Wawrzinek, E.-C. Wamhoff, J. Lefebre, M. Rentzsch, G. Bachem, G. Domeniconi, J. Schulze, F. F. Fuchsberger, H. Zhang, C. Modenutti, L. Schnirch, M. A. Marti, O. Schwardt, M. Bräutigam, M. Guberman, D. Hauck, P. H. Seeberger, O. Seitz, A. Titz, B. Ernst and C. Rademacher, J. Am. Chem. Soc., 2021, 143, 18977–18988 CrossRef CAS PubMed.
- The 1,2-dicarbomethoxycyclohexane-4,5-diol moiety of 1 is conformationally locked to mimic a 1,2-disubstituted mannose residue, i.e. the reducing end of the Manα1–2Man disaccharide.
- D. Budhadev, E. Poole, I. Nehlmeier, Y. Liu, J. Hooper, E. Kalverda, U. S. Akshath, N. Hondow, W. B. Turnbull, S. Pöhlmann, Y. Guo and D. Zhou, J. Am. Chem. Soc., 2020, 142, 18022–18034 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d2cc00121g |
| ‡ These authors contributed equally to this paper. |
| This journal is © The Royal Society of Chemistry 2022 |
