 Open Access Article
Open Access ArticleA peptide-crosslinking approach identifies HSPA8 and PFKL as selective interactors of an actin-derived peptide containing reduced and oxidized methionine†
Aaron
Maurais
and
Eranthie
Weerapana
 *
*
Department of Chemistry, Boston College, Chestnut Hill, MA 02467, USA. E-mail: eranthie@bc.edu
First published on 15th September 2022
Abstract
The oxidation of methionine to methionine sulfoxide occurs under conditions of cellular oxidative stress, and modulates the function of a diverse array of proteins. Enzymatic systems that install and reverse the methionine sulfoxide modifications have been characterized, however, little is known about potential readers of this oxidative modification. Here, we apply a peptide-crosslinking approach to identify proteins that are able to differentially interact with reduced and oxidized methionine-containing peptides. Specifically, we generated a photo-crosslinking peptide derived from actin, which contains two sites of methionine oxidation, M44 and M47. Our proteomic studies identified heat shock proteins, including HSPA8, as selective for the reduced methionine-containing peptide, whereas the phosphofructokinase isoform, PFKL, preferentially interacts with the oxidized form. We then demonstrate that the favored interaction of PFKL with oxidized methionine is also observed in the full-length actin protein, suggesting a role of methionine oxidation in regulating the actin-PFKL interaction in cells. Our studies demonstrate the potential to identify proteins that can differentiate between reduced and oxidized methionine and thereby mediate downstream protein functions under conditions of oxidative stress. Furthermore, given that numerous sites of methionine oxidation have now been identified, these studies set the stage to identify putative readers of methionine oxidation on other protein targets.
Introduction
Oxidative post-translational modifications (PTMs) play an important role in regulating protein function. Cells produce low levels of reactive oxygen species (ROS) as a byproduct of metabolism, and these ROS can regulate protein function and modulate signaling pathways.1 Cysteine is traditionally thought of as the primary ROS target due to its highly reactive thiol side chain.2,3 However, reversible oxidation of methionine (Met) to Met sulfoxide can also occur in the presence of ROS. Met sulfoxide that is generated spontaneously in the presence of high levels of cellular ROS results in a mixture of diastereomers, Met-S-sulfoxide and Met-R-sulfoxide.4 In contrast, enzymes such as the molecule interacting with CasL (Mical) monooxygenases can stereospecifically oxidize Met to form Met-R-sulfoxide. In humans, the Mical family consists of 3 members that each contain a flavoprotein monooxygenase domain thought to be responsible for direct oxidation of Met within proteins, including actin.4 In humans, Met oxidation is reversed by a family of Met sulfoxide reductases, including (MsrA) and B (MsrB).5 MsrA is selective for Met-S-sulfoxide and can act on both the free amino acid, as well as protein substrates. MsrB1, 2 and 3 are specific for protein Met-R-sulfoxide, and are localized to the cytoplasm and nucleus, mitochondria, and endoplasmic reticulum, respectively.6Targeted, reversible Met oxidation can directly regulate protein function.6 For example, Met oxidation has been shown to regulate ROS sensing transcription factors in E. Coli,7 activation of CaMKII8 and the activity of calmodulin.9,10 Of particular interest is the role of Met oxidation in regulating actin dynamics. Actin filaments provide mechanical support, are the driving force for cell movement, and are critical for trafficking intracellular vesicles.11 Eukaryotic actin can exist in either a soluble form (G-actin) or filamentous form (F-actin). After the initial identification of Mical as a cytoskeleton-interacting protein in Drosophila,12 it was shown that Mical can catalyze the stereospecific oxidation of two Met residues in actin, M44 and M47, which are conserved across both vertebrates and invertebrates.13 Mical-catalyzed oxidization of M44 and M47 to Met-R-sulfoxide promotes actin filament disassembly.13,14 MsrB1 reduces both methionine sulfoxide residues and facilitates filament assembly.15
Given the characterized role of Met oxidation in regulating protein function, it is important to fully understand the mechanisms by which this modification is installed, removed and further translated to a downstream functional outcome. Although the role of Mical enzymes in oxidation, and MsrA/B enzymes in reduction of Met sulfoxide, little is known about potential ‘readers’ of the Met sulfoxide modification. We therefore aimed to identify proteins that have the ability to differentially interact with oxidized and reduced Met. One method for identifying ‘readers’ of PTMs is through the chemical synthesis of modified and unmodified peptides16,17 or large scale combinatorial peptide libraries18 to capture proteins which interact with a specific modification state in vitro. In addition, genetic code expansion approaches19 have facilitated the incorporation of PTMs in the context of live cells.20 For labile modifications that are unlikely to persist within the cell, non-hydrolyzable, or more stable analogs have been generated as a proxy for the native modification.21 In the case of Met sulfoxide, the ubiquitous presence of Mical and Msr enzymes will likely either non-specifically install the modification, or reverse the modification from the protein of interest when expressed in cells. Additionally, there is no analog of Met sulfoxide that is resistant to enzymatic reduction. Therefore, we applied a peptide based photo-crosslinking approach17 to identify proteins that differentially interact with Met and Met sulfoxide in vitro. Specifically, we generated a peptide-based photo-crosslinking probe derived from the well characterized Met sulfoxide sites in actin, and demonstrate that HSPA8 and PFKL selectively interact with the Met and Met sulfoxide peptides, respectively.
Results and discussion
Photo-crosslinking probes containing Met and Met sulfoxide show differential protein labeling
Previous studies have used peptide-based photo-crosslinking probes to identify proteins that interact with trimethylated,17 and succinylated lysine.22 We adapted a similar strategy to identify proteins that differentially interact with Met and Met sulfoxide. We chemically synthesized two peptide-based photo-crosslinking probes (Fig. 1A) that spanned the sequence from H42 to K52 in human actin B (ACTB). This region of ACTB contains the two sites of Met oxidation, M46 and M49. Probe 1 contained reduced Met at both positions, whereas probe 2 contained Met sulfoxide at both positions. The Met sulfoxide that was incorporated was a mixture of Met-S-sulfoxide and Met-R-sulfoxide. Additionally, the two probes contained the following components; (1) a benzophenone photo-crosslinker, which upon UV irradiation forms a covalent adduct with proximal proteins, enabling the capture of transient probe-protein interactions; and (2) a biotin tag, which facilitates detection by biotin blot, or affinity enrichment using streptavidin-agarose beads. The two peptides were synthesized using standard solid-phase peptide synthesis (SPPS), purified by high performance liquid chromatography (HPLC), and confirmed to contain the reduced and oxidized methionine residues by LC/MS analysis (Fig. S1, ESI†).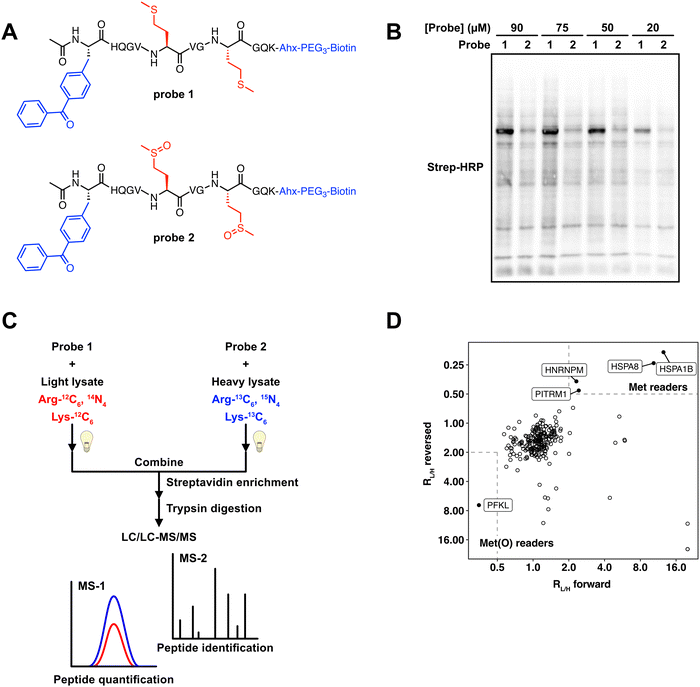 | ||
| Fig. 1 Photo-crosslinking peptide probes to identify readers of methionine oxidation. (A) Structures of actin probes 1 and 2. (B) Probes 1 and 2 label MCF7 lysates in a concentration-dependent manner and show distinct labeling patterns. Uncropped gel image provided in Fig. S5 (ESI†). (C) Workflow used to identify proteins that interact selectively with oxidized or reduced methionine. 50 μM probes 1 or 2 are crosslinked in isotopically heavy or light MCF7 lysates. After UV crosslinking, equal amounts of heavy and light lysate are combined. Proteins that are bound to the probe are enriched on streptavidin agarose beads, trypsinized, and subjected to LC–MS/MS analysis. (D) 2D plot showing SILAC RL/H for each protein in the “forward” (x-axis) and “reversed” (y-axis) experiments. Proteins that interact preferentially with probe 1 are in the upper right portion of the plot and preferential interactors of probe 2 are in the bottom left portion. | ||
A significant limitation of using a peptide-based crosslinking approach is that the peptide can only duplicate the primary amino acid sequence in actin between H42 and K52. Secondary and tertiary structural elements will not be preserved. Therefore, protein interactions with the actin peptide probes may not necessarily translate to the full-length protein. To determine if these peptide-based probes can interact with proteins known to interact with this specific region of actin, we tested the oxidase activity of Mical1 against probe 1. Mical redox enzymes have been shown to induce actin disassembly through targeted oxidation of M46 and M49.13–15,23 Although MICALs have some basal level of NADPH consumption even in the absence of substrate,23,24 they are known to show an increased rate of NADPH consumption in the presence of F-actin.13,14,23,24 We treated the reduced peptide probe 1 with the purified catalytic domain of human Mical1, and monitored the appearance of the oxidized form of this peptide. We observed the formation of oxidized product by LC/MS (Fig. S2, ESI†), suggesting that although our peptide probes are not perfect representations of full-length actin, they do have the ability to interact with interacting proteins known to recognize that specific region of actin.
To test the cross-linking ability of probes 1 and 2, the probes were added to MCF7 cell lysates at varying concentrations. Upon irradiation of the probe-treated cell lysates, cross-linked proteins were visualized by biotin blot. Both probes displayed concentration-dependent covalent labeling of proteins within the MCF7 lysates. The majority of labeled protein bands were shared across the two probes, suggesting that the most interacting proteins were ambivalent as to the oxidation state of the Met residues within the peptide. However, a high intensity protein band was observed at ∼75 kDa in the probe 1 samples (Fig. 1B), suggesting the presence of proteins that could potentially interact selectively with either the Met or Met sulfoxide probes. We next sought to identify these protein interactors using a mass-spectrometry (MS) proteomics workflow.
SILAC-MS experiments identify HSPA8 and PFKP as selective interactors with Met and Met sulfoxide
To identify protein interactors of probes 1 and 2, we applied a stable isotope labeling by amino acids in cell culture (SILAC)-MS experiment. We generated light and heavy MCF7 cell lysates which had been SILAC labeled with isotopically light or heavy arginine and lysine (Fig. S3, ESI†). These lysates were treated with either probe 1 or probe 2, and subjected to UV irradiation. The resulting biotinylated proteins were enriched on streptavidin beads, and subsequent on-bead trypsin digestion afforded a mixture of peptides for MS analysis. In a “forward” experiment, probe 1 was added to light lysate and probe 2 was added to heavy lysate. In a parallel “reverse” experiment, the probes were switched, whereby probe 1 was added to heavy lysate and probe 2 was added to light lysate. In each case the light and heavy lysates were mixed together prior to MS analysis, and the abundance of each peptide was quantified based on the relative intensity of the isotopically light and heavy variants. The resulting data were used to generate a plot where the light to heavy ratios (RL/H) for the forward experiment is plotted on the x-axis, and the RL/H for the reversed experiment is plotted on the y-axis. Proteins that were equally enriched in both light and heavy samples are clustered around a RL/H of ∼1 on both axes, and signify proteins that interact with the actin-derived peptide regardless of the oxidation status of the Met residues. In contrast, proteins that selectively interact with either the Met or Met sulfoxide-containing peptides will be clustered at the top right or bottom left corners respectively (Fig. 1D). Our SILAC-MS analysis resulted in 238 proteins identified in both the “forward” and “reverse” experiments (Table S1, ESI†). Of these 238 proteins, 233 displayed RL/H of ∼1, suggesting that these proteins were agnostic to the oxidation status of the peptide. There were 5 proteins that differentially interacted with either the reduced or oxidized Met probes. Of these, 4 proteins (HSPA8, HSPA1B, HNRNPM, and PITRM1) interacted more strongly with the reduced Met probe, and one protein, PFKL, appeared to selectively interact with the oxidized Met probe.HSPA8 and HSPA1B are members of the heat shock protein (HSP) family that serve a wide array of functions related to maintaining cellular protein homeostasis.25 These HSPs contain a central ATP/ADP binding site, and undergo a conformational change upon the hydrolysis of ATP. Although smaller HSPs are known to interact with actin,26 a direct interaction between HSPA8 and HSPA1B with actin has not been documented. Given that the molecular weights of HSPA8 and HSPA1B are within the ∼70 kDa range, the protein band visualized in the gel analysis (Fig. 1B) to be enriched in the probe 1 treated lysates is likely attributed HSPA8 and HSPA1B. HNRNPM is a heterogeneous nuclear riboprotein that regulates splicing thorough pre-mRNA binding, and PITRM1 is a mitochondrial matrix-localized metalloendopeptidase. Give that the cellular localization of these two proteins precludes interaction with actin and the cytoskeleton, it is unlikely that HNRNPM and PITRM1 are direct interactors with actin.
PFKL is the liver isoform of phosphofructokinase (PFK) and was found to selectively interact with the oxidized Met probe. We identified peptides that are common to all 3 PFK isoforms, however PFKL is the only family member that contained unique peptide sequences in our datasets. Glycolytic enzymes, including PFK, have long been known to directly interact with F-actin filaments, whereupon actin binding serves to increase the rate of glycolytic activity.27 Enrichment studies with antibodies against specific peptide sequences within actin, indicated that PFK can bind to actin at multiple distinct sites, including between residues spanning the peptide sequences used for our crosslinking studies.28 However, preferential interaction of PFK with oxidized actin has not been previously reported.
We focused our attention on HSPA8 and PFKL as putative proteins that selectively interact with reduced and oxidized Met-containing actin, respectively. To confirm our MS data and demonstrate selective interaction of HSPA8 and PFKL with reduced and oxidized Met, we treated cell lysates with each of the probes, performed the UV crosslinking and avidin enrichment. The enriched proteins were then eluted from the beads and assessed by western blotting to monitor the presence of HSPA8 and PFKL. In agreement with the SILAC-MS data, western blotting confirmed that HSPA8 selectively binds to probe 1 containing reduced Met residues. In contrast, PFKL preferentially bound to probe 2 containing oxidized Met residues (Fig. 2A). PFKL did show binding to the reduced Met probe as well, but displayed a 2.5-fold preference for the oxidized form.
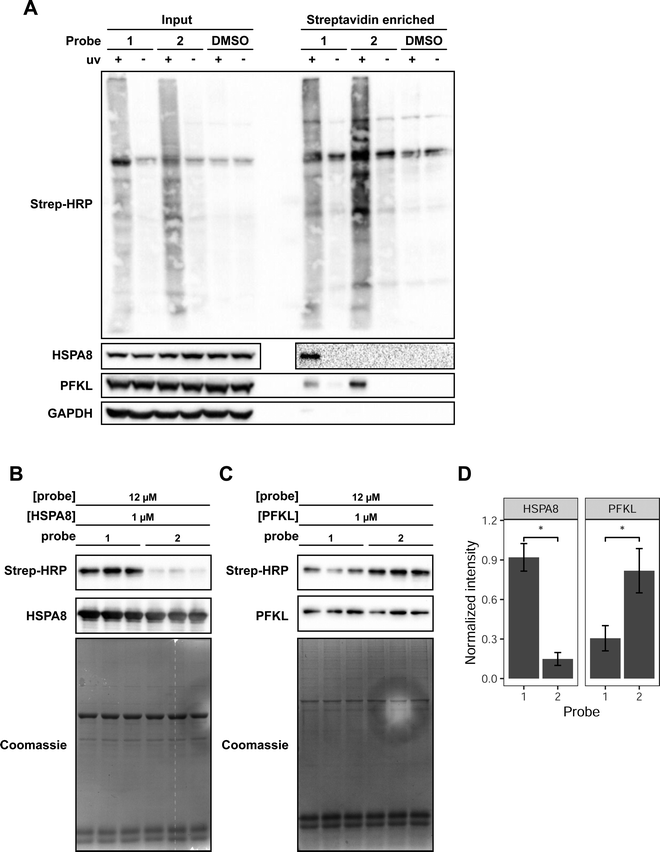 | ||
| Fig. 2 Validation of HSPA8 and PFKP as selective binders for Met and Met sulfoxide-containing peptide probes. (A) Detection of endogenous PFKL and HSPA8 in MCF7 lysates enriched with probe 1 or 2. MCF7 lysates were crosslinked with 75 μM of the indicated probe. An aliquot of the labeled lysate was taken and run as the “input” fraction. The labeled proteins were enriched on streptavidin-agarose beads. The bound proteins were eluted by heating and treatment with 50 μM biotin to obtain the “enriched” fraction. Each fraction was analyzed by western blot using antibodies for HSPA8 and PFKL. Streptavidin-HRP was used to detect probe-labeled proteins. Uncropped gel images provided in Fig. S6 (ESI†). (B) 12 μM of the indicated probe was combined with 1 μM purified HSPA8 and (C) PFKL in vitro. Each lane is a biological replicate performed with a new preparation of protein. Uncropped gel images provided in Fig. S7 (ESI†). (D) Mean densitometry measurements of 3 biological replicates shown in B and C. Band intensities from the streptavidin blot were normalized to the intensity of the HSPA8 or PFKL band and then normalized intensity of the most intense replicate, such that the intensity of the most intense replicate is set to 1. Replicates were statistically analyzed by Student's t test (*p < 0.005). Error bars represent the standard deviation of the 3 measurements. | ||
Upon confirming the selective interaction of probe 1 and 2 with HSPA8 and PFKP in cell lysates, we then proceeded to monitor the ability of probes 1 and 2 to bind to recombinant purified HSPA8 and PFKL. HSPA8 with an N-terminal HA tag was expressed and purified from E. coli. PFKL was expressed and purified from sf9 insect cells via baculovirus transduction.29 To verify that PFKL produced from sf9 cells was active, we performed a spectrophotometric coupled enzyme assay,30 which demonstrated that the sf9-produced PFKL was active with a Vmax of 2.45 × 10−3 s−1, and Km,F6P of 2.3 μM (Fig. S4, ESI†).
Upon obtaining purified HSPA8 and PFKL protein, crosslinking reactions with recombinant protein were performed with 1 μM HSPA8 or PFKL and a 12 molar excess of probes 1 or 2. We also included 0.2 mg mL−1 casein in the reaction mixture as a non-specific protein interactor. The purified protein and casein mixtures were treated with the probes and irradiated to induce the covalent crosslink. Covalent protein labeling of HSPA8 and PFKL by probes 1 and 2 was monitored by biotin blot. Western blotting for recombinant HSPA8 (anti-HA) and PFKL (anti-PFKL) confirmed equal protein loading in each lane. The experiment was performed in 3 biological replicates, and both probes showed a statistically significant preference for the expected protein target. Probe 1 showed a ∼6 fold selectivity for recombinant HSPA8 and probe 2 showed a ∼3 fold selectivity for recombinant PFKL (Fig. 2B–D).
PFKL interacts preferentially with oxidized actin (ACTB)
Next, we sought to investigate if HSPA8 and PFKL can preferentially interact with full-length reduced and oxidized ACTB, respectively. We first evaluated if HSPA8 and PFKL are able to interact with endogenous ACTB in cells. ACTB interactions with HSPA8 and PFKL were investigated by immunoprecipitating ACTB from cell lysates followed by subsequent western blotting for HSPA8 and PFKL. These immunoprecipitation studies showed that PFKL stably interacts with ACTB in MCF7 lysates (Fig. 3A). Both ACTB and PFKL showed some background non-specific binding to the beads, but both signals were significantly enhanced in the presence of the anti-ACTB antibody. In contrast, HSP8 did now show any noticeable signal in the co-immunoprecipitation studies, suggesting that there was no stable interaction between endogenous HSPA8 and ACTB. We then selectively enriched F-actin from cell lysates using biotin-phalloidin.31 Again, PFKL, and not HSPA8, was shown to associate with F-actin (Fig. 3B).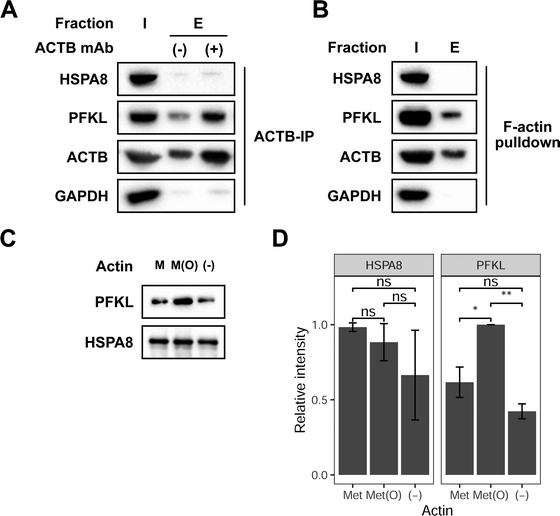 | ||
| Fig. 3 PFKL preferentially interacts with oxidized full-length actin. (A) Immunoprecipitation of ACTB from MCF7 lysates was followed by western blotting of the input (I) and bead-bound (E) fractions with anti-HSPA8, PFKL and GAPDH antibodies. Uncropped gel images provided in Fig. S8 (ESI†). (B) 25 μg biotin-phalloidin was added to MCF7 lysates and then incubated with streptavidin-agarose beads. The input (I) and bead bound (E) fractions were analyzed by western blot with anti-HSPA8, PFKL and GAPDH antibodies. Uncropped gel images are provided in Fig. S9 (ESI†). (C) 50 μg reduced or Mical oxidized G-actin was labeled with biotin-NHS then bound to streptavidin-agarose beads. 6 μM of recombinant HSPA8 and PFKL were combined and incubated with the immobilized actin. The bead bound fractions for reduced (M), oxidized (M(O)), and no actin (−) was analyzed by western blot using anti-PFKL and HA (HSPA8) antibodies. A representative of 3 biological replicates is shown. Uncropped gel images shown in Fig. S10 (ESI†). (D) Mean densitometry measurements of 3 biological replicates of the experiment shown in replicates were statistically analyzed by Student's t test (*p < 0.05, **p < 0.005). Error bars represent the standard deviation of the 3 measurements. | ||
To better understand how the oxidation state of ACTB affects HSPA8 and PFKL binding, reduced and oxidized ACTB was treated with recombinant HSPA8 and PFKL, and the levels of HSPA8 and PFKL that interact with ACTB were monitored upon immunoprecipitation of ACTB. Similar to the interaction we observed with the ACTB-derived peptide probes, we show that PFKL interacts with both reduced and oxidized ACTB, but interacts to a greater extent with the oxidized form (Fig. 3C and D). As before, we could not detect any interaction of HSPA8 with either reduced or oxidized ACTB, suggesting that HSPA8 only stably interacted wtih the truncated ACTB-based peptide probes and not the full-length protein.
Conclusion
In summary, we generated actin-derived crosslinking peptides containing reduced and oxidized Met residues at positions M46 and M49. These crosslinking probes were applied to cell lysates with the goal of identifying proteins that differentially interact with either the reduced or oxidized Met forms. Our SILAC-MS studies identified 4 proteins (HSPA8, HSPA1B, HNRNPM, and PITRM1) that selectively interacted with the reduced Met peptide, and one protein (PFKL) that selectively interacted with the oxidized Met peptide. We focused our attention on HSPA8 and PFKL as selective interactors of reduced and oxidized Met, respectively. Differential interaction of the reduced and oxidized peptide probes was confirmed by crosslinking studies using recombinant HSPA8 and PFKL. Importantly, the preference of PFKP to bind to the oxidized Met peptide was found to be conserved in the full-length actin protein, whereby the interaction between PFKP with oxidized full-length actin was enhanced relative to reduced full-length actin. It is well established that PFK isoforms interact with cytoskeletal filaments,32–34 and several recent studies have also started to unveil a possible interplay between cell metabolism and cytoskeletal organization.35–37 Additionally, PFK has been shown to be downregulated through polyubiquitination by TRIM21 upon cytoskeletal remodelling.38 The preferential interaction between PFKL and oxidized actin could suggest a possible mechanism by which PFKL is selectively recruited and degraded at the cytoskeletal matrix under conditions of oxidative stress. Future experiments will further evaluate the role of the oxidized actin-PFKL interaction in regulating both actin and PFKL function in cells.In contrast to PFKL, although HSPA8 showed robust and selective labeling by probe 1, this interaction could not be recapitulated at the protein level. There are two possible reasons for this; (1) the interaction between HSPA8 and actin is transient and not strong enough to withstand immunoprecipitation; or, (2) HSPA8 only interacts with the actin-derived peptide probe, but not the full-length protein. Additional experiments are warranted to further characterize the biological consequences of methionine oxidation on the dynamics between HSPA8 and actin.
Together, our studies apply photo-crosslinking peptide-based probes to identify proteins that preferentially interact with reduced and oxidized methionine. Although similar strategies have been widely applied to numerous PTMs, methionine oxidation has not been previously investigated using this approach. We note several limitations of this approach to identify bona fide Met/Met sulfoxide interactors. First, as demonstrated for HSPA8, interaction with the peptide probes do not necessarily translate to interactions with the full-length protein. Additionally, Met sulfoxide exists as Met-S-sulfoxide and Met-R-sulfoxide, and it is unclear if reader proteins exist that can differentiate between both these isomers. In our studies we used a mixture of Met-S-sulfoxide and Met-R-sulfoxide, but resolution into the individual isomers may lead to other protein targets that show stereochemical preference for this oxidative modification. Lastly, it is likely that no general Met sulfoxide reader protein exists, and instead each unique Met sulfoxide site may have a dedicated reader protein associated with it. Therefore, a next step is to repeat these studies with peptide probes derived from other known functionally relevant sites of Met oxidation. Future studies will aim to address these limitations and broaden our understanding of how Met oxidation can regulate protein function through selective reader protein-mediated interactions.
Cell lines
MCF7 cells were purchased from ATCC.Conflicts of interest
The authors declare no competing financial interests.Acknowledgements
This work was funded by NIH grant R35GM134964 to E. W. We thank members of the Weerapana Lab for helpful discussions.References
- J. R. Stone and S. Yang, Hydrogen peroxide: a signaling messenger, Antioxid. Redox Signaling, 2006, 8(3-4), 243–270 CrossRef CAS PubMed.
- T. H. Truong and K. S. Carroll, Redox regulation of epidermal growth factor receptor signaling through cysteine oxidation, Biochemistry, 2012, 51(50), 9954–9965 CrossRef CAS PubMed.
- D. W. Bak and E. Weerapana, Cysteine-mediated redox signalling in the mitochondria, Mol. BioSyst., 2015, 11(3), 678–697 RSC.
- B. Manta and V. N. Gladyshev, Regulated methionine oxidation by monooxygenases, Free Radical Biol. Med., 2017, 109, 141–155 CrossRef CAS PubMed.
- B. C. Lee, A. Dikiy, H. Y. Kim and V. N. Gladyshev, Functions and evolution of selenoprotein methionine sulfoxide reductases, Biochim. Biophys. Acta, 2009, 1790(11), 1471–1477 CrossRef CAS.
- A. Kaya, B. C. Lee and V. N. Gladyshev, Regulation of protein function by reversible methionine oxidation and the role of selenoprotein MsrB1, Antioxid. Redox Signaling, 2015, 23(10), 814–822 CrossRef CAS PubMed.
- A. Drazic, H. Miura, J. Peschek, Y. Le, N. C. Bach, T. Kriehuber and J. Winter, Methionine oxidation activates a transcription factor in response to oxidative stress, Proc. Natl. Acad. Sci. U. S. A., 2013, 110(23), 9493–9498 CrossRef CAS PubMed.
- J. R. Erickson, M. L. Joiner, X. Guan, W. Kutschke, J. Yang, C. V. Oddis, R. K. Bartlett, J. S. Lowe, S. E. O'Donnell, N. Aykin-Burns, M. C. Zimmerman, K. Zimmerman, A. J. Ham, R. M. Weiss, D. R. Spitz, M. A. Shea, R. J. Colbran, P. J. Mohler and M. E. Anderson, A dynamic pathway for calcium-independent activation of CaMKII by methionine oxidation, Cell, 2008, 133(3), 462–474 CrossRef CAS PubMed.
- J. C. Lim, G. Kim and R. L. Levine, Stereospecific oxidation of calmodulin by methionine sulfoxide reductase A, Free Radical Biol. Med., 2013, 61, 257–264 CrossRef CAS PubMed.
- M. Marimoutou, D. A. Springer, C. Liu, G. Kim and R. L. Levine, Oxidation of Methionine 77 in Calmodulin Alters Mouse Growth and Behavior, Antioxidants, 2018, 7(10), 140 CrossRef.
- C. Wilson, J. R. Terman, C. Gonzalez-Billault and G. Ahmed, Actin filaments-A target for redox regulation, Cytoskeleton, 2016, 73(10), 577–595 CrossRef CAS PubMed.
- J. R. Terman, T. Mao, R. J. Pasterkamp, H. H. Yu and A. L. Kolodkin, MICALs, a family of conserved flavoprotein oxidoreductases, function in plexin-mediated axonal repulsion, Cell, 2002, 109(7), 887–900 CrossRef CAS PubMed.
- R. J. Hung, U. Yazdani, J. Yoon, H. Wu, T. Yang, N. Gupta, Z. Huang, W. J. van Berkel and J. R. Terman, Mical links semaphorins to F-actin disassembly, Nature, 2010, 463(7282), 823–827 CrossRef CAS.
- R. J. Hung, C. W. Pak and J. R. Terman, Direct redox regulation of F-actin assembly and disassembly by Mical, Science, 2011, 334(6063), 1710–1713 CrossRef CAS PubMed.
- B. C. Lee, Z. Peterfi, F. W. Hoffmann, R. E. Moore, A. Kaya, A. Avanesov, L. Tarrago, Y. Zhou, E. Weerapana, D. E. Fomenko, P. R. Hoffmann and V. N. Gladyshev, MsrB1 and MICALs regulate actin assembly and macrophage function via reversible stereoselective methionine oxidation, Mol. Cell, 2013, 51(3), 397–404 CrossRef CAS PubMed.
- M. Vermeulen, H. C. Eberl, F. Matarese, H. Marks, S. Denissov, F. Butter, K. K. Lee, J. V. Olsen, A. A. Hyman, H. G. Stunnenberg and M. Mann, Quantitative Interaction Proteomics and Genome-wide Profiling of Epigenetic Histone Marks and Their Readers, Cell, 2010, 142(6), 967–980 CrossRef CAS PubMed.
- X. Li and T. M. Kapoor, Approach to profile proteins that recognize post-translationally modified histone “tails”, J. Am. Chem. Soc., 2010, 132(8), 2504–2505 CrossRef CAS PubMed.
- A. L. Garske, S. S. Oliver, E. K. Wagner, C. A. Musselman, G. LeRoy, B. A. Garcia, T. G. Kutateladze and J. M. Denu, Combinatorial profiling of chromatin binding modules reveals multisite discrimination, Nat. Chem. Biol., 2010, 6(4), 283–290 CrossRef CAS PubMed.
- H. Xiao, A. Chatterjee, S. H. Choi, K. M. Bajjuri, S. C. Sinha and P. G. Schultz, Genetic incorporation of multiple unnatural amino acids into proteins in mammalian cells, Angew. Chem., Int. Ed., 2013, 52(52), 14080–14083 CrossRef CAS PubMed.
- H. Chen, S. Venkat, P. McGuire, Q. Gan and C. Fan, Recent Development of Genetic Code Expansion for Posttranslational Modification Studies, Molecules, 2018, 23(7), 1662 CrossRef.
- D. T. Rogerson, A. Sachdeva, K. Wang, T. Haq, A. Kazlauskaite, S. M. Hancock, N. Huguenin-Dezot, M. M. Muqit, A. M. Fry, R. Bayliss and J. W. Chin, Efficient genetic encoding of phosphoserine and its nonhydrolyzable analog, Nat. Chem. Biol., 2015, 11(7), 496–503 CrossRef CAS PubMed.
- K. A. Kalesh and E. W. Tate, A succinyl lysine-based photo-cross-linking peptide probe for Sirtuin 5, Org. Biomol. Chem., 2014, 12(25), 4310–4313 RSC.
- E. E. Grintsevich, H. G. Yesilyurt, S. K. Rich, R. J. Hung, J. R. Terman and E. Reisler, F-actin dismantling through a redox-driven synergy between Mical and cofilin, Nat. Cell Biol., 2016, 18(8), 876–885 CrossRef CAS PubMed.
- H. Wu, H. G. Yesilyurt, J. Yoon and J. R. Terman, The MICALs are a Family of F-actin Dismantling Oxidoreductases Conserved from Drosophila to Humans, Sci. Rep., 2018, 8(1), 937 CrossRef PubMed.
- F. Stricher, C. Macri, M. Ruff and S. Muller, HSPA8/HSC70 chaperone protein: structure, function, and chemical targeting, Autophagy, 2013, 9(12), 1937–1954 CrossRef CAS.
- B. M. Doshi, L. E. Hightower and J. Lee, HSPB1, actin filament dynamics, and aging cells, Ann. N. Y. Acad. Sci., 2010, 1197, 76–84 CrossRef CAS PubMed.
- S. J. Roberts and G. N. Somero, Properties of the Interaction between Phosphofructokinase and Actin, Arch. Biochem. Biophys., 1989, 269(1), 284–294 CrossRef CAS.
- C. Mejean, F. Pons, Y. Benyamin and C. Roustan, Antigenic probes locate binding sites for the glycolytic enzymes glyceraldehyde-3-phosphate dehydrogenase, aldolase and phosphofructokinase on the actin monomer in microfilaments, Biochem. J., 1989, 264(3), 671–677 CrossRef CAS PubMed.
- B. A. Webb, A. M. Dosey, T. Wittmann, J. M. Kollman and D. L. Barber, The glycolytic enzyme phosphofructokinase-1 assembles into filaments, J. Cell Biol., 2017, 216(8), 2305–2313 CrossRef PubMed.
- B. A. Webb, F. Forouhar, F. E. Szu, J. Seetharaman, L. Tong and D. L. Barber, Structures of human phosphofructokinase-1 and atomic basis of cancer-associated mutations, Nature, 2015, 523(7558), 111 CrossRef CAS PubMed.
- J. P. Clarke and K. M. Mearow, Cell stress promotes the association of phosphorylated HspB1 with F-actin, PLoS One, 2013, 8(7), e68978 CrossRef CAS PubMed.
- T. El-Bacha, M. S. de Freitas and M. Sola-Penna, Cellular distribution of phosphofructokinase activity and implications to metabolic regulation in human breast cancer, Mol. Genet. Metab., 2003, 79(4), 294–299 CrossRef CAS.
- M. Sola-Penna, D. Da Silva, W. S. Coelho, M. M. Marinho-Carvalho and P. Zancan, Regulation of mammalian muscle type 6-phosphofructo-1-kinase and its implication for the control of the metabolism, IUBMB Life, 2010, 62(11), 791–796 CrossRef CAS PubMed.
- J. H. Lee, R. Liu, J. Li, C. Zhang, Y. Wang, Q. Cai, X. Qian, Y. Xia, Y. Zheng, Y. Piao, Q. Chen, J. de Groot, T. Jiang and Z. Lu, Stabilization of phosphofructokinase 1 platelet isoform by AKT promotes tumorigenesis, Nat. Commun., 2017, 8(1), 949 CrossRef PubMed.
- H. Hu, A. Juvekar, C. A. Lyssiotis, E. C. Lien, J. G. Albeck, D. Oh, G. Varma, Y. P. Hung, S. Ullas, J. Lauring, P. Seth, M. R. Lundquist, D. R. Tolan, A. K. Grant, D. J. Needleman, J. M. Asara, L. C. Cantley and G. M. Wulf, Phosphoinositide 3-Kinase Regulates Glycolysis through Mobilization of Aldolase from the Actin Cytoskeleton, Cell, 2016, 164(3), 433–446 CrossRef CAS PubMed.
- J. L. Bays, H. K. Campbell, C. Heidema, M. Sebbagh and K. A. DeMali, Linking E-cadherin mechanotransduction to cell metabolism through force-mediated activation of AMPK, Nat. Cell Biol., 2017, 19(6), 724 CrossRef CAS PubMed.
- W. J. Sullivan, P. J. Mullen, E. W. Schmid, A. Flores, M. Momcilovic, M. S. Sharpley, D. Jelinek, A. E. Whiteley, M. B. Maxwell, B. R. Wilde, U. Banerjee, H. A. Coller, D. B. Shackelford, D. Braas, D. E. Ayer, T. Q. D. Vallim, W. E. Lowry and H. R. Christofk, Extracellular Matrix Remodeling Regulates Glucose Metabolism through TXNIP Destabilization, Cell, 2018, 175(1), 117 CrossRef CAS PubMed.
- J. S. Park, C. J. Burckhardt, R. Lazcano, L. M. Solis, T. Isogai, L. Li, C. S. Chen, B. Gao, J. D. Minna, R. Bachoo, R. J. DeBerardinis and G. Danuser, Mechanical regulation of glycolysis via cytoskeleton architecture, Nature, 2020, 578(7796), 621–626 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Experimental procedures, supplemental figures, and supplemental tables. See DOI: https://doi.org/10.1039/d2cb00183g |
| This journal is © The Royal Society of Chemistry 2022 |
