Open Access Article
Open Access ArticleOne-step asparaginyl endopeptidase (OaAEP1)-based protein immobilization for single-molecule force spectroscopy
Xuan
Ding†
 ab,
Ziyi
Wang†
c,
Bin
Zheng†
c,
Shengchao
Shi
c,
Yibing
Deng
c,
Hanyang
Yu
ab,
Ziyi
Wang†
c,
Bin
Zheng†
c,
Shengchao
Shi
c,
Yibing
Deng
c,
Hanyang
Yu
 a and
Peng
Zheng
a and
Peng
Zheng
 *c
*c
aDepartment of Biomedical Engineering, College of Engineering and Applied Sciences, Nanjing University, 163 Xianlin Road, Nanjing, Jiangsu 210023, P. R. China
bState Key Laboratory of Analytical Chemistry for Life Science, Nanjing University, Nanjing, Jiangsu 210023, P. R. China
cState Key Laboratory of Coordination Chemistry, Chemistry and Biomedicine Innovation Center (ChemBIC), School of Chemistry and Chemical Engineering, Nanjing University, Nanjing, Jiangsu 210023, P. R. China. E-mail: pengz@nju.edu.cn
First published on 30th August 2022
Abstract
Enzymatic protein ligation has become the most powerful and widely used method for high-precision atomic force microscopy single-molecule force spectroscopy (AFM-SMFS) study of protein mechanics. However, this methodology typically requires the functionalization of the glass surface with a corresponding peptide sequence/tag for enzymatic recognition and multiple steps are needed. Thus, it is time-consuming and a high level of experience is needed for reliable results. To solve this problem, we simplified the procedure using two strategies both based on asparaginyl endopeptidase (AEP). First, we designed a heterobifunctional peptide-based crosslinker, GL-peptide-propargylglycine, which links to an N3-functionalized surface via the click reaction. Then, the target protein with a C-terminal NGL sequence can be immobilized via the AEP-mediated ligation. Furthermore, we took advantage of the direct ligation between primary amino in a small molecule and protein with C-terminal NGL by AEP. Thus, the target protein can be immobilized on an amino-functionalized surface via AEP in one step. Both approaches were successfully applied to the AFM-SMFS study of eGFP, showing consistent single-molecule results.
Introduction
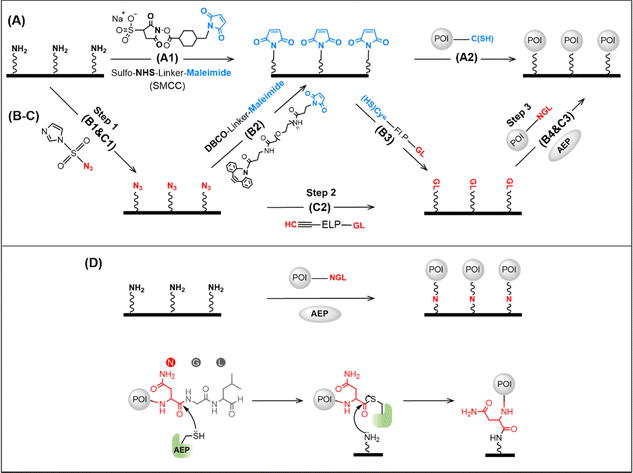
Protein immobilization can be critical for the application and fundamental research of proteins, such as protein detection, characterization, and single-molecule study.1–8 Among many methods, site-specific protein immobilization has become the most powerful method for high-precision atomic force microscopy single-molecule force spectroscopy (AFM-SMFS) study of protein mechanics. For example, the reaction between cysteine in the protein and the maleimide-coated surface is a widely used approach for protein immobilization (Fig. 1A).9,10 However, it suffers from an uncontrolled reaction if several cysteines are present, leading to different immobilization conditions.11–14 Recently, enzymatic ligation has been used, which recognizes and connects two specific peptide sequences/tags, and leads to efficient, site-specific protein immobilization. Nevertheless, it requires the two enzymatic recognition peptide sequences in the target protein and on the surface, respectively.15–17 Typically, the peptide-coated surface is achieved using two steps, starting from an amino-coated glass surface. First, a maleimide-coated surface is obtained using a chemical crosslinker containing an amino-reactive group N-hydroxysuccinimide (NHS) and a cysteine-reactive maleimide group (Fig. 1A, step 1). Then, a heterofunctional peptide linker with cysteine and the target peptide sequence is applied (Fig. 1B, step 3). Finally, the protein with the target peptide sequence can be immobilized (Fig. 1B, step 4).18 Here, the NHS is quite reactive, and multiple steps are needed. Thus, a high level of experience is needed for reliable results, and the whole procedure is time-consuming. Recently, we developed a click reaction-based method to avoid the use of NHS, and a much-improved success rate has been obtained (Fig. 1B, steps 1 and 2).19 However, more steps are introduced, and the maleimide group is always needed as the bridge.To solve this problem, we developed two strategies, both using oldenlandia affinis asparaginyl endopeptidases 1 with mutation C247A (abbreviated as OaAEP1, or AEP) for enzymatic ligation.20 First, a heterofunctional peptide-based crosslinker, glycine–leucine (GL)-peptide-propargylglycine (Pra) was designed. As a result, the target GL sequence can be directly added to the N3-functionalized surface without using the cysteine–maleimide coupling reaction (Fig. 1C, step 2). Furthermore, a more efficient enzymatic protein immobilization was achieved by taking advantage of the AEP-mediated direct ligation between the amino group and target protein with a C-terminal NGL sequence (Fig. 1D). Thus, the target protein can be immobilized on an amino-functionalized surface in one step.21
We tested and applied these methods to atomic force microscopy-based single-molecule force spectroscopy. Force spectroscopy is a powerful tool that can mechanically manipulate a single molecule and study its corresponding conformational changes or interactions.22–36 In the AFM-SMFS experiment of proteins, the protein of interest (POI) is immobilized and stretched from two points between the AFM tip and the protein-deposited surface, revealing a wealth of information of protein (un)folding, stability, and interactions.37–46 Thus, new methods generating site-specific and strong protein immobilization can be invaluable for obtaining reliable and efficient single-molecule results.47–52 We demonstrated these two methods by immobilizing eGFP and studied its unfolding process by AFM-SMFS, both showing ideal results.
Results and discussion
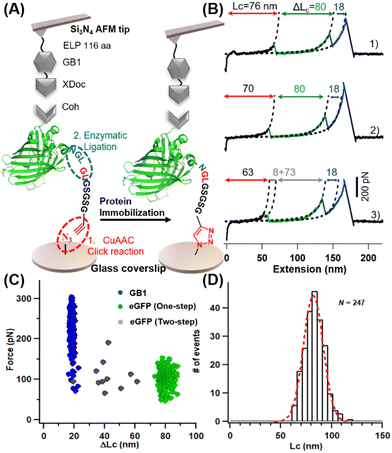
The principle and procedures of the two methods are as follows. For heterofunctional peptide method, an amino-functionalized glass surface was converted to azide first, which takes about one hour (Fig. 1C). Then, heterofunctional peptide-based crosslinker GL-peptide-Pra was added via the Copper-catalyzed azide-alkyne cycloaddition (CuAAC) click reaction, which takes ∼8 hours catalyzed by ascorbic acid. Finally, the protein with an NGL sequence can be immobilized to the GL-coated surface via the OaAEP1-mediate enzymatic ligation for half an hour.18,53,54To demonstrate the applicability of this method for the AFM-SMFS study, a characterized protein, enhanced Green Fluorescent Protein (eGFP), was chosen as a model protein.55 First, a seven-amino acid (aa) length peptide GL-GSGSG-Pra was designed as the heterofunctional crosslinker, which reacted with the N3-functionalized coverslip (Fig. 2A). Then, a fused protein Cohesion (Coh)-eGFP-NGL containing eGFP was designed for subsequent high-precision AFM measurement. It was ligated to the immobilized peptide. Consequently, it can be probed by an ELPn-GB1-Xmodule-dockerin (XDoc) functionalized AFM tip for the protein unfolding experiment (Fig. 2A). Here, protein domain GB1 with a known ΔLc of 18 nm was used as a single-molecule fingerprint.56–58 ELP is a flexible yet mechanically labile elastin-like polypeptides functioning as a spacer to eliminate the non-specific interaction.59 XDoc and Coh form a reversible [Coh:XDoc] protein–protein interaction with a high rupture force of ∼500 pN for efficient single-molecule pick-up.60
Upon stretching, the fused protein unfolded sequentially as observed from force-extension curves (Fig. 2B). Besides the 18 nm-length unfolding peak from GB1 (colored in blue), peak(s) from one-step (traces 1 and 2, in green) and two-step unfolding (trace 3, in gray) of eGFP with a (summed) ΔLc of ∼80 nm were observed. This value agrees well with the theoretical ΔLc of eGFP unfolding in which 227 aa were extended (227 aa × 0.36 nm aa−1 −2.4 nm = 79.3 nm). Moreover, the multiple unfolding pathways (Fig. 2C, n = 247) are similar to previous single-molecule results of GFP.61 A long featureless curve (81.9 ± 13.8 nm, in red) appeared before the first peak, which is from the initial extension of the mechanically labile peptide in the protein system (the seven aa-length peptide crosslinker plus the 145 aa-length ELPn and other 28 aa-length peptide in the fused protein).
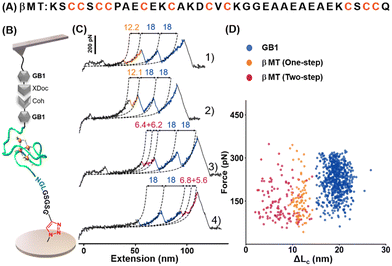
We chose the β domain of metallothionein (βMT), which binds to zinc, as another model protein. As a small size protein domain with only 37 amino acids, βMT contains 11 endogenous cysteines (Fig. 3A).62 It has been studied by AFM-SMFS using a non-specific immobilization method before.63,64 Thus, it serves as an excellent example of this cysteine/maleimide-free method for AFM measurement. A fused protein Coh-GB1-βMT-NGL was constructed and immobilized into the system using the seven aa-length peptide crosslinker accordingly. And the same ELPn-GB1-XDoc functionalized AFM tip was used for probing (Fig. 3B). Upon stretching, two GB1 unfolding peaks were observed as expected (Fig. 3C). Moreover, peak(s) from one-step (traces 1 and 2, in orange) and two-step unfolding (traces 3 and 4, in pink) of βMT with a (summed) ΔLc of ∼12 nm were observed (n = 205, Fig. 3D). This value agrees with the theoretical ΔLc of βMT unfolding in which 37 aa were extended. Moreover, this small protein domain's previously observed multiple unfolding pathways were detected and verified using our new method.63,64
Although the heterofunctional peptide method is one step less, the procedure is still tedious and time-consuming. A recent protein labeling work showed that OaAEP1(C247A) could directly catalyze the attachment of a small molecule with a primary amine to a protein via its C-terminal NGL sequence.21 Thus, we hypothesize that a surface covered with primary amino may also react with properly designed target protein catalyzed by AEP, and a much more convenient one-step protein immobilization may be achieved.
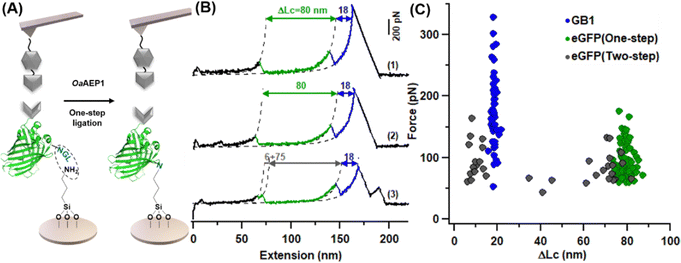
We then pipetted 50 μL of protein mixture containing 6 mg mL−1 Coh-eGFP-NGL and 0.2 mg mL−1 AEP on an amino-functionalized coverslip, which was incubated under 25 °C for half an hour. After washing with high-salt Tris buffer (1 M NaCl, 100 Tris, pH = 7.4) several times, the coverslip was used for AFM-SMFS measurement directly (Fig. 4A). As expected, eGFP was immobilized as the characteristic eGFP unfolding peak with ΔLc of ∼80 nm, including both one-step and two-step unfolding pathways were observed (Fig. 4B). Moreover, we analyzed the mechanical stability of immobilized eGFP in this new method. The unfolding force was 93.6 ± 27.1 pN (n = 78), consistent with previous experiments (Fig. 4C). Therefore, this one-step enzymatic protein immobilization is well-suited for AFM measurement. Moreover, with purified proteins and amino-functionalized coverslip, this one-step method only needs less than an hour for the whole protein immobilization procedure and saves time.
In this work, we demonstrated the application of AEP to efficient protein immobilization for high-precision AFM measurement of protein mechanics. Thanks to its efficient and site-specific ligation, several enzymes have been used for this purpose, including sortase, sfp, haloalkane dehydrogenase, formyl glycine generating enzyme, etc.17,48,59,65 However, all these enzymes, including AEP, require surface functionalization with the corresponding peptide/tag, which leads to multiple steps and lengthy procedures. Here, we optimized the AEP-based protein immobilization from two directions. The first strategy uses a heterofunctional peptide-based crosslinker, connecting the enzymatic recognition site and click reaction group in a single molecule.44,48,65 As a result, one step is removed.
Compared to cysteine-engineered protein coupling or immobilization, this method avoids disadvantages of the cysteine–maleimide or cysteine–cysteine reaction. For example, previous work using cysteine engineering/coupling to study GFP cannot stretch the protein from N and C terminus, possibly due to intramolecular disulfide bond formation. Our enzymatic method avoids it and makes this stretching direction possible. In addition, the previous cysteine coupling-based immobilization method cannot prepare a suitable metallothionein sample for single molecule studies, as there were so many endogenous cysteines and formation of intramolecular disulfide bond messes up the measurement.
Most importantly, we then demonstrated that AEP could immobilize protein with a C-terminal NGL sequence on an amino-functionalized surface in one step. This method only needs an incubation of half an hour for the ligation as the only step, which significantly saves time and simplifies the protein immobilization procedure. In addition, only a C–N covalent bond forms between the target protein and the surface as the linkage using this method. This bond is similar to the peptide backbone of proteins with a much higher mechanical stability compared with other linkages such maleimide–thiol adduct (∼1100 pN after hydrolysis) and 1,2,3-triazole (<1800 pN)19. Thus, we believe this new method not only simplifies the enzymatic protein immobilization but also broadens the systems that can be studied by AFM-SMFS.
Experimental material and procedure
Materials
All reagents were purchased from commercial suppliers and used accordingly. Peptide GL-ELPn-Propargylglycine and C-ELP20-NGL were purchased from GenScript. (3-Aminopropyl) tiethoxysilane (APTES, Sigma-Aldrich), Imidazole-1-sulfonyl azide hydrochloride (ImSO2N3·HCl; Abydos Scientific, China), and DBCO-(polyethylene glycol)4-Mal (DBCO-PEG4-Mal; Biocone, China) were stored and used in the dark. Vitamin C, K2CO3, and CuSO4 were from Aladdin. Solvents, including toluene, ethanol, and DMSO were purchased from Sinopharm Chemical Reagent Co. Ltd. Aqueous solutions were configured with Milli-Q water (18.2 MΩ cm−1, 0.22 μm filter). Microporous membrane filters (0.22 and 0.45 μm) were used for further purification (Jet Biofil, China). For the culture of E. coli, Luria–Bertani (LB) medium and agar plates (Sangon Biotech) were used. Plasma cleaner was used to activate the glass surface. Protein concentrations were routinely determined by Nanodrop 2000.Surface preparation
The glass coverslip (Sail Brand) and silicon nitride-based MLCT-BIO-DC AFM cantilever (Bruker Corp.) were first cleaned by plasma for 10 min. Then, they were amino-silanized and azide-functionalized according to published protocols.19 The peptide C-ELP20-NGL was immobilized on the AFM cantilever according to literature too.Different from the cantilever, coverslips were modified using the heterofunctional peptide-based crosslinker. A ∼30 μL solution of GL-ELPn-Pra (mixed aqueous solution of 2 mM GL-ELPn-Pra, 1 mM CuSO4, 100 mM Vitamin C, and 0.5 × PBS) was sandwiched between two azide-functionalized glass coverslip surfaces, and incubated for 8 hours at room temperature in the dark.
Finally, both cantilevers and coverslips were washed with 50 mL high-salt buffer (100 mM Tris, 1 M NaCl, pH 7.4), rinsed by H2O, and then dried by N2 stream or filter paper. The ELP-coated glass coverslips and cantilevers were immediately used or kept at −20 °C.
AFM-SMFS experiment
A 50 μL mixed solution of 30 μM fusion protein Coh-eGFP-NGL and 1 μM enzyme AEP was pipetted on the GL-functionalized coverslips, and the NGL-functionalized cantilevers with ELP20 as linker were incubated with 50 μL mixed solution of 60 μM GL-GB1-XDoc and 1 μM AEP in the measurement buffer (100 mM Tris, 100 mM NaCl, pH 7.4). The OaAEP1-mediate protein ligation was performed in the measurement buffer, forming a covalent NGL linkage between the C-terminal NGL and N-terminal GL or an amino-functionalized glass surface (Fig. 1).We used a Nanowizard 4 atomic force microscope to perform the AFM-SMFS experiment. After calibration, the AFM tip was pushed onto the surface, and the target protein was captured through the specific [Coh:XDoc] protein-protein interaction. Then, moving the tip up at a constant velocity (1000 nm s−1), the fused protein was stretched and unfolded. Finally, the data was filtered using data processing software (JPK) and analyzed by Igor Pro 6.12 (Wavemetrics).
Funding
This work was funded by Natural Science Foundation of Jiangsu Province (No. BK20200058), the National Natural Science Foundation of China (Grant no. 21771103 and 21977047) and Fundamental Research Funds for the Central Universities (Grant no. 14380259).Conflicts of interest
The authors declare no competing interests.References
- C. Garcia-Galan, Á. Berenguer-Murcia, R. Fernandez-Lafuente and R. C. Rodrigues, Adv. Synth. Catal., 2011, 353, 2885–2904 CrossRef CAS.
- T. D. Becke, S. Ness, S. Sudhop, H. E. Gaub, M. Hilleringmann, A. F. Schilling and H. Clausen-Schaumann, JoVE, 2018, e58167 Search PubMed.
- P. López-García, A. D. de Araujo, A. E. Bergues-Pupo, I. Tunn, D. P. Fairlie and K. G. Blank, Angew. Chem., Int. Ed., 2021, 60, 232–236 CrossRef PubMed.
- L. F. Milles, K. Schulten, H. E. Gaub and R. C. Bernardi, Science, 2018, 359, 1527–1533 CrossRef CAS PubMed.
- H. Fu, Y. Jiang, D. Yang, F. Scheiflinger, W. P. Wong and T. A. Springer, Nat. Commun., 2017, 8, 324 CrossRef PubMed.
- Y. Zhang, Z. Wang, T. B. Kouznetsova, Y. Sha, E. Xu, L. Shannahan, M. Fermen-Coker, Y. Lin, C. Tang and S. L. Craig, Nat. Chem., 2021, 13, 56–62 CrossRef CAS PubMed.
- M. Leitner, J. Brummeir, G. O. Plaimer, I. Kefer, A. Poturnayova, T. Hianik and A. Ebner, Methods, 2022, 197, 54–62 CrossRef CAS PubMed.
- H. E. Morgan, W. B. Turnbull and M. E. Webb, Chem. Soc. Rev., 2022, 51, 4121–4145 RSC.
- J. L. Zimmermann, T. Nicolaus, G. Neuert and K. Blank, Nat. Protoc., 2010, 5, 975–985 CrossRef CAS PubMed.
- D. Sluysmans, N. Willet, J. Thevenot, S. Lecommandoux and A.-S. Duwez, Nanoscale Horiz., 2020, 5, 671–678 RSC.
- M. Synakewicz, D. Bauer, M. Rief and L. S. Itzhaki, Sci. Rep., 2019, 9, 13820–13830 CrossRef PubMed.
- R. Barattin and N. Voyer, Chem. Commun., 2008, 1513–1532 RSC.
- B. P. Duckworth, J. Xu, T. A. Taton, A. Guo and M. D. Distefano, Bioconjugate Chem., 2006, 17, 967–974 CrossRef CAS PubMed.
- W. Zhang and X. Zhang, Prog. Polym. Sci., 2003, 28, 1271–1295 CrossRef CAS.
- W. Ott, E. Durner and H. E. Gaub, Angew. Chem., Int. Ed., 2018, 57, 12666–12669 CrossRef CAS PubMed.
- Y. Deng, B. Zheng, Y. Liu, S. Shi, J. Nie, T. Wu and P. Zheng, JoVE, 2020, e60774 CAS.
- S. Garg, G. S. Singaraju, S. Yengkhom and S. Rakshit, Bioconjugate Chem., 2018, 29, 1714–1719 CrossRef CAS PubMed.
- Y. Deng, T. Wu, M. Wang, S. Shi, G. Yuan, X. Li, H. Chong, B. Wu and P. Zheng, Nat. Commun., 2019, 10, 2775–2785 CrossRef PubMed.
- S. Shi, Z. Wang, Y. Deng, F. Tian, Q. Wu and P. Zheng, CCS Chem., 2022, 4, 598–604 CrossRef CAS.
- R. Yang, Y. H. Wong, G. K. T. Nguyen, J. P. Tam, J. Lescar and B. Wu, J. Am. Chem. Soc., 2017, 139, 5351–5358 CrossRef CAS PubMed.
- F. B. H. Rehm, T. J. Tyler, K. Yap, S. J. de Veer, D. J. Craik and T. Durek, J. Am. Chem. Soc., 2021, 143, 19498–19504 CrossRef CAS PubMed.
- J. Oroz, A. Galera-Prat, R. Hervas, A. Valbuena, D. Fernandez-Bravo and M. Carrion-Vazquez, Sci. Rep., 2019, 9, 13306 CrossRef PubMed.
- T. Hoffmann, K. M. Tych, M. L. Hughes, D. J. Brockwell and L. Dougan, Phys. Chem. Chem. Phys., 2013, 15, 15767–15780 RSC.
- M. Mora, A. Stannard and S. Garcia-Manyes, Chem. Soc. Rev., 2020, 49, 6816–6832 RSC.
- R. Petrosyan, S. Patra, N. Rezajooei, C. R. Garen and M. T. Woodside, Proc. Natl. Acad. Sci. U. S. A., 2021, 118, e2010213118 CrossRef CAS PubMed.
- M. Yu, Z. Zhao, Z. Chen, S. Le and J. Yan, Nat. Commun., 2020, 11, 4476 CrossRef CAS PubMed.
- X. Ma, M. Zhu, J. Liu, X. Li, L. Qu, L. Liang, W. Huang, J. Wang, N. Li and J.-H. Chen, et al. , Bioconjugate Chem., 2019, 30, 2998–3006 CrossRef CAS PubMed.
- M. Muddassir, B. Manna, P. Singh, S. Singh, R. Kumar, A. Ghosh and D. Sharma, Chem. Commun., 2018, 54, 9635–9638 RSC.
- R. X. Yao, X. Li, N. Xiao, W. G. Weng and W. K. Zhang, Nano Res., 2021, 14, 2654–2658 CrossRef CAS.
- P. Sonar, L. Bellucci, A. Mossa, P. O. Heidarsson, B. B. Kragelund and C. Cecconi, Biophys. J., 2020, 119, 1821–1832 CrossRef CAS PubMed.
- P. Zhao, C.-Q. Xu, C. Sun, J. Xia, L. Sun, J. Li and H. Xu, Polym. Chem., 2020, 11, 7087–7093 RSC.
- M. Zhao and M. T. Woodside, Nat. Chem. Biol., 2021, 17, 975–981 CrossRef CAS PubMed.
- S. Zhang, H.-j Qian, Z. Liu, H. Ju, Z.-y Lu, H. Zhang, L. Chi and S. Cui, Angew. Chem., Int. Ed., 2019, 58, 1659–1663 CrossRef CAS PubMed.
- H. Xing, Z. Li, W. Wang, P. Liu, J. Liu, Y. Song, Z. L. Wu, W. Zhang and F. Huang, CCS Chem., 2020, 2, 513–523 CrossRef CAS.
- Z. L. Guo, H. Y. Hong, G. Yuan, H. Qian, B. Li, Y. Cao, W. Wang, C. X. Wu and H. Chen, Phys. Rev. Lett., 2020, 125, 198101 CrossRef CAS PubMed.
- E. M. Mulhall, A. Ward, D. Yang, M. A. Koussa, D. P. Corey and W. P. Wong, Nat. Commun., 2021, 12, 849 CrossRef CAS PubMed.
- M.-A. LeBlanc, M. R. Fink, T. T. Perkins and M. C. Sousa, Proc. Natl. Acad. Sci. U. S. A., 2021, 118, e2019566118 CrossRef CAS PubMed.
- J. Perales-Calvo, D. Giganti, G. Stirnemann and S. Garcia-Manyes, Sci. Adv., 2018, 4, eaaq0243 CrossRef PubMed.
- A. Alonso-Caballero, J. Schonfelder, S. Poly, F. Corsetti, D. De Sancho, E. Artacho and R. Perez-Jimenez, Nat. Commun., 2018, 9, 2758 CrossRef PubMed.
- C. Suay-Corredera, M. R. Pricolo, D. Velazquez-Carreras, D. Pathak, N. Nandwani, C. Pimenta-Lopes, D. Sanchez-Ortiz, I. Urrutia-Irazabal, S. Vilches and F. Dominguez, et al. , ACS Nano, 2021, 15, 10203–10216 CrossRef PubMed.
- Z. Wang, J. Nie, S. Shi, G. Li and P. Zheng, Chem. Commun., 2021, 57, 11489–11492 RSC.
- F. Tian, G. Li, B. Zheng, Y. Liu, S. Shi, Y. Deng and P. Zheng, Chem. Commun., 2020, 56, 3943–3946 RSC.
- W. Cao, C. Dong, S. Kim, D. Hou, W. Tai, L. Du, W. Im and X. F. Zhang, Biophys. J., 2021, 120, 1011–1019 CrossRef CAS PubMed.
- J. Y. Nie, F. Tian, B. Zheng, Z. Y. Wang and P. Zheng, Chem. Lett., 2021, 50, 1667–1675 CrossRef CAS.
- F. Rico, A. Russek, L. Gonzalez, H. Grubmuller and S. Scheuring, Proc. Natl. Acad. Sci. U. S. A., 2019, 116, 6594–6601 CrossRef CAS PubMed.
- D. J. Müller, A. C. Dumitru, C. Lo Giudice, H. E. Gaub, P. Hinterdorfer, G. Hummer, J. J. De Yoreo, Y. F. Dufrêne and D. Alsteens, Chem. Rev., 2021, 121, 11701–11725 CrossRef PubMed.
- G. Song, F. Tian, H. Liu, G. Li and P. Zheng, J. Phys. Chem. Lett., 2021, 12, 3860–3867 CrossRef CAS PubMed.
- B. Yang, Z. Liu, H. Liu and M. A. Nash, Front. Mol. Biosci., 2020, 7, 85 CrossRef CAS PubMed.
- Y. Liu, F. Tian, S. Shi, Y. Deng and P. Zheng, J. Phys. Chem. Lett., 2021, 12, 10914–10919 CrossRef CAS PubMed.
- Q. Li, D. Apostolidou and P. E. Marszalek, Methods, 2022, 197, 39–53 CrossRef CAS PubMed.
- D. R. Jacobson and T. T. Perkins, Proc. Natl. Acad. Sci. U. S. A., 2021, 118, e2020083118 CrossRef CAS PubMed.
- J. Nie, Y. Deng, F. Tian, S. Shi and P. Zheng, Nano Res., 2022, 15, 4251–4257 CrossRef CAS PubMed.
- F. B. H. Rehm, T. J. Tyler, J. Xie, K. Yap, T. Durek and D. J. Craik, ChemBioChem, 2021, 22, 2079–2086 CrossRef CAS PubMed.
- T. J. Harmand, N. Pishesha, F. B. H. Rehm, W. Ma, W. B. Pinney, Y. J. Xie and H. L. Ploegh, ACS Chem. Bio., 2021, 16, 1201–1207 CrossRef CAS PubMed.
- Z. Ganim and M. Rief, Proc. Natl. Acad. Sci. U. S. A., 2017, 114, 11052–11056 CrossRef CAS PubMed.
- Y. Cao, C. Lam, M. Wang and H. Li, Angew. Chem., Int. Ed., 2006, 45, 642–645 CrossRef CAS PubMed.
- P. Zheng, S. J. Takayama, A. G. Mauk and H. Li, J. Am. Chem. Soc., 2012, 134, 4124–4131 CrossRef CAS PubMed.
- P. Zheng and H. Li, J. Am. Chem. Soc., 2011, 133, 6791–6798 CrossRef CAS PubMed.
- W. Ott, M. A. Jobst, M. S. Bauer, E. Durner, L. F. Milles, M. A. Nash and H. E. Gaub, ACS Nano, 2017, 11, 6346–6354 CrossRef CAS PubMed.
- S. W. Stahl, M. A. Nash, D. B. Fried, M. Slutzki, Y. Barak, E. A. Bayer and H. E. Gaub, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 20431–20436 CrossRef CAS PubMed.
- H. Dietz, M. Bertz, M. Schlierf, F. Berkemeier, T. Bornschlogl, J. P. Junker and M. Rief, Nat. Protoc., 2006, 1, 80–84 CrossRef CAS PubMed.
- M. D. Peris-Diaz, R. Guran, C. Domene, V. de los Rios, O. Zitka, V. Adam and A. Krezel, J. Am. Chem. Soc., 2021, 143, 16486–16501 CrossRef CAS PubMed.
- G. Yuan, Q. Ma, T. Wu, M. Wang, X. Li, J. Zuo and P. Zheng, Sci. Rep., 2019, 9, 10518 CrossRef PubMed.
- G. Yuan, F. Curtolo, Y. Deng, T. Wu, F. Tian, Q. Ma, Y. Liu, J. Zuo, G. M. Arantes and P. Zheng, Research, 2021, 2021, 9756945 CrossRef CAS PubMed.
- I. Popa, R. Berkovich, J. Alegre-Cebollada, C. L. Badilla, J. A. Rivas-Pardo, Y. Taniguchi, M. Kawakami and J. M. Fernandez, J. Am. Chem. Soc., 2013, 135, 12762–12771 CrossRef CAS PubMed.
Footnote |
| † These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2022 |