Open Access Article
Open Access ArticleChemical proteomic analysis of bile acid-protein targets in Enterococcus faecium†
Xinglin
Yang‡
 a,
Xiaohui
Zhao‡
a,
Xiaohui
Zhao‡
 a,
Victor
Chen‡
a,
Victor
Chen‡
 b and
Howard C.
Hang
b and
Howard C.
Hang
 *ac
*ac
aDepartment of Immunology and Microbiology, Scripps Research, La Jolla, California 92037, USA. E-mail: hhang@scripps.edu
bLaboratory of Chemical Biology and Microbial Pathogenesis, The Rockefeller University, New York, New York 10065, USA
cDepartment of Chemistry, Scripps Research, La Jolla, California 92037, USA
First published on 20th September 2022
Abstract
Bile acids are important gut microbiota metabolites that regulate both host and microbial functions. To identify the direct protein targets of bile acids in Enterococcus, we synthesized and validated the activity of a lithocholic acid (LCA) photoaffinity reporter, x-alk-LCA-3. Chemical proteomics of x-alk-LCA-3 in E. faecium Com15 reveals many candidate LCA-interacting proteins, which are involved in cell well synthesis, transcriptional regulation and metabolism. To validate the utility of bile acid photoaffinity labeling, we characterized a putative bile salt hydrolase (BSH) crosslinked by x-alk-LCA-3, and demonstrated that this BSH was effective in converting taurolithocholic acid (TLCA) to LCA in E. faecium and in vitro. Chemical proteomics should afford new opportunities to characterize bile acid-protein targets and mechanisms of action in the future.
Introduction
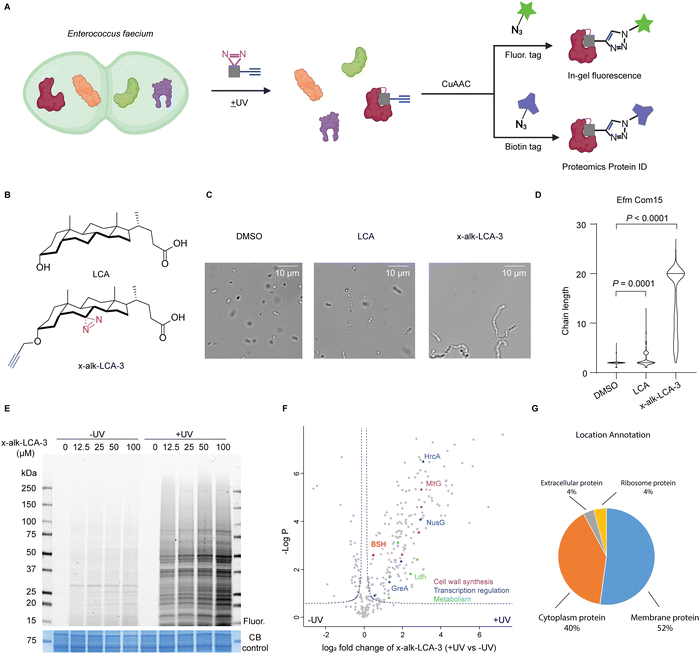
Human microbiota generate structurally diverse small molecules that play important roles in the regulation of host physiology,1,2 as well as serve as signal molecules to gut commensals and pathogens.3 Bile acids are one class of important gut microbiota metabolites. Primary bile acids are synthesized in the liver from cholesterol, and modified with glycine or taurine to form conjugated bile acids that are stored in the gall bladder. These conjugated bile acids can then be secreted into intestine and metabolized (e.g., deconjugation, oxidation and reduction) by gut microbiota to form structurally diverse secondary bile acids.4 These bile acids can directly regulate host physiology through engaging host proteins, such as nuclear receptors5,6 and GPCRs.7,8 In addition, bile acids are important signaling molecules to regulate gut microbiota.9,10 As biological detergents, bile salts may induce membrane damage and affect cellular homeostasis of intestinal bacteria.9 Moreover, bile acids can directly regulate pathogen virulence.11–14 However, the direct identification of bile acid-protein targets and mechanisms of action in bacteria has been challenging.The development of bile acid-based chemical probes has provided important new tools to characterize novel interacting proteins and enzymes. For example, activity-based probes for bile acid hydrolases (BSH) with different reactive warheads such as acyloxymethylketone,15 α-fluoromethyl ketone16 and β-lactam or acrylamide17 have been developed to profile the gut microbiota-associated bile salt hydrolase activity. Alternatively, photoaffinity reporters for bile acids and other microbiota metabolites can be employed to characterize other protein targets in mammalian cells and microbes.18 Of note, photoaffinity reporters were developed for primary bile acid cholic acid and enabled the identification of cholic acid-binding proteins in mammalian cell lines19 and E. coli.20 In addition, our chemoproteomic analysis in Salmonella revealed anti-infective bile acids such as chenodeoxycholic acid can bind and inactivate a transcriptional regulator of Salmonella virulence HilD.21 Furthermore, chemical proteomic analysis of bile acid targets in Clostridium difficile showed that lithocholic acid (LCA) can bind and modulate the function of a stress response transcriptional regulator BapR.22 In this study, we employ bile acid photoaffinity chemical proteomics to investigate their protein targets in Enterococcus faecium (Fig. 1A), a prominent commensal and drug-resistant bacterial species in humans.
E. faecium and E. faecalis are common commensals in humans, but are also prevalent causes of hospital-acquired infections.23,24 The primary location of enterococci in humans is small and large intestine, where the bacteria encounter high concentration of bile acids ranging from low micromolar to low millimolar. Thus, the ability of these intestinal bacteria to adapt to bile is important for their survival and colonization in intestine. A previous study demonstrated that sublethal concentration of bile exposure increases the survival of E. faecalis to the bile challenge.25 To understand the molecular mechanisms responsible for bile acid responses, transcriptome26–28 and proteome29–31 studies of bile-treated E. faecalis were carried out, which revealed the up-regulation of fatty acid- and phospholipid pathways. In addition, screening of transposon mutagenesis of E. faecium and E. faecalis identified key genes required for bile acid resistance,32,33 such as gltK8, a glutamate/aspartate permease and sagA, which encodes a secreted peptidoglycan hydrolase.34 In addition, LCA has been reported to induce diplococci-chaining and increased biofilm formation in vancomycin-resistant strains of E. faecium.35 These studies revealed key pathways involved in Enterococcus responses to bile acids, but did not identify specific bile acid-binding protein targets.
To identify the bile acid-interacting proteins in E. faecium, we employed a LCA photoaffinity reporter for chemoproteomic studies. Our proteomic analysis in E. faecium Com15 revealed many candidate bile acid-interacting proteins, including key metabolic enzymes, transcriptional regulators and cell well synthesis factors. We validated these LCA reporter proteomic studies by biochemical characterization of a E. faecium Com15 bile salt hydrolase (BSH), demonstrating this approach can directly reveal bile acid-interacting proteins and enzymes.
Results and discussion
Chemical proteomics of LCA protein targets in E. faecium Com15
We first evaluated the activity of five free bile acids on E. faecium Com15, a well-characterized commensal strain. Bacteria were grown overnight with DMSO or bile acids and then harvested for microscopy analysis. Consistent with previous studies,35 LCA most effectively induce chaining of E. faecium (Fig. S1, ESI†). Importantly, our LCA photoaffinty reporter (x-alk-LCA-3) (Fig. 1B) showed similar and even slightly higher activity than LCA in E. faecium diplococci chaining (Fig. 1C and D). We therefore used x-alk-LCA-3 to perform chemical proteomics in E. faecium.To identify LCA-interacting proteins in E. faecium, bacteria were treated with different concentrations of x-alk-LCA-3 for 1 hour, followed by UV irradiation. As shown in Fig. 1E, x-alk-LCA-3 photo-crosslinks proteins in dose- and UV-dependent manners. Since 25 μM of x-alk-LCA-3 significantly induces chaining of E. faecium Com15, we used this concentration for chemical proteomics. Without UV irradiation, x-alk-LCA-3 enriched 17 protein hits compared to DMSO control, which suggests x-alk-LCA-3 may covalently modify these proteins. These potential covalent modified proteins include enzymes, such as cytochrome P450, phosphoglucomutase, ATP-dependent Clp protease, and transporter and carrier, such as major intrinsic protein and C4-dicarboxylate anaerobic carrier (Fig. S2, ESI†). To identify proteins which non-covalently interact with x-alk-LCA-3, we compare protein hits under UV irradiation to those without UV irradiation. As shown in the volcano plot (Fig. 1F), 182 protein hits are enriched with UV-dependent manner. These proteins are located in the cell membrane (52%), cytoplasm (40%), ribosome (4%) and extracellular region (4%) (Fig. 1G). Further analysis reveals different types of proteins are enriched, including: (i) metabolic enzymes, such as Ldh which is L-lactate dehydrogenase, (ii) transcription regulation related proteins, such as transcription termination protein NusG, (iii) cell wall synthesis related protein, such as endolytic murein transglycosylase MltG. Since LCA induces chaining of E. faecium, it was proposed that LCA may interact and interfere with the function of cell wall synthesis-related proteins, which can be further validated in the future. Meanwhile, a putative bile salt hydrolase (BSH, EFWG_00531) was also enriched in our proteomics dataset. BSH in E. faecium shares 82.35% identity with a reported highly active BSH from Enterococcus faecalis.36,37 BSHs hydrolyse taurine- or glycine-conjugated bile acids to free bile acids. The deconjugation step is the gateway reaction for following transformations of primary bile acids to divergent secondary bile acids. There is no reported BSHs from E. faecium Com15, therefore, we decided to validate the interaction of this potential BSH with LCA.
Validation of E. faecium bile salt hydrolase LCA target
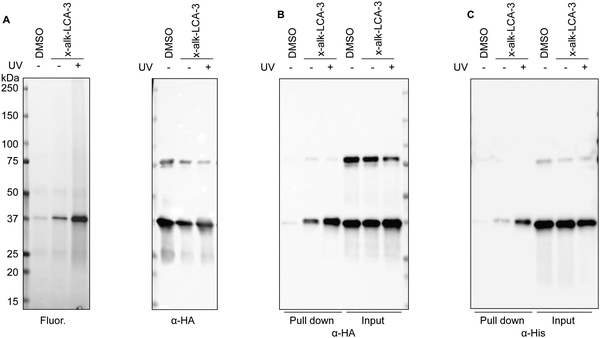
To validate BSH's interaction with LCA, we generated a C-terminally HA-tagged bsh construct in the E. faecium genome using dsDNA recombineering methods we previously reported38 (Fig. S3A, ESI†). Transformants were selected on chloramphenicol plates and assayed for bsh-HA and cat insertion via PCR 1 kb upstream and downstream of the bsh gene. Gel shifts were observed corresponding to the insertion size of about 800 bp when compared to the wild-type, indicating correct insertion of HA at the C-terminus of BSH (Fig. S3B, ESI†). Nanopore sequencing of the PCR products further validated the correct insertion into the genome. To validate if BSH-HA is properly being expressed, western blot analysis was performed on wild-type and BSH-HA E. faecium. A single band appears at the predicted size of BSH-HA in E. faecium Com15 BSH-HA but not in wild-type bacteria, indicating BSH-HA is being expressed in these cells (Fig. S3C, ESI†). E. faecium BSH-HA was treated with x-alk-LCA-3. After UV irradiation, bacteria were lysed and reacted with fluorescence or biotin tags through Click Chemistry (Fig. 1A). We found that x-alk-LCA-3 photo-crosslinks BSH-HA in a UV-dependent manner (Fig. 2A). BSHs usually form homo-tetramer assembly in solution39 which is also confirmed by the crystal structure.16,37 Interestingly, we found potential dimer in the anti-HA blot for BSH-HA strain (Fig. 2A–C), but the formation of potential dimer depends on Cu-catalyzed click reaction (Fig. S4, ESI†). Importantly, we found the chromosomally expressed BSH-HA is pulled down by x-alk-LCA-3 in a UV-dependent manner (Fig. 2B). Pull-down of the overexpressed BSH-His6 by x-alk-LCA-3 further confirms the interaction between BSH and x-alk-LCA-3 (Fig. 2C).Activity of bile salt hydrolase in E. faecium
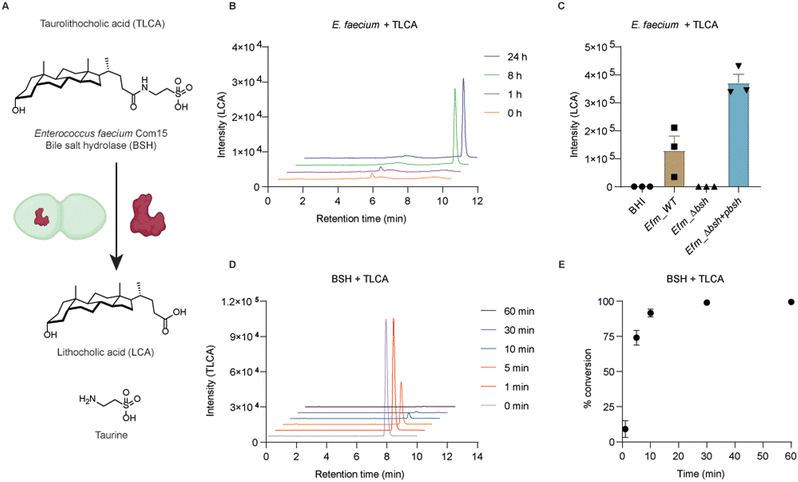
To examine if conjugated secondary bile acids are able to be transformed to free bile acids by this putative BSH (Fig. 3A), we cultured E. faecium with taurolithocholic acid (TLCA). First, wild-type bacteria were grown in BHI media supplemented with TLCA up to 24 h. At different time points, bacterial cultures were collected for extracting bile acids from the supernatants. Liquid chromatography-mass spectrometry (LC-MS) was used to determine the formation of LCA. We could detect LCA after 8 hours of culturing E. faecium with TLCA, which indicates the transformation of TLCA in E. faecium (Fig. 3B and Fig. S7A, ESI†). To evaluate the specific activity of BSH in E. faecium, we performed knockout of bsh using dsDNA recombineering. E. faecium bsh was replaced with cat flanked by loxP sites to generate E. faecium Com15 Δbsh::cat (Efm_Δbsh) (Fig. S5A, ESI†). Gene replacement of bsh was confirmed via sanger sequencing of the PCR product. We cultured wild-type (Efm_WT) and BSH knock-out (Efm_Δbsh) strains in TLCA supplemented BHI media, separately. We observed the loss of capability in converting TLCA to LCA by Efm_Δbsh strain (Fig. 3C). In addition, we transformed a plasmid in Efm_Δbsh that is able to express a C-terminus His6-tagged BSH (Efm_Δbsh + pbsh, Fig. S6, ESI†). Importantly, complementation of BSH enzyme in Efm_Δbsh recovered the capability and produced more LCA than the wild-type E. faecium (Fig. 3C). The incapability of producing LCA by Efm_Δbsh and recovery of LCA production in Efm_Δbsh + pbsh are not caused by defective growth of respective strains (Fig. S7B and C, ESI†).Biochemical activity of E. faecium bile salt hydrolase in vitro
To evaluate the enzymatic activity of E. faecium BSH, we expressed and purified recombinant BSH for in vitro studies (Fig. 3A). The C-terminal His6-tagged BSH was purified as a 37 kDa protein (Fig. S7D, ESI†). Incubation of E. faecium BSH with TLCA in PBS buffer at pH 7.5 and 37 °C resulted in formation of LCA after 1 minute (Fig. S7E, ESI†) and total conversion of TLCA in 30 minutes (Fig. 3D and E). Next, we performed enzyme kinetics analysis of the deconjugation reaction. TLCA of increasing concentrations (1, 5, 10, 20, 50, 100, 200, 500 μM) were incubated with 100 nM BSH for 1 min. The input of TLCA and production of LCA were examined by LC-MS (Fig. S7F and G, ESI†). The substrate saturation curve (Fig. S7H, ESI†) revealed an approximate maximum enzyme velocity (Vmax) value of 13.2 μM min−1 and Km value of around 15 μM. It is of interest to note that high concentrations of TLCA decrease the enzymatic activity of E. faecium BSH. This effect was also reported in a previous characterized BSH from Bacteroides fragilis.40 These experiments demonstrate x-alk-LCA-3 crosslinked E. faecium Com15 (EFWG_00531) bile salt hydrolase that can covert TLCA to free secondary bile acid LCA.Conclusions
Bile acids are a group of important metabolites that modulate human physiology and activity of other microbes. Identification of bile acid-interacting proteins in host cells and microbes is important for understanding the mechanisms of host–microbiota interactions. In this study, we employed an active bile acid photoaffinity reporter (x-alk-LCA-3) for chemical proteomic analysis of LCA-interacting proteins in Enterococcus. These studies identify many candidate LCA-interacting proteins, including a putative bile salt hydrolase that was validated in E. faecium by gene deletion, overexpression and in vitro with purified recombinant enzyme using activity assays. Our study highlights the utility of chemical proteomics for investigating the protein targets of microbiota metabolites.Author contributions
H. C. H conceived the project. X. Y. designed and synthesized the bile acid reporter, performed proteomics and validated the BSH-bile acid interaction. V. C. tested the activity of bile acids and bile acid reporter and generated mutated strains of E. faecium. X. Z. evaluated the activity of E. faecium BSH. H. C. H, X. Y., X. Z. and V. C. wrote the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We thank The Rockefeller Proteomics Resource Center for analysis. V. C. acknowledges support from The Rockefeller University Graduate Program and the National Institutes of Health (T32 A1070084). H. C. H. acknowledges support from the National Institutes of Health (R01AI172915). Some figures were created with http://BioRender.com.Notes and references
- M. S. Donia and M. A. Fischbach, Science, 2015, 349, 1254766 CrossRef.
- S. V. Lynch, E. G. Phimister and O. Pedersen, N. Engl. J. Med., 2016, 375, 2369–2379 CrossRef CAS.
- A. J. Baumler and V. Sperandio, Nature, 2016, 535, 85–93 CrossRef CAS PubMed.
- A. F. Hofmann and L. R. Hagey, J. Lipid Res., 2014, 55, 1553–1595 CrossRef CAS PubMed.
- M. Makishima, A. Y. Okamoto, J. J. Repa, H. Tu, R. M. Learned, A. Luk, M. V. Hull, K. D. Lustig, D. J. Mangelsdorf and B. Shan, Science, 1999, 284, 1362–1365 CrossRef CAS.
- D.-J. Shin and L. Wang, in Bile Acids and Their Receptors, ed. S. Fiorucci and E. Distrutti, Springer International Publishing, Cham, 2019, pp. 51–72 DOI:10.1007/164_2019_236.
- Y. Kawamata, R. Fujii, M. Hosoya, M. Harada, H. Yoshida, M. Miwa, S. Fukusumi, Y. Habata, T. Itoh, Y. Shintani, S. Hinuma, Y. Fujisawa and M. Fujino, J. Biol. Chem., 2003, 278, 9435–9440 CrossRef CAS PubMed.
- H. Yu, T. Zhao, S. Liu, Q. Wu, O. Johnson, Z. Wu, Z. Zhuang, Y. Shi, L. Peng, R. He, Y. Yang, J. Sun, X. Wang, H. Xu, Z. Zeng, P. Zou, X. Lei, W. Luo and Y. Li, eLife, 2019, 8, e48431 CrossRef CAS PubMed.
- M. Begley, C. G. Gahan and C. Hill, FEMS Microbiol. Rev., 2005, 29, 625–651 CrossRef CAS PubMed.
- K. S. Gipson, K. P. Nickerson, E. Drenkard, A. Llanos-Chea, S. K. Dogiparthi, B. B. Lanter, R. M. Hibbler, L. M. Yonker, B. P. Hurley and C. S. Faherty, Infect. Immun., 2020, 88, e00865-19 CrossRef PubMed.
- D. T. Hung and J. J. Mekalanos, Proc. Natl. Acad. Sci. U. S. A., 2005, 102, 3028–3033 CrossRef CAS.
- M. Yang, Z. Liu, C. Hughes, A. M. Stern, H. Wang, Z. Zhong, B. Kan, W. Fenical and J. Zhu, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 2348–2353 CrossRef CAS.
- S. Alavi, J. D. Mitchell, J. Y. Cho, R. Liu, J. C. Macbeth and A. Hsiao, Cell, 2020, 181, 1533–1546 CrossRef CAS e1513.
- J. Tam, S. Icho, E. Utama, K. E. Orrell, R. F. Gomez-Biagi, C. M. Theriot, H. K. Kroh, S. A. Rutherford, D. B. Lacy and R. A. Melnyk, Proc. Natl. Acad. Sci. U. S. A., 2020, 117, 6792–6800 CrossRef CAS.
- B. Parasar, H. Zhou, X. Xiao, Q. Shi, I. L. Brito and P. V. Chang, ACS Cent. Sci., 2019, 5, 867–873 CrossRef CAS PubMed.
- A. A. Adhikari, T. C. M. Seegar, S. B. Ficarro, M. D. McCurry, D. Ramachandran, L. Yao, S. N. Chaudhari, S. Ndousse-Fetter, A. S. Banks, J. A. Marto, S. C. Blacklow and A. S. Devlin, Nat. Chem. Biol., 2020, 16, 318–326 CrossRef CAS PubMed.
- K. R. Brandvold, C. J. Miller, R. F. Volk, B. J. Killinger, C. Whidbey and A. T. Wright, ChemBioChem, 2021, 22, 1448–1455 CrossRef CAS PubMed.
- X. Zhao, X. Yang and H. C. Hang, Biochemistry, 2022 DOI:10.1021/acs.biochem.1c00758.
- S. Zhuang, Q. Li, L. Cai, C. Wang and X. Lei, ACS Cent. Sci., 2017, 3, 501–509 CrossRef CAS.
- B. Liu, S. Zhuang, R. Tian, Y. Liu, Y. Wang, X. Lei and C. Wang, ACS Chem. Biol., 2022, 17, 2461–2470 CrossRef CAS PubMed.
- X. Yang, K. R. Stein and H. C. Hang, Nat. Chem. Biol., 2022 DOI:10.1038/s41589-022-01122-3.
- E. Forster, X. Yang, H. C. Hang and A. Shen, 2022, bioRxiv DOI:10.1101/2022.05.26.493666.
- F. Lebreton, W. van Schaik, A. M. McGuire, P. Godfrey, A. Griggs, V. Mazumdar, J. Corander, L. Cheng, S. Saif, S. Young, Q. Zeng, J. Wortman, B. Birren, R. J. Willems, A. M. Earl and M. S. Gilmore, mBio, 2013, 4, e00534-13 CrossRef PubMed.
- K. Dubin, G. Pamer Eric, A. Britton Robert and D. Cani Patrice, Microbiol. Spectrum, 2017, 5, 5.6.01 CrossRef.
- A. Rincé, Y. Le Breton, N. Verneuil, J.-C. Giard, A. Hartke and Y. Auffray, Int. J. Food Microbiol., 2003, 88, 207–213 CrossRef.
- M. Solheim, A. Aakra, H. Vebo, L. Snipen and I. F. Nes, Appl. Environ. Microbiol., 2007, 73, 5767–5774 CrossRef CAS PubMed.
- L. Zhou, L. Wang, P. Tian, T. Bao, L. Li and X. Zhao, Front. Microbiol., 2019, 10, 1048 CrossRef PubMed.
- M. Salze, J.-C. Giard, E. Riboulet-Bisson, T. Hain, A. Rincé and C. Muller, Arch. Microbiol., 2020, 202, 233–246 CrossRef CAS PubMed.
- S. Flahaut, J. Frere, P. Boutibonnes and Y. Auffray, Appl. Environ. Microbiol., 1996, 62, 2416–2420 CrossRef CAS.
- S. Flahaut, A. Hartke, J.-c Giard, A. Benachour, P. Boutibonnes and Y. Auffray, FEMS Microbiol. Lett., 1996, 138, 49–54 CrossRef CAS PubMed.
- L. A. Bøhle, E. M. Færgestad, E. Veiseth-Kent, H. Steinmoen, I. F. Nes, V. G. H. Eijsink and G. Mathiesen, Proteome Sci., 2010, 8, 37 CrossRef PubMed.
- Y. L. Breton, A. Mazé, A. Hartke, S. Lemarinier, Y. Auffray and A. Rincé, Curr. Microbiol., 2002, 45, 0434–0439 CrossRef PubMed.
- X. Zhang, D. Bierschenk, J. Top, I. Anastasiou, M. J. M. Bonten, R. J. L. Willems and W. van Schaik, BMC Genomics, 2013, 14, 299 CrossRef CAS.
- F. Teng, M. Kawalec, G. M. Weinstock, W. Hryniewicz and B. E. Murray, Infect. Immun., 2003, 71, 5033–5041 CrossRef CAS PubMed.
- P. T. McKenney, J. Yan, J. Vaubourgeix, S. Becattini, N. Lampen, A. Motzer, P. J. Larson, D. Dannaoui, S. Fujisawa, J. B. Xavier and E. G. Pamer, Cell Host Microbe, 2019, 25, 695–705 CrossRef CAS PubMed e695.
- D. Chand, S. Ramasamy and C. G. Suresh, Process Biochem., 2016, 51, 263–269 CrossRef CAS.
- D. Chand, P. Panigrahi, N. Varshney, S. Ramasamy and C. G. Suresh, Biochim. Biophys. Acta, Proteins Proteomics, 2018, 1866, 507–518 CrossRef CAS PubMed.
- V. Chen, M. E. Griffin, P. Maguin, A. Varble and H. C. Hang, Appl. Environ. Microbiol., 2021, 87, e00844-21 CrossRef PubMed.
- M. Rossocha, R. Schultz-Heienbrok, H. von Moeller, J. P. Coleman and W. Saenger, Biochemistry, 2005, 44, 5739–5748 CrossRef CAS PubMed.
- E. J. Stellwag and P. B. Hylemon, Biochim. Biophys. Acta, Enzymol., 1976, 452, 165–176 CrossRef CAS.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2cb00178k |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2022 |