Open Access Article
Open Access ArticleA light-initiated chemical reporter strategy for spatiotemporal labeling of biomolecules†
Feifei
Wang
a,
Hao
Kong
a,
Xiangfeng
Meng
a,
Xiao
Tian
a,
Changjiang
Wang
a,
Lei
Xu
bc,
Xiang
Zhang
bc,
Lei
Wang
*bc and
Ran
Xie
 *ad
*ad
aState Key Laboratory of Coordination Chemistry, School of Chemistry and Chemical Engineering, Nanjing University, Nanjing 210023, China. E-mail: ranxie@nju.edu.cn
bDepartment of Gastroenterology, Nanjing Drum Tower Hospital, The Affiliated, Hospital of Nanjing University Medical School, Nanjing 210008, China. E-mail: leiwang9631@nju.edu.cn
cDepartment of Gastroenterology, Nanjing Drum Tower Hospital, Drum Tower, Clinical Medical College of Nanjing Medical University, Nanjing 210008, China
dChemistry and Biomedicine Innovation Center (ChemBIC), Nanjing University, Nanjing 210023, China
First published on 30th March 2022
Abstract
The emergence of optochemical approaches has had a diverse impact over a broad range of biological research due to spatiotemporal regulation. Herein, we integrate this feature into the bioorthogonal chemical reporter strategy, which enables visible light-controlled spatiotemporal labeling of cell-surface glycans, lipids, and proteins. The metabolic precursors were first incorporated into live cells, and next the bioorthogonal reaction converted the azide/alkyne into a photo-active functional group, which allowed for subsequent photo-click reaction. We demonstrated this strategy by specifically labeling sialome, mucin-type O-glycome, phospholipids and newly-synthesized membrane proteins, respectively. The sequential photoirradiation-orthogonal reporter tagging (SPORT) should facilitate the probing of biomolecules in complex biological systems with high spatiotemporal precision.
The past 25 years have witnessed the emergence, development and utilization of novel chemical biology methods to probe and manipulate biomolecules. Of note is the bioorthogonal chemical reporter strategy.1,2 By introducing non-native functionality (i.e., chemical reporter), through metabolic incorporation or protein engineering, followed by a selective covalent chemical reaction (i.e., bioorthogonal reaction), this two-step approach greatly expanded the scope of biomolecules analyzed within their native environs. Proteins,3 glycans,4 lipids,5 DNA/RNA6 and small molecule metabolites have all been fashioned with chemical handles (e.g., azide, alkyne) in cells and animals, and subsequently ligated with a reactive labeling probe, making it an essential tool to perturb, perceive and profile their biological functions.7 One challenge that has arisen for this method is the lack of conditional control over biological functions, which naturally occur in a strict spatially and temporally regulated fashion on the cellular and organism level. To this end, optochemical approaches using photoirradiation as a conditional stimulus for spatiotemporal regulation of biomolecules have found a myriad of applications for photocatalytic-activation,8,9 photo-caging,10 photo-crosslinking11–13 and photo-switching14 of small molecules, peptides and oligonucleotides.15,16 In addition, the emergence of light-induced click reactions (i.e., photo-click reaction) endowed bioorthogonal chemistry with exogenous spatiotemporal control and the absence of an activating reagent/catalyst.17–20 By introducing photo-controllable functional groups into metabolic precursors through chemical synthesis, several labs were able to exploit the metabolic synthetic pathway for non-proteinaceous biomolecules, to metabolically incorporate photo-responsive biomolecules within cells.21–24 For instance, monosaccharide analogs containing photo-crosslinkable diazirine moieties have been recently developed and successfully incorporated into cell-surface glycans for the capture and identification of glycan-binding counterparts.25,26 Other labs have developed strategies for light-controlled metabolic labeling of sialoglycans based on photo-caged azidosugars.27,28 However, to implement these approaches, caution is needed in the design of photo-responsive precursors because bulky substituents on the backbone of metabolic precursors will greatly reduce the incorporation and expression level, or even result in the failure of metabolic incorporation.29
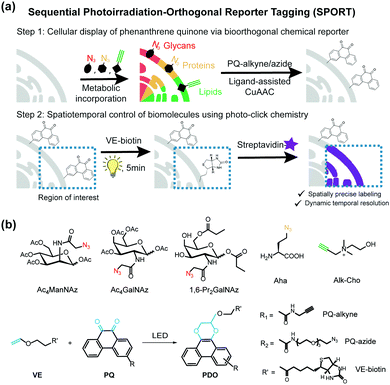
Herein we present a general and versatile methodology, namely sequential photoirradiation-orthogonal reporter tagging (SPORT), to clearly differentiate it from the bioorthogonal chemical reporter strategy (Fig. 1). In this strategy, metabolic precursors were first incorporated into the newly-synthesized glycans, proteins and lipids, and next a thermally activated bioorthogonal reaction converted the azide/alkyne to a photo-active functionality, which allowed for a subsequent visible light-induced click reaction (Fig. 1a). Of several light induced photo-click chemistry reactions,30–32 we chose a recently reported visible light-triggered reaction, 9,10-phenanthrenequinone (PQ) and electron-rich vinyl ether (VE) fluorogenic [4+2] cycloaddition, as the second step visible light-induced click reaction (Fig. 1b).33 This reaction is fast, biocompatible, inert to alkyne/azide, and can be initiated using an LED lamp.
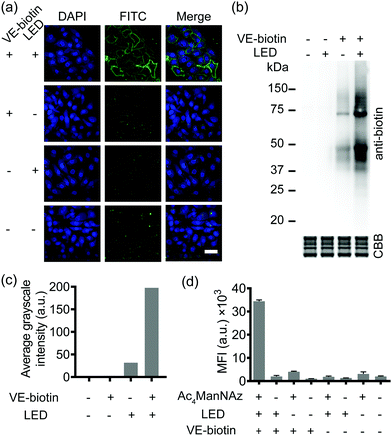
SPORT was first demonstrated for imaging of glycans. By hijacking the glycan biosynthesis pathway, peracetylated N-azidoacetylmannosamine (Ac4ManNAz), a classical chemical reporter for sialoglycans, has been used to metabolically introduce azide into cell-surface sialoglycoconjugates.34 We hypothesized that the display level of cell-surface PQ is the crucial determinant for the PQ–VE photo-click labeling, given that (1) bioorthogonal reaction would yield stoichiometric attachment of one PQ per cell-surface azide,35 and (2) different cell lines vary in the efficiency of metabolic incorporation of unnatural sugars.36 Because HCCC-9810 (human cholangiocarcinoma cell line) overexpressed sialylation, whereas A549 (human lung adenocarcinoma cell line) displayed medium sialoglycan density on the cell membrane, we chose these two cell types to demonstrate the feasibility of SPORT. Conventionally, micromolar concentrations of protected sialic acid analogues are needed in order to achieve a significant level of metabolic labeling. We treated HCCC-9810 cells with 200 μM Ac4ManNAz for 48 h at 37 °C. The cells were then washed, and labeled with PQ-alkyne (Fig. S1 and S2, ESI†) using BTTAA-assisted copper(I) azide–alkyne cycloaddition (CuAAC) to conjugate the PQ moiety onto the cell-surface.37 The cells were then washed, labeled with VE-biotin (Fig. S3 and S4, ESI†), irradiated with an LED lamp, stained with Alexa Flour 488-streptavidin, and analyzed using confocal microscopy. Robust fluorescence was observed in the presence of metabolic precursor, VE-biotin and light (Fig. 2a, first row), whereas weak fluorescence was observed when illumination is absent (Fig. 2a, second row). The background signal could presumably be due to the increased hydrophobic interaction between the cell-surface and Alexa Flour 488-streptavidin when PQ was coated. Though we also observed a photo-triggered side reaction between the biotin moiety and PQ by HPLC, only minimal fluorescence labeling was observed if the unnatural sugar, and/or illumination were omitted from the reaction conditions (Fig. 2a, third and fourth rows and Fig. S5, S6, ESI†). We also confirmed that similar labeling results can be achieved in A549 cells (Fig. S7, ESI†). Thus, all reagents and visible light are necessary and sufficient for SPORT to occur in sialoglycan-containing live cells.
Subsequently, in order to investigate whether the PQ–VE reaction in SPORT indeed occurred at the glycoprotein level, we incubated the Ac4ManNAz-treated cell lysates with PQ-alkyne for a CuAAC ligation, conjugated VE-biotin under illumination, and characterized the SDS-PAGE resolved samples. As expected, more significant labeling of sialylated proteins was observed in the SPORT group than in the control group, albeit with a visible background signal in the LED (−) group (Fig. 2b). Faint bands were observed on the Western blot at ∼55 kDa and ∼65 kDa for the VE-biotin (−) group, presumably due to innate biotinylation on certain proteins. Densitometric analysis indicated about 50% signal intensity on the blot for the LED (−) group compared to the LED (+) group when VE-biotin was added (Fig. 2c), implying the existence of a light-independent side reaction (Fig. S5, ESI†), yet with proper controls the photo-click signal is distinguishable. Similar results were obtained with A549 cells (Fig. S8, ESI†).
We then quantified the total labeling efficiency of SPORT using flow cytometry (FACS). We first optimized the exposure time of SPORT using Ac4ManNAz-treated HCCC-9810 cells as the study case. Ac4ManNAz-treated HCCC-9810 cells were ligated with PQ-alkyne, reacted with VE-biotin upon photo-irradiation for various reaction times, and analyzed using flow cytometry (FACS). Previous study indicated a time-dependent manner for the PQ–VE photocycloaddition spanning from 0 to 30 min in protein modification.33 Surprisingly, we observed a linear increase to saturation (0–5 min), and a drastic decrease afterwards (7–9 min) when the reaction occurred on the cellular surface (Fig. S9, ESI†). Therefore, we chose 5 min as the optimized irradiation time and therefore used this time throughout the experiment in vitro. HCCC-9810 cells displayed about 8- to 36-fold higher labeling than control groups (Fig. 2d). Similar fluorescence intensity contrast was observed in A549 cells (Fig. S10, ESI†). We further confirmed that the SPORT labeling in the LED(−) group resulted in minimal or low levels of fluorescent labeling, presumably through the light-independent side reaction, in a cell-type dependent manner. The SPORT labeling efficiency was calculated by comparing the mean fluorescence intensity between cells treated with and without LED light for photoillumination (Fig. S11, ESI†). More importantly, they all showed adequate contrast, and the effect from cell-surface sialic acid expression, photoirradiation, photo-click efficiency and its mechanism-of-action may be an integrated result of SPORT efficiency. Together, these results prove the success of SPORT for installing photo-reactive probes, implying the possibility of visualizing sialoglycans with light-triggered spatiotemporal control.
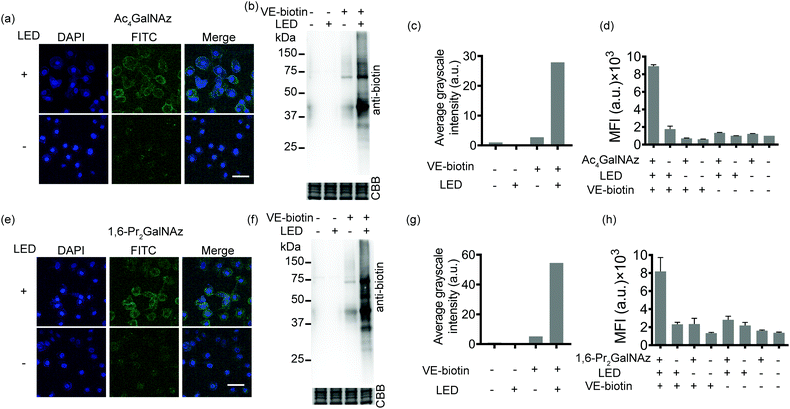
Given the availability of a variety of azide-containing monosaccharide analogues,38 the SPORT methodology can be readily applied to visualize other types of glycosylation processes. Identification of mucin-type O-linked glycosylation and O-GlcNAcylation has been realized using azido-containing N-acetylgalactosamine (GalNAc) and N-acetylglucosamine (GlcNAc) derivatives.39,40 The metabolic crosstalk between O-GlcNAcylation and the GalNAc salvage pathway enables the labeling of O-GlcNAc substrates with azidosugars when N-azidoacetylgalactosamine (GalNAz) is introduced.41 Recent reports also discovered and deciphered an artificial protein S-glyco-modification side reaction when peracetylated monosaccharide precursors were used.42–44 As additional showcases of SPORT imaging of glycans using azidosugars, we selected peracetylated GalNAz (Ac4GalNAz)39 and 1,6-di-O-propioylated GalNAz (1,6-Pr2GalNAz)42 as azidosugar reporters. Confocal fluorescence microscopy, flow cytometric analysis, and an anti-biotin western blot were conducted according to the above-mentioned SPORT procedures on both HCCC-9810 and A549 cells, respectively (Fig. 3 and Fig. S12–S14, ESI†). Notably, the confocal imaging and FACS results may only reflect the mucin-type O-linked glycosylation level, while Western blot analysis should additionally include the expression level on O-GlcNAcylation because labeling of cytosolic and nucleic O-GlcNAcylated proteins requires fixation and permeabilization for the SPORT reagents to pass through the cell membrane.45 In agreement with our hypothesis, both Ac4GalNAz and 1,6-Pr2GalNAz are compatible with SPORT, as quantified via flow cytometric analysis (Fig. S15 and S16, ESI†). Interestingly, the fluorescent signal of Ac4GalNAz appears to be 30% higher than that of 1,6-Pr2GalNAz under the same flow cytometric conditions, probably reflecting the differences in azide expression level generated from additional S-glyco-modification (Fig. S17, ESI†).
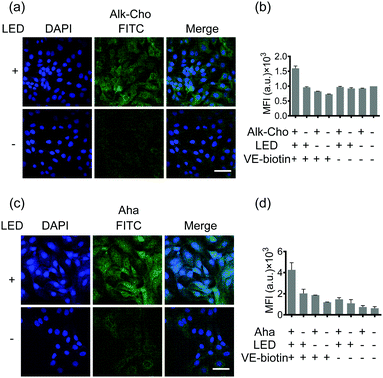
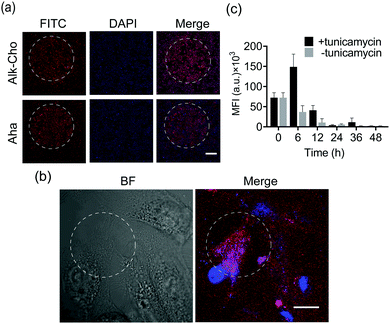
Aiming to demonstrate the broad applicability, we next sought to extend SPORT to other classes of biomolecules. To visualize lipids, we used alkynylated propargylcholine (Alk-Cho), which is metabolically incorporated into the cell membrane in lieu of phosphatidylcholine.46 Cells were treated with 500 μM propargylcholine for 12 h, washed, and labeled with PQ-azide (Fig. S18 and S19, ESI†) and VE-biotin to form phenanthrodioxine (PDO) upon illumination (Fig. 4a and Fig. S20, S21, ESI†). Confocal imaging and FACS analysis on both A549 and HCCC-9810 cells qualitatively and quantitatively exhibited that cell-surface alkynyl handles can be detected and visualized via the click converter and subsequent photo-click reaction. We also treated A549 cells and HCCC-9810 cells with azidohomoalanine (Aha), an azide-containing non-canonical amino acid serving as a surrogate for methionine.47 Newly-synthesized proteins can also be monitored and analyzed in a light-dependent manner (Fig. 4c and d). We also quantifiably compared the SPORT labeling efficiency among various metabolite precursors in the same cell based on their flow cytometry data (Fig. S17 and S22, ESI†). On these grounds, we concluded that the versatile SPORT platform is compatible with a myriad of metabolic precursors.
Finally, we explored the feasibility of spatiotemporal labeling over biomolecules using SPORT. To exemplify, HCCC-9810 cells were cultured on a glass slide and fed with either Ac4ManNAz, Alk-Cho, or Aha. A pinhole (2 mm in diameter) was set for a selected circular region prior to LED irradiation for SPORT reaction. We found that 5 min of illumination bestowed optimized signal contrast upon SPORT labeling, in accordance with the flow cytometry results (Fig. 5a and Fig. S9, S23, ESI†), further proving that light irradiation is indeed needed for SPORT to occur. We then pursued the possibility of lipid labeling with single-cell spatial resolution. Obviously, SPORT can discriminate the illumination region; only the cellular region within the photo-illumination spotlight was robustly labeled, although background fluorescence can be also observed for cells outside the irradiation zone (Fig. 5b and Fig. S24, S25, Video S1, ESI†). Next, we performed pulse-chase experiments using SPORT to probe the turn-over dynamics of newly-synthesized cell-surface sialome under different conditions. The antibiotic tunicamycin blocks the transfer of GlcNAc-1-P from UDP-GlcNAc to dolichol phosphate, thereby blocking the synthesis of N-linked oligosaccharide chains on glycoproteins.48 Sialic acid (Neu5Ac) is biosynthesized from UDP-GlcNAc, converted to precursor N-acetylmannosamine (ManNAc) via UDP-GlcNAc 2-epimerase, and then converted to cytidine-5′-monophosphate Neu5Ac (CMP-Neu5Ac), the nucleotide sugar donor used by sialyltransferases for transferring Neu5Ac onto sialoglycans. HCCC-9810 cells were treated with Ac4ManNAz for 48 h, and then the culture medium was replaced with fresh medium, or medium containing tunicamycin. The cells were chased for 0–48 h, followed by flow cytometric analysis (Fig. 5c). Remarkably, addition of tunicamycin exhibited a significant increase at 6 h, indicating an accelerated sialylation from cytidine-5′-monophosphate Neu5Az (CMP-Neu5Az, the unnatural counterpart for CMP-Neu5Ac) to cell-surface sialoglycans (Fig. 5c). These results suggest that the dynamics of sialoglycan biosynthesis may be regulated in a spatiotemporally controlled manner, and that the turn-over rate of sialoglycans is regulated by tunicamycin. These results, collectively, proved that SPORT can label cellular components with temporal and spatial resolution in physiological environments.
Conclusions
In summary, we developed a general methodology for spatiotemporal labeling of biomolecules in live cells. Light-activated functionalities can be facilely introduced into metabolic precursors via a classical bioorthogonal chemical reporter strategy, allowing for the fast, specific, and catalyst-free photocycloaddition reaction under LED light. Our method employs transcendent small molecule precursors, which can minimize the perturbation while maximizing the display density on the cell surface. This proof-of-concept work revealed undesired background noise when SPORT was executed, somewhat hampering the application of SPORT. However, the availability of various bioorthogonal chemistry and visible-light photo-click chemistry reactions with better kinetics (e.g., TCO-tetrazine reaction)49 should address this hiccup, thus optimizing the overall labeling efficiency.Ideally, appending either VE or PQ functionalities onto the metabolic precursor could directly incorporate photo-active metabolic precursors into cellular components. So far, only limited photo-clickable monosaccharide chemical reporters with intricate design on the N-acetyl position have been proven to tolerate the sialic acid biosynthetic pathway, and ultimately be incorporated into sialoglycans with high efficiency.50,51 Indeed, our trial experiment using peracetylated N-vinyl ether mannosamine (Ac4ManNVE) and 9-phenanthrenequinone-sialic acid (9PQ-Neu5Ac) as the sialic acid derivative were unsuccessful (Fig. S26–S28, ESI†). We attributed the failure of incorporation to the acid-sensitivity of the VE moiety during the glycan biosynthetic processes,52 and the limited expression level of 9PQ-Neu5Ac by steric hinderance.29 SPORT can circumvent the “one functionality, one design” dilemma in the design of metabolic precursors, and greatly expand the chemical toolbox to perform, compare, and apply versatile choices of photo-induced reactions onto cellular components at different cells for temporal and spatial control of specific biomolecules, and thus to regulate their corresponding function.
On the other hand, versatile bioorthogonal functional groups (e.g., azide, alkyne, tetrazine) allow for simultaneous, orthogonal introduction of various photo-active moieties onto multiple biomolecules. A simultaneous, spatiotemporal visualization of multiple biomolecules via different photocycloaddition reactions that are regulated with different wavelengths for conditional photo-click reaction is currently being pursued in our laboratory.
Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
We thank Prof. Yan Zhang (Nanjing University) for guidance with the PQ–VE photo-cycloaddition experiment. We gratefully acknowledge support from the National Natural Science Foundation of China (No. 2207070006), the Natural Science Foundation of Jiangsu Province (No. BK20202299), and the Programs for High-level Entrepreneurial and Innovative Talents Introduction of Jiangsu Province (Individual and Group Program).Notes and references
- J. A. Prescher and C. R. Bertozzi, Nat. Chem. Biol., 2005, 1, 13–21 CrossRef CAS PubMed.
- M. Grammel and H. C. Hang, Nat. Chem. Biol., 2013, 9, 475–484 CrossRef CAS PubMed.
- K. Lang and J. W. Chin, Chem. Rev., 2014, 114, 4764–4806 CrossRef CAS PubMed.
- N. J. Pedowitz and M. R. Pratt, RSC Chem. Biol., 2021, 2, 306–321 RSC.
- E. Thinon, A. Percher and H. C. Hang, ChemBioChem, 2016, 17, 1800–1803 CrossRef CAS PubMed.
- D. Ganz, D. Harijan and H.-A. Wagenknecht, RSC Chem. Biol., 2020, 1, 86–97 RSC.
- J. M. Baskin and C. R. Bertozzi, QSAR Comb. Sci., 2007, 26, 1211–1219 CrossRef CAS.
- Z. Huang, Z. Liu, X. Xie, R. Zeng, Z. Chen, L. Kong, X. Fan and P. R. Chen, J. Am. Chem. Soc., 2021, 143, 18714–18720 CrossRef CAS PubMed.
- A. Jemas, Y. Xie, J. E. Pigga, J. L. Caplan, C. W. Am Ende and J. M. Fox, J. Am. Chem. Soc., 2022, 144, 1647–1662 CrossRef CAS PubMed.
- Y. Liu, R. Zeng, R. Wang, Y. Weng, R. Wang, P. Zou and P. R. Chen, Proc. Natl. Acad. Sci. U. S. A., 2021, 118, e2025299118 CrossRef CAS PubMed.
- Y. Tanaka, M. R. Bond and J. J. Kohler, Mol. BioSyst., 2008, 4, 473–480 RSC.
- M. Suchanek, A. Radzikowska and C. Thiele, Nat. Methods, 2005, 2, 261–268 CrossRef CAS PubMed.
- J. B. Geri, J. V. Oakley, T. Reyes-Robles, T. Wang, S. J. McCarver, C. H. White, F. P. Rodriguez-Rivera, D. L. Parker Jr, E. C. Hett and O. O. Fadeyi, Science, 2020, 367, 1091–1097 CrossRef CAS PubMed.
- M. Zhu and H. Zhou, Org. Biomol. Chem., 2018, 16, 8434–8445 RSC.
- Y. Tian and Q. Lin, CHIMIA, 2018, 72, 758–763 CrossRef CAS PubMed.
- J. Li, H. Kong, C. Zhu and Y. Zhang, Chem. Sci., 2020, 11, 3390–3396 RSC.
- M. A. Tasdelen and Y. Yagci, Angew. Chem., Int. Ed., 2013, 52, 5930–5938 CrossRef CAS PubMed.
- G. S. Kumar and Q. Lin, Chem. Rev., 2020, 121, 6991–7031 CrossRef PubMed.
- B. D. Fairbanks, L. J. Macdougall, S. Mavila, J. Sinha, B. E. Kirkpatrick, K. S. Anseth and C. N. Bowman, Chem. Rev., 2021, 121, 6915–6990 CrossRef CAS PubMed.
- S. Arumugam, S. V. Orski, N. E. Mbua, C. McNitt, G.-J. Boons, J. Locklin and V. V. Popik, Pure Appl. Chem., 2013, 85, 1499–1513 CrossRef CAS.
- J.-i. Mihara and K. Fujimoto, Org. Biomol. Chem., 2021, 19, 9860–9866 RSC.
- J. Flores, B. M. White, R. J. Brea, J. M. Baskin and N. K. Devaraj, Chem. Soc. Rev., 2020, 49, 4602–4614 RSC.
- J. A. Frank, H. G. Franquelim, P. Schwille and D. Trauner, J. Am. Chem. Soc., 2016, 138, 12981–12986 CrossRef CAS PubMed.
- R. K. Lim and Q. Lin, Acc. Chem. Res., 2011, 44, 828–839 CrossRef CAS PubMed.
- H. Wu and J. Kohler, Curr. Opin. Chem. Biol., 2019, 53, 173–182 CrossRef CAS PubMed.
- L. Feng, S. Hong, J. Rong, Q. You, P. Dai, R. Huang, Y. Tan, W. Hong, C. Xie and J. Zhao, J. Am. Chem. Soc., 2013, 135, 9244–9247 CrossRef CAS PubMed.
- B. Cheng, Y. Wan, Q. Tang, Y. Du, F. Xu, Z. Huang, W. Qin and X. Chen, Chin. J. Chem., 2022, 40, 806–812 CrossRef CAS.
- H. Wang, R. Wang, K. Cai, H. He, Y. Liu, J. Yen, Z. Wang, M. Xu, Y. Sun and X. Zhou, Nat. Chem. Biol., 2017, 13, 415–424 CrossRef CAS PubMed.
- S. J. Luchansky, S. Goon and C. R. Bertozzi, ChemBioChem, 2004, 5, 371–374 CrossRef CAS PubMed.
- G. S. Kumar, S. Racioppi, E. Zurek and Q. Lin, J. Am. Chem. Soc., 2022, 144, 57–62 CrossRef CAS PubMed.
- S. Jiang, X. Wu, H. Liu, J. Deng, X. Zhang, Z. Yao, Y. Zheng, B. Li and Z. Yu, ChemPhotoChem, 2020, 4, 327–331 CrossRef CAS.
- X. Zhang, X. Wu, S. Jiang, J. Gao, Z. Yao, J. Deng, L. Zhang and Z. Yu, Chem. Commun., 2019, 55, 7187–7190 RSC.
- J. Li, H. Kong, L. Huang, B. Cheng, K. Qin, M. Zheng, Z. Yan and Y. Zhang, J. Am. Chem. Soc., 2018, 140, 14542–14546 CrossRef CAS PubMed.
- E. Saxon and C. R. Bertozzi, Science, 2000, 287, 2007–2010 CrossRef CAS PubMed.
- V. O. Rodionov, S. I. Presolski, D. Díaz Díaz, V. V. Fokin and M. Finn, J. Am. Chem. Soc., 2007, 129, 12705–12712 CrossRef CAS PubMed.
- S. J. Luchansky, H. C. Hang, E. Saxon, J. R. Grunwell, C. Yu, D. H. Dube and C. R. Bertozzi, Methods in enzymology, Elsevier, 2003, vol. 362, pp. 249–272 Search PubMed.
- C. Besanceney-Webler, H. Jiang, T. Zheng, L. Feng, D. Soriano del Amo, W. Wang, L. M. Klivansky, F. L. Marlow, Y. Liu and P. Wu, Angew. Chem., Int. Ed., 2011, 50, 8051–8056 CrossRef CAS PubMed.
- B. Cheng, Q. Tang, C. Zhang and X. Chen, Annu. Rev. Anal. Chem., 2021, 14, 363–387 CrossRef CAS PubMed.
- H. C. Hang, C. Yu, D. L. Kato and C. R. Bertozzi, Proc. Natl. Acad. Sci. U. S. A., 2003, 100, 14846–14851 CrossRef CAS PubMed.
- D. J. Vocadlo, H. C. Hang, E.-J. Kim, J. A. Hanover and C. R. Bertozzi, Proc. Natl. Acad. Sci. U. S. A., 2003, 100, 9116–9121 CrossRef CAS PubMed.
- M. Boyce, I. S. Carrico, A. S. Ganguli, S.-H. Yu, M. J. Hangauer, S. C. Hubbard, J. J. Kohler and C. R. Bertozzi, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 3141–3146 CrossRef CAS PubMed.
- K. Qin, H. Zhang, Z. Zhao and X. Chen, J. Am. Chem. Soc., 2020, 142, 9382–9388 CrossRef CAS PubMed.
- Y. Hao, X. Fan, Y. Shi, C. Zhang, D.-e. Sun, K. Qin, W. Qin, W. Zhou and X. Chen, Nat. Commun., 2019, 10, 1–13 CrossRef PubMed.
- W. Qin, K. Qin, X. Fan, L. Peng, W. Hong, Y. Zhu, P. Lv, Y. Du, R. Huang, M. Han, B. Cheng, Y. Liu, W. Zhou, C. Wang and X. Chen, Angew. Chem., Int. Ed., 2018, 57, 1817–1820 CrossRef CAS PubMed.
- W. Lin, L. Gao and X. Chen, ChemBioChem, 2015, 16, 2571–2575 CrossRef CAS PubMed.
- C. Y. Jao, M. Roth, R. Welti and A. Salic, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 15332–15337 CrossRef CAS PubMed.
- D. C. Dieterich, A. J. Link, J. Graumann, D. A. Tirrell and E. M. Schuman, Proc. Natl. Acad. Sci. U. S. A., 2006, 103, 9482–9487 CrossRef CAS PubMed.
- S. P. Guarnaccia, J. H. Shaper and R. L. Schnaar, Proc. Natl. Acad. Sci. U. S. A., 1983, 80, 1551–1555 CrossRef CAS PubMed.
- J. C. Carlson, L. G. Meimetis, S. A. Hilderbrand and R. Weissleder, Angew. Chem., Int. Ed., 2013, 52, 6917–6920 CrossRef CAS PubMed.
- V. F. Schart, J. Hassenrück, A. K. Späte, J. E. Dold, R. Fahrner and V. Wittmann, ChemBioChem, 2019, 20, 166–171 CrossRef CAS PubMed.
- Y. Wu, J. Zheng, D. Xing and T. Zhang, Nanoscale, 2020, 12, 10361–10368 RSC.
- M. Finnveden, S. Brännström, M. Johansson, E. Malmström and M. Martinelle, RSC Adv., 2018, 8, 24716–24723 RSC.
Footnote |
| † Electronic supplementary information (ESI) available. See https://doi.org/10.1039/d2cb00072e |
| This journal is © The Royal Society of Chemistry 2022 |