Open Access Article
Open Access ArticleRecent advances in DNA-encoded dynamic libraries
Bingbing
Shi
a,
Yu
Zhou
*b and
Xiaoyu
Li
 *bc
*bc
aDepartment of Biochemistry and Molecular Biology, College of Basic Medicine, Jining Medical University, Jining, Shandong 272067, P. R. China
bDepartment of Chemistry and State Key Laboratory of Synthetic Chemistry, The University of Hong Kong, Pokfulam Road, Hong Kong SAR, China. E-mail: yuchow@hku.hk; xiaoyuli@hku.hk
cLaboratory for Synthetic Chemistry and Chemical Biology Limited, Health@InnoHK, Innovation and Technology Commission, Units 1503-1511, 15/F., Building 17W, Hong Kong SAR, China
First published on 17th February 2022
Abstract
The DNA-encoded chemical library (DEL) has emerged as a powerful technology platform in drug discovery and is also gaining momentum in academic research. The rapid development of DNA-/DEL-compatible chemistries has greatly expanded the chemical space accessible to DELs. DEL technology has been widely adopted in the pharmaceutical industry and a number of clinical drug candidates have been identified from DEL selections. Recent innovations have combined DELs with other legacy and emerging techniques. Among them, the DNA-encoded dynamic library (DEDL) introduces DNA encoding into the classic dynamic combinatorial libraries (DCLs) and also integrates the principle of fragment-based drug discovery (FBDD), making DEDL a novel approach with distinct features from static DELs. In this Review, we provide a summary of the recently developed DEDL methods and their applications. Future developments in DEDLs are expected to extend the application scope of DELs to complex biological systems with unique ligand-discovery capabilities.
1. Introduction
1.1 DNA-encoded chemical library (DEL)
The identification of small-molecule ligands that can modulate the functions of biological targets is central to chemical, biological, and pharmaceutical sciences. For decades, high-throughput screening (HTS) has been a major ligand-discovery modality;1 however, HTS platforms are complex, prohibitively expensive, and are limited to several millions of compounds. In the past two decades, the emergence of the DNA-encoded library (DEL) technology, also called DELT, has replaced spatial encoding with DNA encoding in chemical libraries, thus overcoming the throughput limit of traditional HTS and being much more affordable and accessible to researchers.2–11 For instance, many companies and institutions are offering DEL selection services: premade DEL kits12 for direct use in selections, and reagent kits13 for custom DEL synthesis are also available to meet the needs of individual researchers. Furthermore, with the recent expansion of DNA-/DEL-compatible reaction toolkits,2,6,7,14–16 DELs can access much greater chemical space, which has been a major driver for its adoption in the pharmaceutical industry.3,11,17–20The concept of the DEL was proposed by Brenner and Lerner in 1992.21 It was originally devised as an approach to improve the traditional one-bead, one-compound (OBOC) combinatorial chemical library by synthesizing an on-bead DNA tag along with the compound. After the screening, the DNA-tagged OBOC library (OBOC-DEL) could be deconvoluted by reading the DNA sequence attached to the isolated beads. The concept was quickly realized by Nielsen, Brenner, and Janda22 as well as by Gallop and co-workers in 1993.23 OBOC-DELs had a practical limit on the library size (e.g., the number of beads that can be handled) and involved the challenging parallel synthesis of chemical compounds and oligonucleotides. Nevertheless, these early studies laid out the technological foundation of DELs, including the encoding strategy, selection method, and decoding techniques. Later, OBOC-DELs became less reported, and until recently, the research groups of Paegel and Kodadek have revived OBOC-DELs with sophisticated bead designs, novel on-bead chemistry, and advanced instrumentation, which have led to a variety of novel applications.7,15,23–29 The details of modern OBOC-DELs fall outside the scope of this review and we refer interested readers to these excellent review articles.9,30,31 In 2004, Neri,32 Liu,33 Harbury,34 Winssinger,35 and their respective co-workers independently reported four types of encoded library: the dual-pharmacophore encoded self-assembling chemical (ESAC) library, the DNA-templated synthesis (DTS) library, the DNA-routing library, and the peptide–nucleic-acid (PNA)-encoded library. The libraries are encoded by either DNA translation, routing, or ligation in the solution phase, thereby overcoming the limitations of OBOC libraries. In a seminal work in 2009, GlaxoSmithKline (GSK) demonstrated the application of DELs at an industrial scale.36 Following these landmark studies, DELs entered a stage of rapid developments. The declining cost of genomic-scale next-generation sequencing (NGS) technologies has made the selection of multi-billion-compound DELs a routine practice. With the rapid development of DEL-compatible chemistry, arguably, the chemical space of DELs is limited by the availability of the building blocks, rather than the reactions to connect them.2,7 Recent innovations have also integrated DEL with many legacy and emerging techniques, such as fragment-based drug discovery (FBDD),32,37–40 the dynamic combinatorial library (DCL),41–47 diversity-oriented synthesis (DOS),48,49 machine learning,50–52 OBOC libraries/microfluidics,24,26,53–55 flow cytometry,23,27,28etc., leading to novel applications in ligand discovery and functional assays.9,30 DEL selections have generated a number of clinical candidates,17 such as GSK2256294, an epoxide hydrolase inhibitor to treat pulmonary disease,17,18 GSK2982772, a first-in-class receptor-interacting protein-1 kinase inhibitor to treat inflammatory diseases,17,19 and X-165, an autotaxin inhibitor as the clinical candidate for pulmonary fibrosis.20 Very recently, researchers selected a 4-billion DEL against the SARS-CoV-2 main protease (Mpro) and obtained potent inhibitors with high potential for clinical development.56
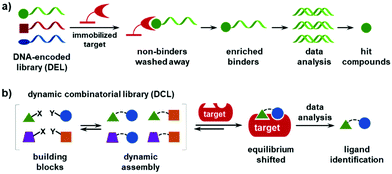
Regardless of whether in solution or on beads, most DELs are “static”, i.e., the population of the library is fixed. Incubation with the target establishes a thermodynamic equilibrium, which is usually driven by the high concentration of the protein (0.4–50 μM),8 and a series of washing steps to remove the non-binders, whereas the “enriched” binders are eluted under denaturing conditions (Fig. 1a). The ideal condition for affinity-based DEL selections should be a balance of the thermodynamic association (Ka) and kinetic disassociation (koff) parameters. However, first, developing an optimal washing protocol is rather difficult as many factors are implicated (the number of washes, time duration, buffers, centrifugation conditions, temperature, etc.). The Neri group provided an excellent protocol as a reference for DEL selections,57 and recent studies have shown that quantitative PCR (qPCR) is an effective tool for quality control.58–62 Second, the binders with low abundance may be missed if their copy numbers fall below the detection threshold after the selection,59,63 which may worsen for very large libraries. Third, the washing-based selections require a purified and immobilized protein, which may limit the target scope. Recent studies have developed several methods for in-solution DEL selections in buffers, in cell lysates, on/within live cells, with whole bacteria, and even in human sera.8 However, DEL selections with immobilized targets remain the mainstream practice.
 | ||
| Fig. 1 Schematic illustrations of (a) the selection of a DNA-encoded library (DEL) against immobilized protein targets and (b) the formation and selection of a dynamic combinatorial library (DCL). | ||
1.2 Dynamic combinatorial library (DCL)
The dynamic combinatorial library (DCL) is a type of combinatorial chemical library, where the library is formed in situ by mixing the building blocks (BBs) that can undergo dynamic exchange through reversible chemical reactions (Fig. 1b).64–78 In contrast to traditional HTS and DEL selection, DCL screening utilizes the protein to template the ligand synthesis. Compared with a static library, first, the DCL allows for the spontaneous formation of the binders without spatial separation; second, the DCL has the advantage of high-affinity but low-abundance binders being amplified from a large excess of a non-binding background, i.e., a higher signal-to-noise ratio; third, DCLs can be feasibly prepared by mixing the BBs (also called fragments) in a single solution without the synthesis of individual compounds; fourth, DCLs are under thermodynamic control, and a comparison of the equilibria before and after target addition is able to identify the binders. In the past decades, DCLs have shown great potential in drug discovery, materials science, and systems chemistry. However, the development and application of DCLs have been significantly hampered by the small library size. Although elegant strategies have been developed to access large dynamic libraries,79–84 most DCLs only contain a few dozen to several hundreds of compounds.68 The major underlying reason is the lack of analytical methods suitable for resolving large libraries and identifying the binders from complex mixtures.68,78,85,86 Typically, liquid chromatography,68,87–91 mass spectrometry,92 NMR,93 and spectroscopic methods81,94,95 are used for hit identification in DCL screening, but these methods lack the resolution and throughput for large libraries. In addition, DCL screening usually requires high protein consumption and involves tedious sample-processing procedures (e.g., the separation of protein from bound ligands prior to analysis).1.3 Concept and advantages of the DNA-encoded dynamic library (DEDL)
The DCL and the DEL share some similarities in that both are “one-pot” libraries and all the compounds are synthesized, processed, and selected simultaneously without spatial separation. However, DELs can contain a very large number of compounds and the library selection can be conducted at a minute scale. Hence, introducing DNA encoding may be a promising strategy to address the long-lasting issue of DCLs. On the other hand, dynamic DELs have the advantages of DCLs. The preparation of dynamic DELs is straightforward by simply mixing the DNA-encoded BBs. The protein target can act as the template to promote the synthesis/enrichment of the binders, which may give a higher signal-to-noise ratio and enable the identification of the high-affinity binders with copy numbers below the reliability threshold in DEL selections (103–104 copies per compound).59,63 Furthermore, the selection of dynamic DELs may be conducted without physical washes, thus realizing selections in solution or with complex biological targets. Indeed, researchers have developed a number of dynamic DEL approaches.96 Although these methods are given different names in different studies, to avoid confusion, they are generally referred as DNA-encoded dynamic libraries (DEDLs) in this Review Article.2. Previous studies leading to the development of DEDLs
2.1 Dual-pharmacophore ESAC library
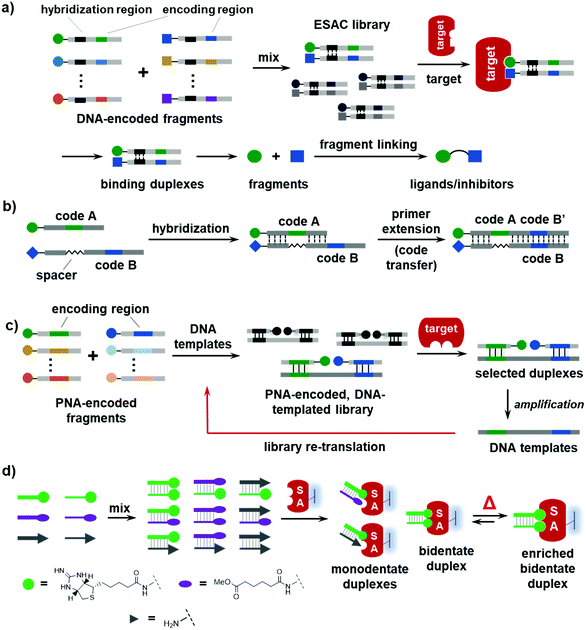
In 2004, the Neri group reported an encoded self-assembling chemical (ESAC) library method in which two sets of partially complementary DNA-encoded BBs are mixed to form a combinatorial library of fragments (Fig. 2a).32,37,97–102 The library is selected against immobilized targets, and the binding BBs can be decoded by sequencing the coding regions. Initially, the two BBs in a binding pair are separately decoded and their correlation is lost; thus, ESAC libraries were mostly used for affinity maturation of known ligands.98,99,103 Recently, an elegant tag-transfer approach was developed to incorporate both sets of codes into one DNA strand (Fig. 2b),37 so that the synergistic BB pairs, instead of individual BBs, could be identified. ESAC libraries have been successfully employed to discover novel ligands against a variety of biological targets.104 The quality of ESAC libraries is very high, since each DNA-conjugated BB is chromatographically purified and there are no truncated/side products as in regular DELs.105–107 In principle, the ESAC library is in line with fragment-based drug discovery (FBDD), where small fragments (i.e., the BBs) that cooperatively bind to the target are identified before being elaborated into high-affinity binders through fragment linking, growing, and/or merging.85 However, ESAC libraries are static and are usually selected against purified and immobilized proteins.2.2 PNA/DNA-directed fragment assembly
PNA (peptide nucleic acid) is a type of nucleic acid that can form duplexes with DNA, RNA, and the PNA itself with a neutral and achiral peptidic backbone. PNA also has a coding system of four canonical bases, but it is more chemically stable and compatible with organic reactions. Previously, PNA has been extensively used to encode small-molecule probes in proteomic profiling, combinatorial libraries, and ligand displays.108–113 However, PNA is not compatible with PCR amplification and DNA sequencing technologies. Winssinger and co-workers developed an elegant strategy combining the advantages of DNA and PNA.39,114 As shown in Fig. 2c, two sets of PNA-encoded fragments are assembled on DNA templates, forming a fragment-pair library. After the selection, the DNA templates of the selected fragment pairs are amplified via PCR and then re-translated into a more focused fragment library for more rounds of selection. Although these PNA/DNA-encoded libraries were also static, as suggested by the authors, this approach may provide a viable encoding strategy for DCLs.392.3 Dynamic DNA ligand assemblies
Beyond duplexes, higher dimensions of fragment assemblies have been constructed with DNA triplexes,32,115,116 quadruplexes,117–121 or even pentaplex structures.122 The first report on using DNA duplexes to dynamically display small molecules was by the Hamilton group in 2005.123 As shown in Fig. 2d, they prepared a 9-member library assembled from two sets of DNA-encoded small molecules containing an iminobiotin, a methyl adipate, and an amino group. The library was exposed to an immobilized streptavidin (SA), a homo-tetrameric protein that can bind to four biotin or biotin derivatives with four binding pockets. Without heating, the library was static (analogous to an ESAC library), and the selection identified both the monodentate and bidentate biotin duplexes. After the library was heated above the melting temperature of the DNA duplexes, the library became dynamic and started exchanging DNA strands, and the selection with SA identified the enrichment of the high-affinity bidentate iminobiotin duplexes. This study has demonstrated that, in a dynamic DNA display, the protein can promote the formation of high-affinity duplex binders at the expense of the non-binders and the moderate binders. This work has established the basic principle for the future development of DEDLs.3. Development of DEDLs
3.1 DNA-encoded dynamic combinatorial chemical library (EDCCL)
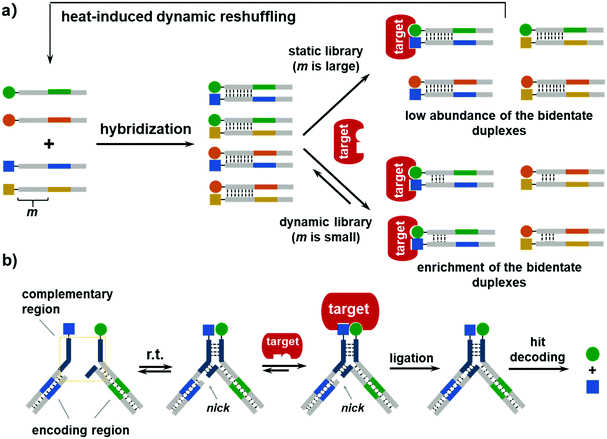
The first DEDL method was reported by Zhang and co-workers in 2015, and was named the DNA-encoded dynamic combinatorial chemical library (EDCCL).41 As shown in Fig. 3a, first, an ESAC-type library with stable DNA duplexes was formed. After the first round of selection with an immobilized target, the flow-through was heated to melt the DNA duplexes and enable the dynamic exchange of the DNA strands. Such a reshuffling process promoted the formation of more high-affinity duplexes, which can be further enriched in more rounds of selection. Several model libraries were prepared by mixing different ratios of the non-binding DNA duplexes and the duplexes with an iminobiotin. The authors also tested different numbers (m) of complementary base pairs to adjust the duplex stability. When m is large (e.g., m = 21), the duplexes were stable, and the library was static. The selection with SA did enrich the high-affinity bidentate iminobiotin duplexes, but the enrichment fold was low due to the low reshuffling rate of the DNA duplexes, i.e., a high kinetic barrier (Fig. 3a). When m was decreased to 6–8, the library became dynamic, and the addition of SA shifted the equilibrium and led to the enrichment of the bidentate duplexes. The enrichment of the dynamic library was more significant than the static library. Notably, the weaker the monovalent binding, the greater is the enrichment of the bidentate duplexes. The authors also showed that this method could identify low- to high-micromolar binders, a typical affinity range for unbiased DEL selections. Furthermore, this approach was extended to heat-induced EDCCLs (hi-EDCCLs), where the affinity selection and the heat-induced recombination process were carried out separately. As shown in Fig. 3a, after the selection of the stable library against the immobilized SA, the flow-through containing the non-binders and weak binders was subjected to a heating/annealing process to reshuffle the DNA strands and form more of the high-affinity bidentate duplexes. The reshuffled library was then subjected to more rounds of selection for further enrichment. EDCCL/hi-EDCCL was the first systematic study demonstrating the preparation and selection strategies of dynamic DELs. Although no chemical and sequence diversities were included in these model libraries, this study has shown that, in principle, DNA encoding could enable the selection of large DCLs, as the bidentate duplex was enriched from a 90![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 600-fold excess of the non-binding background.
600-fold excess of the non-binding background.
The hi-EDCCL suffers the issue of strong heating (up to 90 °C), which may be detrimental to the protein and/or the compounds. In 2019, Zhang and co-workers improved the EDCCL method by using a Y-shaped DNA architecture (Y-EDCCL).42 As shown in Fig. 3b, the Y-EDCCL is assembled from two sets of partially complementary DNA-encoded BBs, and the short hybridization region makes the dynamic assembly/reshuffling possible at room temperature. The selection of Y-EDCCLs was also conducted with an immobilized target, and the high-affinity duplexes bound to the target stabilized the Y-shaped DNA complex. The DNA codes of the two strands were enzymatically ligated for decoding (Fig. 3b). The ligation step may provide an additional mechanism for reducing the background arising from single BB binding since the corresponding DNA codes could not be ligated. Notably, the selection of Y-EDCCLs gave a significantly higher enrichment of the bidentate binders in comparison with either the Y-shaped or the linear static library (>26-fold higher enrichment in SA selection). Although only model libraries with a limited diversity were tested (a 285-member Y-EDCCL was selected against carbonic anhydrase II, CA-2), Y-EDCCL is compatible with the standard hit decoding scheme of DELs, suggesting that large DEDLs could be built and selected with this method. Collectively, these studies have nicely shown that DNA encoding can indeed be used to prepare, process, and select large dynamic libraries against proteins. In addition, the EDCCL has been adapted to an automated microfluidic platform with improved efficiency,124 and the technology has also been commercialized and is being sold as a selection kit (DyNAbind®) for fragment discovery.12
For DCL selections, after the equilibrium is shifted to a new state induced by the target, an equilibrium-locking procedure is carried out to stop the dynamic exchange, so that the “locked” equilibrium may be compared with that without target addition (or other controls) for hit identification. Equilibrium locking can be accomplished either by changing the experimental condition (e.g., pH, temperature, or solvent composition) or by using additives (e.g., using NaBH3CN to reduce imine to amine or lowering the pH to stop disulfide exchange). Ideally, the “locking” mechanism should be very fast or at least significantly faster than the dynamic BB exchange, so that the locking process itself has a minimal effect on the position of the equilibrium.65,66 In Hamilton's dynamic duplex display and EDCCL, there was no equilibrium locking, and the binders and non-binders were separated via washes off the immobilized targets. In the EDCCL, a model library was selected against non-immobilized SA using a selective digestion strategy,125 but this was still a virtual washing procedure and nuclease digestion may perturb the equilibrium.65 Hence, it is desirable to develop DEDL methods that can reliably lock the equilibrium after target addition, so that the dynamic libraries can be applied to more diverse types of biological target.
3.2 Photo-crosslinking-mediated selection of DEDLs
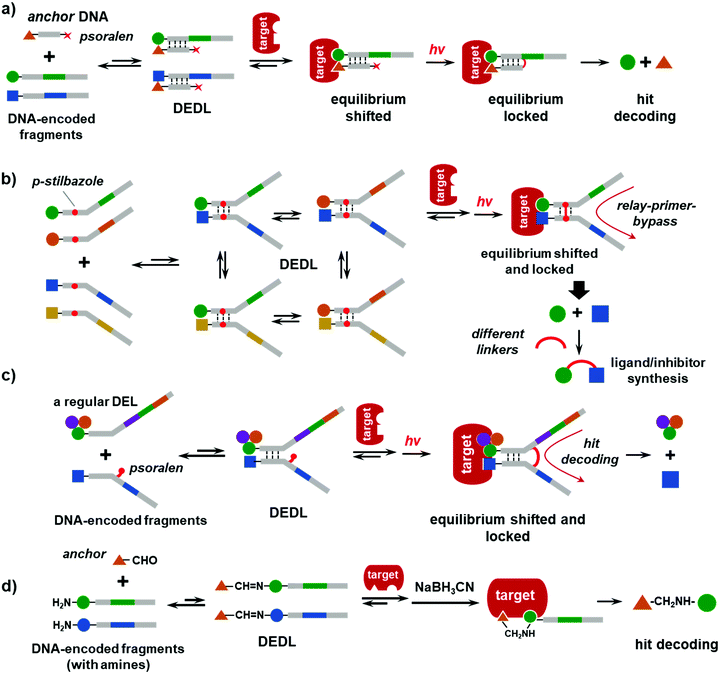
In 2015, the Li group reported a DEDL method using photo-crosslinking to lock the equilibrium.43 As shown in Fig. 4a, a set of DNA-encoded fragments is hybridized with an “anchor” DNA, which bears a known ligand for the target and a photo-crosslinker (psoralen) at the distal end of the DNA strand. When the DNA-encoded fragments are in excess, they compete for hybridization with the anchor DNA. Due to the short length (6–7 nt), the duplexes are unstable and constantly exchange DNA strands, thereby forming a DEDL of “fragment–anchor” pairs. Upon target addition, the equilibrium shifts to form more high-affinity bidentate duplexes. Next, the photo-reactive psoralen group crosslinks to the opposite DNA strand upon UV irradiation, which stops the dynamic exchange and locks the equilibrium. The crosslinked duplexes can be isolated for hit identification by reading the encoding DNA sequence (Fig. 4a). Notably, the photo-crosslinking step is very fast (30–60 s) and can be easily imposed/withdrawn without any additives and little perturbation to the equilibrium, i.e., like taking a photo snapshot. Although the efficiency of the photo-crosslinking reaction is low in aqueous solution, this was not a major issue because the crosslinked duplexes could be amplified via the highly sensitive PCR. Actually, this might be an advantage because of the low background signal arising from the non-binding duplexes. The authors have carefully verified that the system was under thermodynamic control, i.e., different starting points led to the same thermodynamic equilibrium, which is an important feature of DCLs.126,127 Moreover, this method completely abolished protein modification/immobilization and physical washes. The selection takes place in one solution and is suitable for unmodified, non-immobilized proteins.125,128,129 This study selected a model library against SA and CA-2, and the SA selection gave a 19.2-fold enrichment of the bidentate desthiobiotin duplex, which is comparable to typical DCL selections.90,130–132 However, this approach can only be used for the “affinity maturation” of known ligands,32,37,97,98 rather than de novo discovery.In 2018, the Li group made a major advancement on the photo-crosslinking-based DEDL method.44 As shown in Fig. 4b, two sets of DNA-encoded fragments formed a combinatorial library of fragment pairs. Again, the short hybridization region resulted in dynamic exchange of the DNA strands, whereas the distal regions of the DNA strands were not complementary and contained the DNA codes. The hybridization region was also embedded with a pair of p-stilbazole molecules, an efficient inter-strand DNA photo-crosslinker orthogonal to natural nucleobases with little impact on the properties of the DNA duplexes.133 After target incubation, UV irradiation crosslinked the duplexes and locked the equilibrium. The crosslinking resulted in an unnatural junction that is unreadable to DNA polymerase. To read the codes, a relay-primer-bypass decoding strategy was used.134 In addition to the regular terminal primers, a relay primer was added to mask the unnatural linkage; polymerase-extension of the primers and enzymatic ligation generated a DNA template that is compatible with PCR amplification (Fig. 4b). A 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000-member DEDL with actual chemical diversity was prepared and selected against four protein targets covering a range of protein families (SIRT3, SAE1, UBC9, and TRIM28). For each target, multiple fragment pairs were enriched, which were further conjugated with a series of linkers of different lengths, flexibilities, and lipophilicities.98 In general, the shorter linkers or no linker (i.e., direct conjugation of the fragments) gave more potent ligands/inhibitors. More recently, Li and co-workers selected the same DEDL against SIRT-1, -2, and -5, as well as the BD1 and BD2 domains of bromodomain (BRD4).135 Despite the moderate library size, a series of isoform-/domain-selective ligands/inhibitors were identified. Collectively, these studies have demonstrated the generality and performance of chemically diverse DEDLs in de novo ligand discovery.
000-member DEDL with actual chemical diversity was prepared and selected against four protein targets covering a range of protein families (SIRT3, SAE1, UBC9, and TRIM28). For each target, multiple fragment pairs were enriched, which were further conjugated with a series of linkers of different lengths, flexibilities, and lipophilicities.98 In general, the shorter linkers or no linker (i.e., direct conjugation of the fragments) gave more potent ligands/inhibitors. More recently, Li and co-workers selected the same DEDL against SIRT-1, -2, and -5, as well as the BD1 and BD2 domains of bromodomain (BRD4).135 Despite the moderate library size, a series of isoform-/domain-selective ligands/inhibitors were identified. Collectively, these studies have demonstrated the generality and performance of chemically diverse DEDLs in de novo ligand discovery.
Typical DELs contain 3–4 sets of BBs; however, most DEDLs are built on DNA duplexes with two sets of BBs, which limits the library size. In addition, the DEDL shown in Fig. 4a requires a pair of unnatural photo-crosslinkers. It is more desirable that regular DELs can be adapted to a DEDL format without library resynthesis. In 2020, the Li group replaced the p-stilbazole with a psoralen group (Fig. 4c).136 Psoralen can be conjugated to the non-base-pairing side of the nucleobases in DNA, is commercially available, and does not require a crosslinking partner in the opposite strand. Thus, in principle, any DEL encoded with single-stranded DNA tags may be adapted to a DEDL and the common primer-binding site can be used for dynamic hybridization.
3.3 Non-fragment-based DNA-encoded dynamic library
All the DEDLs discussed above are fragment-based libraries; thus, they can only be used for the discovery of synergistic fragment pairs or affinity maturation of the known ligands.137 A fragment-linking step is required to obtain the full ligands. In fact, fragment linking is a frequently used strategy to evolve fragments into high-affinity/high-specificity ligands.138–141 Ideally, the linked fragments should bind to the target more strongly than the sum of individual ones;142–144 however, the linker has profound effects on the binding properties.145–148 Previously, Seitz, Winssinger, Neri, and their respective co-workers have elegantly shown that small fragments selected from DELs or DNA-display libraries could be elaborated into potent binders by screening different linkers37,38,98,99,149–151 or using rigid scaffolds.146 Nevertheless, fragment linking is still a tedious and empirical procedure with lots of trial-and-error.144,145 In fact, fragment linking takes considerably more effort than identifying the fragment itself and is a major issue in fragment-based ligand discovery.85,147,152Most DCLs are also fragment libraries; however, since the protein target templates the synthesis of the full ligands in situ, fragment linking is no longer necessary. In most DEDLs, there is no chemical reaction between the fragments and the dynamic interaction only takes place between the DNA strands. In 2020, the Li group partially addressed in the issue.45 As shown in Fig. 4d, a regular DEL encoded with a single-stranded DNA tag bearing a primary amine is incubated with an “anchor”, which is a known ligand of the target with an aldehyde group. Imine formation via aldehyde–amine condensation is a widely used dynamic combinatorial chemistry (DCC) step in DCLs and is compatible with physiological conditions.76 The dynamic imine library is selected against the non-immobilized target. The shifted equilibrium is locked by NaBH3CN reduction to amines. The binders could be isolated from the mixture using an N-hydroxysuccinimide (NHS) ester, which has a lower activity for secondary amines. A more selective reagent, such as o-phthaldialdehyde (OPA),153 may be used to improve the specificity. The anchor could be flexibly changed to convert an unbiased DEL to a focused one. Existing DELs prepared using other methods or encoded with double-stranded DNA tags may be used, as long as the library contains a primary amine. The method was tested with five proteins, i.e., CA-2, BD1 and BD2 of BRD4, acetylcholinesterase (AChE), and the X-linked inhibitor of apoptosis protein (XIAP), and it identified the binders of a wide range of affinities (low-nM to mid-μM). However, the molecular weight of the library compounds had significant effects on the selection results. The selection of a small, 2-BB library (67![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 compounds) identified many more potent and selective ligands than the large 3-BB library (17
000 compounds) identified many more potent and selective ligands than the large 3-BB library (17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 576–one million compounds), presumably because the large compound size offsets the anchor's directing effect.154 This DEDL method more resembles classic DCLs, as the dynamic interaction occurs between the anchor and the library compounds, and the protein templates the in situ synthesis of the ligands; thus, fragment linking is not necessary. However, this approach requires a known ligand of the target, which may limit its applications.
576–one million compounds), presumably because the large compound size offsets the anchor's directing effect.154 This DEDL method more resembles classic DCLs, as the dynamic interaction occurs between the anchor and the library compounds, and the protein templates the in situ synthesis of the ligands; thus, fragment linking is not necessary. However, this approach requires a known ligand of the target, which may limit its applications.
3.4 PNA-encoded/conjugated dynamic library
PNA has been widely employed to encode small-molecule probes in proteomic profiling, combinatorial libraries, and ligand displays.108–113 PNA–small-molecule (especially glycans) conjugates have been used to tailor the valency, distance, and geometry of the ligands in the assembly.112,155,156 In principle, many of the PNA-encoded systems could be adapted to be dynamic and lead to novel applications;39 however, so far, the examples are scarce. In 2017, the Winssinger group introduced dynamic hybridization in a multivalent assembly of PNA–glycan conjugates, which achieved an exceptional affinity gain and inhibitory activity against bacteria lectins, as compared with the monovalent ligands (Fig. 5a).47 More recently, this approach of supramolecular dynamic assembly has been developed into a library format, called a PNA-based dynamic combinatorial library (PDCL) with >6000 assemblies.46 The PDCL is synthesized in a “split-and-mix” fashion wherein the PNA, linker, and glycan were conjugated step-wise as three BBs in a combinatorial library. The library contains not only the structural diversity, but also the positional diversity of the glycan ligands. Rather than an encoding tag, the PNA serves as a handle for dynamic assembly of the conjugates (Fig. 5b). Because of the high stability of PNA–PNA duplexes, a short 4-mer PNA is sufficient for hybridization; thus, hit conjugates can be directly identified using MALDI mass spectrometry. The library was selected against two bacteria lectins (AFL and RSL), and a series of bivalent conjugates have been identified with significantly higher affinity than the monovalent glycans. In addition, although not directly interacting with the protein and not connecting the two glycans, the linker motif appeared to be very important in ligand binding, as its structural properties are implicated in balancing the enthalpy and entropy of the binding event.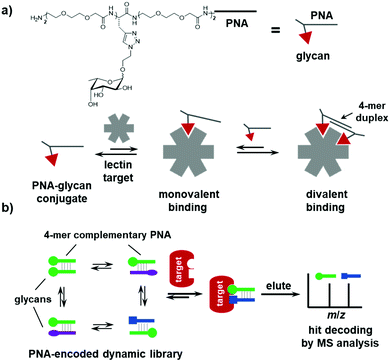 | ||
| Fig. 5 (a) Multivalent dynamic assembly of PNA–glycan conjugates on a lectin target. (b) Selection of PNA-based dynamic combinatorial libraries (PDCLs). | ||
4. Conclusions
Since the milestone study by Lehn and Hun in 1997, which demonstrated that small-molecule ligands can be identified from a dynamic chemical population against proteins,88 DCLs have been exploited as a useful tool for ligand discovery.64–78 However, after decades of research, DCLs still face two main obstacles: the limited scope of dynamic combinatorial chemistry and the small library size. The latter is a particularly acute problem: unlike in materials science and systems chemistry, for ligand discovery, the library size (chemical diversity) needs to be very large to increase the chance of identifying potent binders. However, with only a few exceptions,79–84,157 most DCLs only have less than a few hundred compounds, due to the lack of analytical methods to resolve complex mixtures.69,86–88 The power of DNA encoding has been well proved in DELs, and the DEDL approaches have effectively addressed the library size issue in DCLs.The past decade has witnessed the significant development of DEDLs. Table 1 summarizes the key information of all reported DEDLs. We anticipate more diverse applications of DEDLs to emerge in the future. For example, the biological targets previously interrogated using traditional DCLs may be revisited through the use of DEDLs with much larger chemical diversity. DEDL selection is compatible with non-immobilized targets, making it an attractive method of choice for interrogating complex biological targets, such as endogenous proteins, membrane proteins on live cells, or even intracellular targets.10,60,158 Furthermore, multivalency is ubiquitous and plays pivotal roles in biology; nature has exploited multivalency to achieve strong, specific, but reversible molecular interactions in biological systems.159 DEDLs may be particularly suitable for discovering novel multivalent binders, because of their reversible, dynamic nature and their programmability in ligand assembly. The recent work on PDCL selection against a bacterial lectin is an excellent example.46
| Library architecture | Selection method | Target(s) | Building block sets | Building blocks in each set | Size | Key reference(s) | |
|---|---|---|---|---|---|---|---|
| ESAC library |

|
Immobilized protein | Alpha-1-acid glycoprotein, carbonic anhydrase IX | 2 | 550 members in sub-library A, 202 members in sub-library B | 111![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 100 |
32,37 and 104 |
| Dynamic assembly of DNA-encoded fragments |

|
Immobilized protein | Streptavidin | 1 | 3 × 3 model library with iminobiotin as positive control | 9 | 123 |
| EDCCL |

|
Immobilized protein | Streptavidin | 1 | Model library with iminobiotin as positive control | 90![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 601 601 |
41 |
| Unmodified, non-immobilized protein | Alpha-1-acid glycoprotein | 1 | Model library with furan derivatives as positive control | 4001 | 41 | ||
| Heat-induced EDCCLs |

|
Immobilized protein | Streptavidin | 1 | Model library with iminobiotin as positive control | 961 | 41 |
| Y-EDCCL |
|
Immobilized protein | Carbonic anhydrase II | 1 | 285 members in sub-library A, and a positive control CBS on sub-library B | 286 | 42 |
| First version of the photo-crosslinking-based DEDL |

|
Unmodified, non-immobilized proteins | Streptavidin, carbonic anhydrase II | 1 | Model library with desthiobiotin as positive control | 1025 | 43 |
| Second version of the photo-crosslinking-based DEDL |
|
Unmodified, non-immobilized proteins | SIRT3, SAE1, UBC9, and TRIM28 | 2 | 100 members in each sub-library | 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
44 |
| Third version of the photo-crosslinking-based DEDL |
|
Unmodified, non-immobilized proteins | Streptavidin | 1 | Model library with desthiobiotin as positive control | 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 001 001 |
136 |
| Anchor-directed DEDL |

|
Unmodified, non-immobilized proteins | Carbonic anhydrase II, BRD4 (BD1 and BD2), AChE, and XIAP | 2 or 3 | 260 × 260 for 2-BB library, 242 × 260 × 260 for 3-BB library | 67![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 600 and 17 600 and 17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 576–one million 576–one million |
45 |
| PNA-based dynamic combinatorial libraries (PDCLs) |
|
Immobilized proteins | Lectins (AFL and RSL) | 2 | 150 members with 5 linkers and 10 glycans | 5000 | 46 |
There are still several technical hurdles to be circumvented in DEDLs. First, in most DEDLs, the dynamic interactions are between the DNA strands, rather than the BBs as in DCLs. Although fragment-based ligand discovery has its advantages,152 post-selection fragment linking is highly challenging, especially considering the large number of fragments that could be identified from DEDL selections. Future developments may aim to incorporate and encode the linker motifs in the library as a diversity element.96 Alternatively, DEDLs may be designed to allow the protein to promote the synthesis of the full ligands in situ. Second, the dynamic nature of DEDLs has the advantage of amplifying strong binders from a large excess of background, which may lead to better signal-to-noise ratios and a lower incidence of false positives than static libraries. However, this point has only been tested with model libraries,41,42 and it remains to be verified with large, chemically diverse libraries, especially at the scale of typical DELs.160 Third, DCL is a thermodynamically controlled system. Adding the target protein induces a shift of the equilibrium to the point of the lowest free-energy position collectively for all compounds; thus, it is possible that a DEDL selection enriches a large number of “mediocre” ligands, whereas the enrichment of the most potent binders might be obscured. This potential issue may be addressed by using kinetic target-guided synthesis (KTGS), in which the protein templates the irreversible reaction between the fragments.161 Although it would not be a dynamic library per se, this strategy may be an alternative to DEDL for identifying the best binders from large fragment libraries. Finally, the canonical duplex architecture limits DEDLs to two sets of fragments. Although a triplex-display library was proposed by Neri and co-workers,32 it has not been practically implemented yet. Such a 3-dimensional library would boost the diversity to a level comparable to regular DELs, but it also presents more challenges in selection decoding and fragment linking, which need to be carefully addressed in future methodology development.
Author contributions
All authors contributed to writing and editing the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work is supported by the Shandong Provincial Natural Science Foundation, China (ZR2021QB152), Faculty start-up funds of Jining Medical University, the college students’ Innovative Entrepreneurial Training Plan Program of Jining Medical University (S201910443002), by the Shenzhen Bay Laboratory, Shenzhen, China (SZBL2020090501008), the Research Grants Council of Hong Kong SAR, China (AoE/P-705/16, 17301118, 17111319, 17303220, 17300321, and C7005-20G), and the NSFC of China (21877093 and 91953119). The authors acknowledge the support from “Laboratory for Synthetic Chemistry and Chemical Biology” under the Health@InnoHK Program by the Innovation and Technology Commission, Hong Kong SAR, China.Notes and references
- R. Macarron, M. N. Banks, D. Bojanic, D. J. Burns, D. A. Cirovic, T. Garyantes, D. V. Green, R. P. Hertzberg, W. P. Janzen, J. W. Paslay, U. Schopfer and G. S. Sittampalam, Nat. Rev. Drug Discovery, 2011, 10, 188–195 CrossRef CAS PubMed.
- D. Conole, J. H. Hunter and M. J. Waring, Future Med. Chem., 2021, 13, 173–191 CrossRef CAS PubMed.
- A. L. Satz, L. Kuai and X. Peng, Bioorg. Med. Chem. Lett., 2021, 39, 127851 CrossRef CAS PubMed.
- D. T. Flood, C. Kingston, J. C. Vantourout, P. E. Dawson and P. S. Baran, Isr. J. Chem., 2020, 60, 1–14 CrossRef.
- V. B. K. Kunig, M. Potowski, M. Klika Skopic and A. Brunschweiger, ChemMedChem, 2021, 16, 1048–1062 CrossRef CAS PubMed.
- K. Götte, S. Chines and A. Brunschweiger, Tetrahedron Lett., 2020, 61, 151889 CrossRef.
- P. R. Fitzgerald and B. M. Paegel, Chem. Rev., 2021, 121, 7155–7177 CrossRef CAS PubMed.
- Y. Huang and X. Li, ChemBioChem, 2021, 22, 2384–2397 CrossRef CAS PubMed.
- T. Kodadek, N. G. Paciaroni, M. Balzarini and P. Dickson, Chem. Commun., 2019, 55, 13330–13341 RSC.
- Y. Song and X. Li, Acc. Chem. Res., 2021, 54, 3491–3503 CrossRef CAS PubMed.
- M. Song and G. T. Hwang, J. Med. Chem., 2020, 63, 6578–6599 CrossRef CAS PubMed.
- DyNAbind; https://www.sigmaaldrich.com/.
- https://www.hitgen.com/en/capabilities-details-22.html .
- S. Patel, S. O. Badir and G. A. Molander, Trends Chem., 2021, 3, 161–175 CrossRef CAS PubMed.
- P. Dickson and T. Kodadek, Org. Biomol. Chem., 2019, 17, 4676–4688 RSC.
- D. T. Flood, C. Kingston, J. C. Vantourout, P. E. Dawson and P. S. Baran, Isr. J. Chem., 2020, 60, 268–280 CrossRef CAS.
- C. C. Arico-Muendel, MedChemComm, 2016, 7, 1898–1909 RSC.
- S. L. Belyanskaya, Y. Ding, J. F. Callahan, A. L. Lazaar and D. I. Israel, ChemBioChem, 2017, 18, 837–842 CrossRef CAS PubMed.
- P. A. Harris, S. B. Berger, J. U. Jeong, R. Nagilla, D. Bandyopadhyay, N. Campobasso, C. A. Capriotti, J. A. Cox, L. Dare, X. Dong, P. M. Eidam, J. N. Finger, S. J. Hoffman, J. Kang, V. Kasparcova, B. W. King, R. Lehr, Y. Lan, L. K. Leister, J. D. Lich, T. T. MacDonald, N. A. Miller, M. T. Ouellette, C. S. Pao, A. Rahman, M. A. Reilly, A. R. Rendina, E. J. Rivera, M. C. Schaeffer, C. A. Sehon, R. R. Singhaus, H. H. Sun, B. A. Swift, R. D. Totoritis, A. Vossenkamper, P. Ward, D. D. Wisnoski, D. Zhang, R. W. Marquis, P. J. Gough and J. Bertin, J. Med. Chem., 2017, 60, 1247–1261 CrossRef CAS PubMed.
- J. W. Cuozzo, M. A. Clark, A. D. Keefe, A. Kohlmann, M. Mulvihill, H. Ni, L. M. Renzetti, D. I. Resnicow, F. Ruebsam, E. A. Sigel, H. A. Thomson, C. Wang, Z. Xie and Y. Zhang, J. Med. Chem., 2020, 63, 7840–7856 CrossRef CAS PubMed.
- S. Brenner and R. A. Lerner, Proc. Natl. Acad. Sci. U. S. A., 1992, 89, 5381–5383 CrossRef CAS PubMed.
- J. Nielsen, S. Brenner and K. D. Janda, J. Am. Chem. Soc., 1993, 115, 9812–9813 CrossRef CAS.
- M. C. Needels, D. G. Jones, E. H. Tate, G. L. Heinkel, L. M. Kochersperger, W. J. Dower, R. W. Barrett and M. A. Gallop, Proc. Natl. Acad. Sci. U. S. A., 1993, 90, 10700–10704 CrossRef CAS PubMed.
- A. L. Hackler, F. G. FitzGerald, V. Q. Dang, A. L. Satz and B. M. Paegel, ACS Comb. Sci., 2020, 22, 25–34 CrossRef CAS PubMed.
- A. B. MacConnell, A. K. Price and B. M. Paegel, ACS Comb. Sci., 2017, 19, 181–192 CrossRef CAS PubMed.
- W. G. Cochrane, M. L. Malone, V. Q. Dang, V. Cavett, A. L. Satz and B. M. Paegel, ACS Comb. Sci., 2019, 21, 425–435 CrossRef CAS PubMed.
- K. R. Mendes, M. L. Malone, J. M. Ndungu, I. Suponitsky-Kroyter, V. J. Cavett, P. J. McEnaney, A. B. MacConnell, T. M. Doran, K. Ronacher, K. Stanley, O. Utset, G. Walzl, B. M. Paegel and T. Kodadek, ACS Chem. Biol., 2017, 12, 234–243 CrossRef CAS PubMed.
- M. H. Shin, K. J. Lee and H. S. Lim, Bioconjugate Chem., 2019, 30, 2931–2938 CrossRef CAS PubMed.
- A. Roy, E. Koesema and T. J. Kodadek, Angew. Chem., Int. Ed., 2021, 60, 11983–11990 CrossRef CAS PubMed.
- A. L. Satz, L. Kuai and X. Peng, Bioorg. Med. Chem. Lett., 2021, 39, 127851 CrossRef CAS PubMed.
- P. Dickson and T. Kodadek, Org. Biomol. Chem., 2019, 17, 4676–4688 RSC.
- S. Melkko, J. Scheuermann, C. E. Dumelin and D. Neri, Nat. Biotechnol., 2004, 22, 568–574 CrossRef CAS PubMed.
- Z. J. Gartner, N. T. Brian, R. Grubina, J. B. Doyon, T. M. Snyder and D. R. Liu, Science, 2004, 305, 1601–1605 CrossRef CAS PubMed.
- D. R. Halpin and P. B. Harbury, PLoS Biol., 2004, 2, 1015–1021 CAS.
- F. Debaene, L. Mejias, J. L. Harris and N. Winssinger, Tetrahedron, 2004, 60, 8677–8690 CrossRef CAS.
- M. A. Clark, R. A. Acharya, C. C. Arico-Muendel, S. L. Belyanskaya, D. R. Benjamin, N. R. Carlson, P. A. Centrella, C. H. Chiu, S. P. Creaser, J. W. Cuozzo, C. P. Davie, Y. Ding, G. J. Franklin, K. D. Franzen, M. L. Gefter, S. P. Hale, N. J. V. Hansen, D. I. Israel, J. W. Jiang, M. J. Kavarana, M. S. Kelley, C. S. Kollmann, F. Li, K. Lind, S. Mataruse, P. F. Medeiros, J. A. Messer, P. Myers, H. O'Keefe, M. C. Oliff, C. E. Rise, A. L. Satz, S. R. Skinner, J. L. Svendsen, L. J. Tang, K. van Vloten, R. W. Wagner, G. Yao, B. G. Zhao and B. A. Morgan, Nat. Chem. Biol., 2009, 5, 647–654 CrossRef CAS PubMed.
- M. Wichert, N. Krall, W. Decurtins, R. M. Franzini, F. Pretto, P. Schneider, D. Neri and J. Scheuermann, Nat. Chem., 2015, 7, 241–249 CrossRef CAS PubMed.
- J. P. Daguer, C. Zambaldo, M. Ciobanu, P. Morieux, S. Barluenga and N. Winssinger, Chem. Sci., 2015, 6, 739–744 RSC.
- J. P. Daguer, M. Ciobanu, S. Alvarez, S. Barluenga and N. Winssinger, Chem. Sci., 2011, 2, 625–632 RSC.
- S. Barluenga, C. Zambaldo, H. A. Ioannidou, M. Ciobanu, P. Morieux, J. P. Daguer and N. Winssinger, Bioorg. Med. Chem. Lett., 2016, 26, 1080–1085 CrossRef CAS PubMed.
- F. V. Reddavide, W. Lin, S. Lehnert and Y. Zhang, Angew. Chem., Int. Ed., 2015, 54, 7924–7928 CrossRef CAS PubMed.
- F. V. Reddavide, M. Cui, W. Lin, N. Fu, S. Heiden, H. Andrade, M. Thompson and Y. Zhang, Chem. Commun., 2019, 55, 3753–3756 RSC.
- G. Li, W. Zheng, Z. Chen, Y. Zhou, Y. Liu, J. Yang, Y. Huang and X. Li, Chem. Sci., 2015, 6, 7097–7104 RSC.
- Y. Zhou, C. Li, J. Peng, L. Xie, L. Meng, Q. Li, J. Zhang, X. D. Li, X. Li, X. Huang and X. Li, J. Am. Chem. Soc., 2018, 140, 15859–15867 CrossRef CAS PubMed.
- Y. Deng, J. Peng, F. Xiong, Y. Song, Y. Zhou, J. Zhang, F. S. Lam, C. Xie, W. Shen, Y. Huang, L. Meng and X. Li, Angew. Chem., Int. Ed., 2020, 59, 14965–14972 CrossRef CAS PubMed.
- L. Farrera-Soler, J. P. Daguer, P. Raunft, S. Barluenga, A. Imberty and N. Winssinger, Bioorg. Med. Chem., 2020, 28, 115458 CrossRef CAS PubMed.
- T. Machida, A. Novoa, E. Gillon, S. Zheng, J. Claudinon, T. Eierhoff, A. Imberty, W. Romer and N. Winssinger, Angew. Chem., Int. Ed., 2017, 56, 6762–6766 CrossRef CAS PubMed.
- E. Lenci, L. Baldini and A. Trabocchi, Bioorg. Med. Chem., 2021, 41, 116218 CrossRef CAS PubMed.
- C. J. Gerry, M. J. Wawer, P. A. Clemons and S. L. Schreiber, J. Am. Chem. Soc., 2019, 141, 10225–10235 CrossRef CAS PubMed.
- L. Guasch, M. Reutlinger, D. Stoffler and M. Wichert, Chimia, 2021, 75, 105–107 CrossRef CAS PubMed.
- A. Martin, C. A. Nicolaou and M. A. Toledo, Commun. Chem., 2020, 3, 127 CrossRef CAS.
- K. McCloskey, E. A. Sigel, S. Kearnes, L. Xue, X. Tian, D. Moccia, D. Gikunju, S. Bazzaz, B. Chan, M. A. Clark, J. W. Cuozzo, M. A. Guie, J. P. Guilinger, C. Huguet, C. D. Hupp, A. D. Keefe, C. J. Mulhern, Y. Zhang and P. Riley, J. Med. Chem., 2020, 63, 8857–8866 CrossRef CAS PubMed.
- A. B. MacConnell, P. J. McEnaney, V. J. Cavett and B. M. Paegel, ACS Comb. Sci., 2015, 17, 518–534 CrossRef CAS PubMed.
- J. Bobers, M. K. Skopic, R. Dinter, P. Sakthithasan, L. Neukirch, C. Gramse, R. Weberskirch, A. Brunschweiger and N. Kockmann, ACS Comb. Sci., 2020, 22, 101–108 CrossRef CAS PubMed.
- J. Castanon, J. P. Roman, T. C. Jessop, J. de Blas and R. Haro, SLAS Discovery, 2018, 23, 387–396 CrossRef CAS PubMed.
- S. Chamakuri, S. Lu, M. N. Ucisik, K. M. Bohren, Y. C. Chen, H. C. Du, J. C. Faver, R. Jimmidi, F. Li, J. Y. Li, P. Nyshadham, S. S. Palmer, J. Pollet, X. Qin, S. E. Ronca, B. Sankaran, K. L. Sharma, Z. Tan, L. Versteeg, Z. Yu, M. M. Matzuk, T. Palzkill and D. W. Young, Proc. Natl. Acad. Sci. U. S. A., 2021, 118, e2111172118 CrossRef CAS PubMed.
- W. Decurtins, M. Wichert, R. M. Franzini, F. Buller, M. A. Stravs, Y. Zhang, D. Neri and J. Scheuermann, Nat. Protoc., 2016, 11, 764–780 CrossRef CAS PubMed.
- Y. Li, G. Zimmermann, J. Scheuermann and D. Neri, ChemBioChem, 2017, 18, 848–852 CrossRef CAS PubMed.
- A. Sannino, E. Gabriele, M. Bigatti, S. Mulatto, J. Piazzi, J. Scheuermann, D. Neri, E. J. Donckele and F. Samain, ChemBioChem, 2019, 20, 955–962 CrossRef CAS PubMed.
- K. E. Denton and C. J. Krusemark, MedChemComm, 2016, 7, 2020–2027 RSC.
- K. E. Denton, S. Wang, M. C. Gignac, N. Milosevich, F. Hof, E. C. Dykhuizen and C. J. Krusemark, SLAS Discovery, 2018, 23, 417–428 CrossRef CAS PubMed.
- A. Sannino, A. Gironda-Martinez, E. M. D. Gorre, L. Prati, J. Piazzi, J. Scheuermann, D. Neri, E. J. Donckele and F. Samain, ACS Comb. Sci., 2020, 22, 204–212 CrossRef CAS PubMed.
- Q. Chen, X. Cheng, L. Zhang, X. Li, P. Chen, J. Liu, L. Zhang, H. Wei, Z. Li and D. Dou, SLAS Discovery, 2020, 25, 523–529 CrossRef PubMed.
- S. Otto, R. L. Furlan and J. K. Sanders, Drug Discovery Today, 2002, 7, 117–125 CrossRef CAS PubMed.
- O. Ramstrom and J. M. Lehn, Nat. Rev. Drug Discovery, 2002, 1, 26–36 CrossRef CAS PubMed.
- P. T. Corbett, J. Leclaire, L. Vial, K. R. West, J. L. Wietor, J. K. Sanders and S. Otto, Chem. Rev., 2006, 106, 3652–3711 CrossRef CAS PubMed.
- J. M. Lehn, Chem. Soc. Rev., 2007, 36, 151–160 RSC.
- S. Ladame, Org. Biomol. Chem., 2008, 6, 219–226 RSC.
- M. F. Schmidt and J. Rademann, Trends Biotechnol., 2009, 27, 512–521 CrossRef CAS PubMed.
- R. A. Hunt and S. Otto, Chem. Commun., 2011, 47, 847–858 RSC.
- F. B. Cougnon and J. K. Sanders, Acc. Chem. Res., 2012, 45, 2211–2221 CrossRef CAS PubMed.
- Y. Jin, C. Yu, R. J. Denman and W. Zhang, Chem. Soc. Rev., 2013, 42, 6634–6654 RSC.
- J. Li, P. Nowak and S. Otto, J. Am. Chem. Soc., 2013, 135, 9222–9239 CrossRef CAS PubMed.
- A. Herrmann, Chem. Soc. Rev., 2014, 43, 1899–1933 RSC.
- Q. Ji, R. C. Lirag and O. S. Miljanic, Chem. Soc. Rev., 2014, 43, 1873–1884 RSC.
- M. Mondal and A. K. Hirsch, Chem. Soc. Rev., 2015, 44, 2455–2488 RSC.
- J. M. Lehn, Angew. Chem., Int. Ed., 2015, 54, 3276–3289 CrossRef CAS PubMed.
- A. Canal-Martin and R. Perez-Fernandez, ACS Omega, 2020, 5, 26307–26315 CrossRef CAS PubMed.
- D. A. Erlanson, A. C. Braisted, D. R. Raphael, M. Randal, R. M. Stroud, E. M. Gordon and J. A. Wells, Proc. Natl. Acad. Sci. U. S. A., 2000, 97, 9367–9372 CrossRef CAS PubMed.
- D. A. Erlanson, J. W. Lam, C. Wiesmann, T. N. Luong, R. L. Simmons, W. L. DeLano, I. C. Choong, M. T. Burdett, W. M. Flanagan, D. Lee, E. M. Gordon and T. O'Brien, Nat. Biotechnol., 2003, 21, 308–314 CrossRef CAS PubMed.
- B. R. McNaughton and B. L. Miller, Org. Lett., 2006, 8, 1803–1806 CrossRef CAS PubMed.
- R. F. Ludlow and S. Otto, J. Am. Chem. Soc., 2008, 130, 12218–12219 CrossRef CAS PubMed.
- B. R. McNaughton, P. C. Gareiss and B. L. Miller, J. Am. Chem. Soc., 2007, 129, 11306–11307 CrossRef CAS PubMed.
- P. C. Gareiss, K. Sobczak, B. R. McNaughton, P. B. Palde, C. A. Thornton and B. L. Miller, J. Am. Chem. Soc., 2008, 130, 16254–16261 CrossRef CAS PubMed.
- D. A. Erlanson, S. W. Fesik, R. E. Hubbard, W. Jahnke and H. Jhoti, Nat. Rev. Drug Discovery, 2016, 15, 605–619 CrossRef CAS PubMed.
- R. Huang and I. K. Leung, Molecules, 2016, 21, 910 CrossRef PubMed.
- V. T. Bhat, A. M. Caniard, T. Luksch, R. Brenk, D. J. Campopiano and M. F. Greaney, Nat. Chem., 2010, 2, 490–497 CrossRef CAS PubMed.
- I. Huc and J. M. Lehn, Proc. Natl. Acad. Sci. U. S. A., 1997, 94, 2106–2110 CrossRef CAS PubMed.
- S. Zameo, B. Vauzeilles and J. M. Beau, Angew. Chem., Int. Ed., 2005, 44, 965–969 CrossRef CAS PubMed.
- B. Shi, R. Stevenson, D. J. Campopiano and M. F. Greaney, J. Am. Chem. Soc., 2006, 128, 8459–8467 CrossRef CAS PubMed.
- O. Ramström and J. M. Lehn, ChemBioChem, 2000, 1, 41–48 CrossRef.
- M. Demetriades, I. K. Leung, R. Chowdhury, M. C. Chan, M. A. McDonough, K. K. Yeoh, Y. M. Tian, T. D. Claridge, P. J. Ratcliffe and E. C. Woon, Angew. Chem., Int. Ed., 2012, 51, 6672–6675 CrossRef CAS PubMed.
- R. Larsson, Z. Pei and O. Ramstrom, Angew. Chem., Int. Ed., 2004, 43, 3716–3718 CrossRef CAS PubMed.
- S. M. Voshell, S. J. Lee and M. R. Gagne, J. Am. Chem. Soc., 2006, 128, 12422–12423 CrossRef CAS PubMed.
- A. Buryak and K. Severin, Angew. Chem., Int. Ed., 2005, 44, 7935–7938 CrossRef CAS PubMed.
- F. V. Reddavide, M. Thompson, L. Mannocci and Y. X. Zhang, Aldrichimica Acta, 2019, 52, 63–74 CAS.
- S. Melkko, Y. Zhang, C. E. Dumelin, J. Scheuermann and D. Neri, Angew. Chem., Int. Ed., 2007, 46, 4671–4674 CrossRef CAS PubMed.
- J. Scheuermann, C. E. Dumelin, S. Melkko, Y. X. Zhang, L. Mannocci, M. Jaggi, J. Sobek and D. Neri, Bioconjugate Chem., 2008, 19, 778–785 CrossRef CAS PubMed.
- G. Zimmermann, U. Rieder, D. Bajic, S. Vanetti, A. Chaikuad, S. Knapp, J. Scheuermann, M. Mattarella and D. Neri, Chem. – Eur. J., 2017, 23, 8152–8155 CrossRef CAS PubMed.
- M. Catalano, M. Moroglu, P. Balbi, F. Mazzieri, J. Clayton, K. H. Andrews, M. Bigatti, J. Scheuermann, S. J. Conway and D. Neri, ChemMedChem, 2020, 15, 1752–1756 CrossRef CAS PubMed.
- S. Oehler, M. Catalano, I. Scapozza, M. Bigatti, G. Bassi, N. Favalli, M. R. Mortensen, F. Samain, J. Scheuermann and D. Neri, Chem. – Eur. J., 2021, 27, 8985–8993 CrossRef CAS PubMed.
- S. Oehler, L. Plais, G. Bassi, D. Neri and J. Scheuermann, Chem. Commun., 2021, 57, 12289–12292 RSC.
- M. Catalano, G. Bassi, G. Rotondi, L. Khettabi, M. Dichiara, P. Murer, J. Scheuermann, M. Soler-Lopez and D. Neri, RSC Med. Chem., 2021, 12, 363–369 RSC.
- J. Scheuermann and D. Neri, Curr. Opin. Chem. Biol., 2015, 26, 99–103 CrossRef CAS PubMed.
- J. C. Faver, K. Riehle, D. R. Lancia, Jr., J. B. J. Milbank, C. S. Kollmann, N. Simmons, Z. Yu and M. M. Matzuk, ACS Comb. Sci., 2019, 21, 75–82 CrossRef CAS PubMed.
- P. Komar and M. Kalinic, ACS Comb. Sci., 2020, 22, 410–421 CrossRef CAS PubMed.
- L. Kuai, T. O'Keeffe and C. Arico-Muendel, SLAS Discovery, 2018, 23, 405–416 CrossRef CAS PubMed.
- N. Winssinger and J. L. Harris, Expert Rev. Proteomics, 2005, 2, 937–947 CrossRef CAS PubMed.
- Z. L. Pianowski and N. Winssinger, Chem. Soc. Rev., 2008, 37, 1330–1336 RSC.
- K. K. Sadhu and N. Winssinger, Chimia, 2013, 67, 905–909 CrossRef CAS PubMed.
- J. L. Harris and N. Winssinger, Chem. – Eur. J., 2005, 11, 6792–6801 CrossRef CAS PubMed.
- S. Barluenga and N. Winssinger, Acc. Chem. Res., 2015, 48, 1319–1331 CrossRef CAS PubMed.
- C. Zambaldo, S. Barluenga and N. Winssinger, Curr. Opin. Chem. Biol., 2015, 26, 8–15 CrossRef CAS PubMed.
- M. Ciobanu, K. T. Huang, J. P. Daguer, S. Barluenga, O. Chaloin, E. Schaeffer, C. G. Mueller, D. A. Mitchell and N. Winssinger, Chem. Commun., 2011, 47, 9321–9323 RSC.
- J. Scheuermann, C. E. Dumelin, S. Melkko and D. Neri, J. Biotechnol., 2006, 126, 568–581 CrossRef CAS PubMed.
- S. Melkko, C. E. Dumelin, J. Scheuermann and D. Neri, Drug Discovery Today, 2007, 12, 465–471 CrossRef CAS PubMed.
- J. Cai, B. A. Rosenzweig and A. D. Hamilton, Chem. – Eur. J., 2009, 15, 328–332 CrossRef CAS PubMed.
- D. M. Tagore, K. I. Sprinz, S. Fletcher, J. Jayawickramarajah and A. D. Hamilton, Angew. Chem., Int. Ed., 2007, 46, 223–225 CrossRef CAS PubMed.
- D. Margulies and A. D. Hamilton, J. Am. Chem. Soc., 2009, 131, 9142–9143 CrossRef CAS PubMed.
- D. Margulies and A. D. Hamilton, Angew. Chem., Int. Ed., 2009, 48, 1771–1774 CrossRef CAS PubMed.
- J. Cai, E. M. Shapiro and A. D. Hamilton, Bioconjugate Chem., 2009, 20, 205–208 CrossRef CAS PubMed.
- B. A. Rosenzweig, N. T. Ross, D. M. Tagore, J. Jayawickramarajah, I. Saraogi and A. D. Hamilton, J. Am. Chem. Soc., 2009, 131, 5020–5021 CrossRef CAS PubMed.
- K. I. Sprinz, D. M. Tagore and A. D. Hamilton, Bioorg. Med. Chem. Lett., 2005, 15, 3908–3911 CrossRef CAS PubMed.
- S. Grunzner, F. V. Reddavide, C. Steinfelder, M. Cui, M. Busek, U. Klotzbach, Y. Zhang and F. Sonntag, Microfluidics, Biomems, and Medical Microsystems XV, 2017, 10061.
- P. Zhao, Z. Chen, Y. Li, D. Sun, Y. Gao, Y. Huang and X. Li, Angew. Chem., Int. Ed., 2014, 53, 10056–10059 CrossRef CAS PubMed.
- B. Shi and M. F. Greaney, Chem. Commun., 2005, 886–888 RSC.
- S. Ladame, A. M. Whitney and S. Balasubramanian, Angew. Chem., Int. Ed., 2005, 44, 5736–5739 CrossRef CAS PubMed.
- L. M. McGregor, T. Jain and D. R. Liu, J. Am. Chem. Soc., 2014, 136, 3264–3270 CrossRef CAS PubMed.
- L. M. McGregor, D. J. Gorin, C. E. Dumelin and D. R. Liu, J. Am. Chem. Soc., 2010, 132, 15522–15524 CrossRef CAS PubMed.
- M. Mondal, N. Radeva, H. Koster, A. Park, C. Potamitis, M. Zervou, G. Klebe and A. K. Hirsch, Angew. Chem., Int. Ed., 2014, 53, 3259–3263 CrossRef CAS PubMed.
- M. Demetriades, I. K. H. Leung, R. Chowdhury, M. C. Chan, M. A. McDonough, K. K. Yeoh, Y.-M. Tian, T. D. W. Claridge, P. J. Ratcliffe, E. C. Y. Woon and C. J. Schofield, Angew. Chem., Int. Ed., 2012, 51, 6672–6675 CrossRef CAS PubMed.
- D. E. Scott, G. J. Dawes, M. Ando, C. Abell and A. Ciulli, ChemBioChem, 2009, 10, 2772–2779 CrossRef CAS PubMed.
- H. Kashida, T. Doi, T. Sakakibara, T. Hayashi and H. Asanuma, J. Am. Chem. Soc., 2013, 135, 7960–7966 CrossRef CAS PubMed.
- A. Litovchick, M. A. Clark and A. D. Keefe, Artif DNA PNA XNA, 2014, 5, e27896 CrossRef PubMed.
- Y. Zhou, W. Shen, J. Peng, Y. Deng and X. Li, Bioorg. Med. Chem., 2021, 45, 116328 CrossRef CAS PubMed.
- Y. Zhou, J. Peng, W. Shen and X. Li, Biochem. Biophys. Res. Commun., 2020, 533, 215–222 CrossRef CAS PubMed.
- D. Neri and R. A. Lerner, Annu. Rev. Biochem., 2018, 87, 479–502 CrossRef CAS PubMed.
- C. W. Murray and D. C. Rees, Nat. Chem., 2009, 1, 187–192 CrossRef CAS PubMed.
- D. A. Erlanson, Top. Curr. Chem., 2012, 317, 1–32 CAS.
- D. A. Erlanson, J. A. Wells and A. C. Braisted, Annu. Rev. Biophys. Biomol. Struct., 2004, 33, 199–223 CrossRef CAS PubMed.
- R. H. Nonoo, A. Armstrong and D. J. Mann, ChemMedChem, 2012, 7, 2082–2086 CrossRef CAS PubMed.
- W. P. Jencks, Proc. Natl. Acad. Sci. U. S. A., 1981, 78, 4046–4050 CrossRef CAS PubMed.
- G. G. Ferenczy and G. M. Keseru, MedChemComm, 2016, 7, 332–337 RSC.
- G. G. Ferenczy and G. M. Keseru, J. Chem. Inf. Model., 2012, 52, 1039–1045 CrossRef CAS PubMed.
- S. Melkko, C. E. Dumelin, J. Scheuermann and D. Neri, Chem. Biol., 2006, 13, 225–231 CrossRef CAS PubMed.
- M. Bigatti, A. Dal Corso, S. Vanetti, S. Cazzamalli, U. Rieder, J. Scheuermann, D. Neri and F. Sladojevich, ChemMedChem, 2017, 12, 1748–1752 CrossRef CAS PubMed.
- S. Chung, J. B. Parker, M. Bianchet, L. M. Amzel and J. T. Stivers, Nat. Chem. Biol., 2009, 5, 407–413 CrossRef CAS PubMed.
- A. W. Hung, H. L. Silvestre, S. Wen, A. Ciulli, T. L. Blundell and C. Abell, Angew. Chem., Int. Ed., 2009, 48, 8452–8456 CrossRef CAS PubMed.
- G. Zimmermann, U. Rieder, D. Bajic, S. Vanetti, A. Chaikuad, S. Knapp, J. Scheuermann, M. Mattarella and D. Neri, Chem. – Eur. J., 2017, 23, 8152–8155 CrossRef CAS PubMed.
- F. Abendroth, A. Bujotzek, M. Shan, R. Haag, M. Weber and O. Seitz, Angew. Chem., Int. Ed., 2011, 50, 8592–8596 CrossRef CAS PubMed.
- M. Bigatti, A. Dal Corso, S. Vanetti, S. Cazzamalli, U. Rieder, J. Scheuermann, D. Neri and F. Sladojevich, ChemMedChem, 2017, 12, 1748–1752 CrossRef CAS PubMed.
- M. Jaegle, E. L. Wong, C. Tauber, E. Nawrotzky, C. Arkona and J. Rademann, Angew. Chem., Int. Ed., 2017, 56, 7358–7378 CrossRef CAS PubMed.
- C. L. Tung, C. T. Wong, E. Y. Fung and X. Li, Org. Lett., 2016, 18, 2600–2603 CrossRef CAS PubMed.
- R. M. Franzini, T. Ekblad, N. Zhong, M. Wichert, W. Decurtins, A. Nauer, M. Zimmermann, F. Samain, J. Scheuermann, P. J. Brown, J. Hall, S. Graslund, H. Schuler and D. Neri, Angew. Chem., Int. Ed., 2015, 54, 3927–3931 CrossRef CAS PubMed.
- F. Diezmann and O. Seitz, Chem. Soc. Rev., 2011, 40, 5789–5801 RSC.
- B. R. Vummidi, L. Farrera-Soler, J. P. Daguer, M. Dockerill, S. Barluenga and N. Winssinger, Nat. Chem., 2021, 1–12 Search PubMed.
- M. T. Cancilla, M. M. He, N. Viswanathan, R. L. Simmons, M. Taylor, A. D. Fung, K. Cao and D. A. Erlanson, Bioorg. Med. Chem. Lett., 2008, 18, 3978–3981 CrossRef CAS PubMed.
- L. K. Petersen, A. B. Christensen, J. Andersen, C. G. Folkesson, O. Kristensen, C. Andersen, A. Alzu, F. A. Slok, P. Blakskjaer, D. Madsen, C. Azevedo, I. Micco and N. J. V. Hansen, J. Am. Chem. Soc., 2021, 143, 2751–2756 CrossRef CAS PubMed.
- C. O. Mellet, J. F. Nierengarten and J. M. G. Fernandez, J. Mater. Chem. B, 2017, 5, 6428–6436 RSC.
- A. L. Satz, R. Hochstrasser and A. C. Petersen, ACS Comb. Sci., 2017, 19, 234–238 CrossRef CAS PubMed.
- D. Bosc, V. Camberlein, R. Gealageas, O. Castillo-Aguilera, B. Deprez and R. Deprez-Poulain, J. Med. Chem., 2020, 63, 3817–3833 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2022 |