Open Access Article
Open Access ArticleStructural insights into inhibition of the drug target dihydroorotate dehydrogenase by bacterial hydroxyalkylquinolines†
Samantha M.
Horwitz
 a,
Tamra C.
Blue
a,
Joseph A.
Ambarian
a,
Tamra C.
Blue
a,
Joseph A.
Ambarian
 a,
Shotaro
Hoshino
b,
Mohammad R.
Seyedsayamdost
a,
Shotaro
Hoshino
b,
Mohammad R.
Seyedsayamdost
 b and
Katherine M.
Davis
b and
Katherine M.
Davis
 *a
*a
aDepartment of Chemistry, Emory University, Atlanta, GA 30322, USA. E-mail: katherine.davis@emory.edu
bDepartment of Chemistry, Princeton University, Princeton, NJ 08544, USA
First published on 7th February 2022
Abstract
Hydroxyalkylquinolines (HAQs) are ubiquitious natural products but their interactions with associated protein targets remain elusive. We report X-ray crystal structures of two HAQs in complex with dihydroorotate dehydrogenase (DHODH). Our results reveal the structural basis of DHODH inhibition by HAQs and open the door to downstream structure–activity relationship studies.
Introduction
Natural products have provided a powerful foundation from which to develop novel therapeutics. In the past 40 years, more than half of the medicines approved by the US Food and Drug Administration were derived from this group of compounds.1,2 Despite their ubiquity, however, obtaining a holistic understanding of their respective mechanisms of action is challenging, due in part to the scarcity of target-bound natural product structures. As drug development often relies on pointed modifications to conserved chemical features, visualizing how these moieties directly interact with their targets is important for understanding and improving a candidate compound's therapeutic effectiveness.3,4 Hydroxyalkylquinolines (HAQs) are one class of natural products that have been studied extensively due to their marked antimicrobial activity. Their growth-inhibitory effect on Gram-positive bacteria was first noted in 1945 in supernatants of Pseudomonas aeruginosa, an observation that later helped explain why P. aeruginosa is able to outcompete Staphylococcus aureus during cystic fibrosis lung infections.5,6 The quinoline substructure in HAQs is a privileged scaffold as it can bind to different macromolecular targets, and small modifications have strong effects on biological activity. Fluoroquinolones, for example, are successful clinical antibiotics that inhibit DNA replication by interfering with DNA gyrase and topoisomerase.7 HAQs, by contrast, act as intraspecies quorum sensing signals and interspecies antibiotics that target energy production and DNA biosynthesis.5,7,8 Given their effects on diverse cellular pathways, there is broad interest in elucidating HAQ mechanisms of action.Although HAQs from P. aeruginosa may be most prominent, similar molecules are generated by a range of proteobacteria. These include Burkholderia thailandensis, which encodes a biosynthetic gene cluster for a subfamily of HAQs that predominantly contain a 3-methyl substituent.9,10 Two B. thailandensis-derived HAQs, 4-hydroxy-3-methyl-2-(2-nonenyl)-quinoline (HMNQ) and 2-heptyl-4-(1H)-quinolone N-oxide (HQNO), are of particular interest, as they were recently shown to exhibit potent synergistic and broad spectrum antibiosis originating from their dual modes of action (Fig. 1A).11 Each congener blocks different steps in energy production, with HMNQ dissipating the proton motive force and HQNO inhibiting the cytochrome bc1 complex.11–13 Both compounds also interfere with de novo pyrimidine biosynthesis by inhibiting dihydroorotate dehydrogenase (DHODH). However, the mechanism by which these disparate HAQs inhibit DHODH is not known, as the structural basis for their interaction with the enzyme has not been determined.
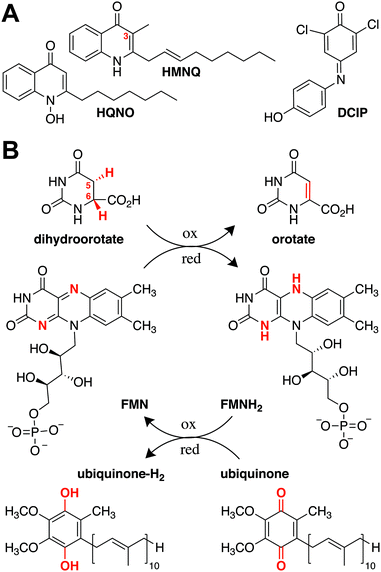 | ||
| Fig. 1 (A) Structures of the natural products HMNQ, HQNO, as well as the common ubiquinone surrogate DCIP. (B) Reactions catalyzed by Class II DHODHs. | ||
DHODHs are ubiquitous enzymes known to catalyze the flavin mononucleotide (FMN)-mediated oxidation of dihydroorotate (DHO) to orotate (ORO) (Fig. 1B).14 They are divided into two classes according to the mechanism by which the FMN prosthetic group is regenerated, with Class I proteins generally found in Gram-positive bacteria and Class II proteins in Gram-negative bacteria, as well as most eukaryotes.15 The latter utilize ubiquinone as a terminal electron acceptor to re-oxidize the catalytically reduced flavin and promote product dissociation (Fig. 1B).16,17 Although both Class I and Class II DHODHs fulfill the same functional role within a cell, they differ in sequence and structure. For example, in addition to the α/β barrel that is conserved across both classes, Class II DHODHs are characterized by a canonical N-terminal extension containing two α-helices, α1 and α2, responsible for associating the enzyme with the membrane.18 It has been suggested that ubiquinone binds at the end of the hydrophobic tunnel between α1 and α2, which terminates near the FMN binding site in the catalytic barrel.18–20
Previous enzymatic assays with the common ubiquinone surrogate 2,6-dichloroindophenol (DCIP) suggest that the basis for HMNQ and HQNO's DHODH-associated activity lies in competitive inhibition of the ubiquinone binding site rather than that of substrate DHO.11 However, this has not been directly demonstrated, nor do we have a firm understanding of how the native quinone interacts with Class II DHODHs. Despite the focus on inhibitors that compete with ubiquinone binding in the Class II human DHODH, which is a drug target for anticancer, immunosuppressive, and antimalarial agents,18,21–23 no structures visualizing the DHODH·ubiquinone complex (or any ubiquinone surrogate complexes) have been solved to date, hindering interpretation of inhibitor binding modes across Class II enzymes. Accordingly, we sought to probe the basis for Escherichia coli DHODH inhibition by HAQs, while simultaneously confirming the ubiquinone binding site via crystallographic characterization of the enzyme in complex with HMNQ and HQNO (Fig. S1, ESI†), and with DCIP for comparison.
Results
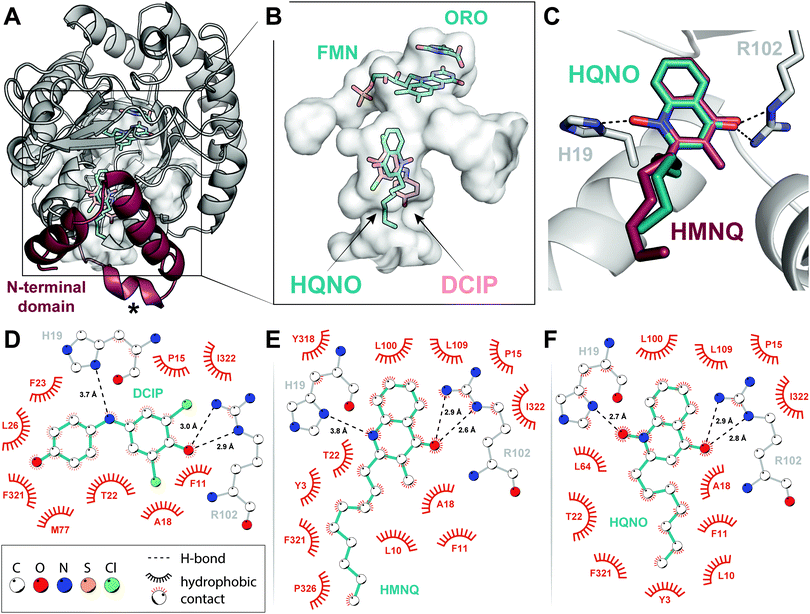
In addition to an improved model for the DHODH holo-enzyme, we herein report X-ray crystal structures of DHODH bound to the quinone surrogate DCIP (2.53 Å), HMNQ (2.63 Å), and HQNO (2.25 Å), respectively. Electron density for all three ligands—DCIP, HMNQ and HQNO—as well as for the prosthetic group FMN and the product ORO was well-defined in each structure (Fig. S2, ESI†), allowing for unambiguous assignment of ligand-enzyme interactions. We initiated our studies with a concerted effort to obtain a ubiquinone-bound DHODH structure, but low solubility of the hydrophobic quinone and its analogues in aqueous solution proved insurmountable. We therefore crystallized DHODH without added ligands, as well as with DCIP, HMNQ, and HQNO, respectively, and solved their structures using molecular replacement (see ESI†).Global structures and FMN/ORO binding
All models overlay well both with each other and with the previously determined structure of E. coli DHODH (Protein Data Bank accession code 1F76), having root-mean-square deviations (rmsds) of the peptide backbone <0.49 Å over 335 Cα atoms. In particular, the catalytic domain containing cofactor and substrate binding sites forms a canonical α/β barrel that is extended by a series of α-helices at the N-terminus (Fig. 2A). The binding modes of FMN and ORO within the primary fold were likewise conserved. The isoalloxazine ring of FMN is stabilized via H-bonding interactions with Lys66, Thr86, Asn172, Lys217, and Asn139, while the phosphate head group makes close contacts with Gly268, Gly297, Tyr318, and Ser319 (Fig. S3A, ESI†). ORO associates with FMN through π–π stacking interactions (Fig. 2B and Fig. S3B, ESI†) and is held in the DHODH binding pocket via a network of H-bonds with the heteroatoms of four asparagines (Asn111, Asn172, Asn177 and Asn246), Thr247 and the amide groups of Gly114 and Phe115 (Fig. S3C, ESI†). This tight system of H-bonding interactions is predicted to persist until re-oxidation of FMN by ubiquinone, allowing ORO to dissociate.13 After observing density for a pyrimidine-shaped ligand bound to the as-purified enzyme, we performed GC–MS extraction assays and confirmed its identity as ORO rather than DHO (Fig. S4, ESI†). These experiments suggest that Class II DHODHs purify with ORO bound. Furthermore, if ubiquinone is not present or is prevented from binding by an inhibitor, the oxidized product remains bound to the enzyme (Fig. S4, ESI†).16 Although subtle, this is a notable detail that may influence future structural and biochemical characterization of these enzymes.HMNQ/HQNO binding
As was biochemically predicted, comparison of the structures confirms that HMNQ and HQNO occupy the same hydrophobic binding pocket as DCIP. More specifically, these molecules are visualized within the N-terminal extension characteristic of Class II enzymes (Fig. 2A), at distances suitable for electron transfer to FMN.18,21,24 The observed HAQ binding pocket is not only consistent with this theory, but also aligns well with similarly classified inhibitors of the human enzyme.18,20All three ligands are anchored in the putative ubiquinone-binding tunnel via similar H-bonding interactions with the guanadino group of Arg102. These H-bonds help to position the DCIP ketone and quinolone carbonyl groups, respectively (Fig. 2C–F). HQNO is additionally secured through an electrostatic association between its N-oxide moiety and His19, stronger than those observed for the imine nitrogen of DCIP or the nitrogen of HMNQ (Fig. 2D–F). This key residue has been proposed to play a role in co-substrate binding via parallel displaced π-stacking with the quinone ring system.24 Furthermore, mutation of the analogous histidine in the human enzyme was demonstrated to reduce activity by a factor of 10 while dramatically impacting binding of brequinar and other similar inhibitors.25 Our structures, by contrast, depict the imidazole ring in an offset, but perpendicular orientation, unfavourable for π–π interactions (Fig. 2C). However, this arrangement is ideally poised to facilitate H-bonding interactions, the strength of which is modulated by ligand identity. The described polar contacts are further supplemented by numerous largely conserved hydrophobic interactions—Phe11, Pro15, Ala18, Thr22, Phe321, Ile322—both within the proximal redox site and throughout the associated tunnel (Fig. 2D–F). What discrepancies exist primarily arise due to the more rigid molecular geometry of the ubiquinone surrogate, which positions the phenol ‘tail’ toward the opposite side of the cavity compared to the more flexible hydrocarbon chains of the HAQs (Fig. 2B).
Docking studies
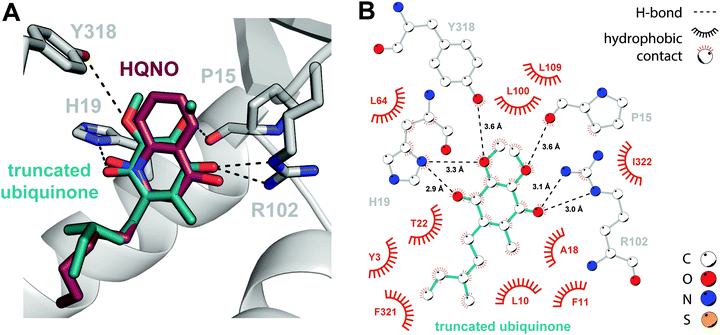
It is qualitatively obvious that HMNQ and HQNO resemble the native ubiquinone co-substrate, while DCIP is structurally distinct, despite its ability to serve as a functional replacement. Thus, in the absence of a ubiquinone-bound crystal structure, we proceeded to conduct a docking study using the SwissDock server for a more representative comparison.26 We first applied the docking protocol to HMNQ and HQNO, the results of which recapitulate crystallographically observed binding modes (Fig. S5, ESI†). Following validation of this approach, we attempted docking of ubiquinone. However, due to challenges associated with placing the isoprenoid tail, which is predicted to traverse the protein–membrane interface to interact with the lipid bilayer,27 a ubiquinone construct truncated in the alkyl chain was ultimately used instead.The resultant docking model arranges the co-substrate in a strikingly similar pose to that observed experimentally for the HAQs, in which the electron acceptor makes virtually identical interactions with the enzyme (Fig. 3). Additions are limited to weaker H-bonding interactions with the backbone carbonyl of Pro15 and the Tyr318 side chain. This high degree of similarity may begin to explain the unexpected consistency in side chain and loop conformations between the holo- and inhibitor-bound enzyme models, uncommon for synthetic inhibition of eukaryotic DHODHs,24,28 as the enzyme is already primed for ubiquinone-binding. The largest observed deviations occur in a flexible loop (residues 30–40, Fig. 2A) proposed to serve a gatekeeping role for the N-terminal tunnel. Although poor electron density in our structures does suggest flexibility in this region, shifts in the associated Cα positions are nonetheless modest, on the order of 1–1.5 Å, and the flanking α-helices retain their original positions upon inhibitor binding.
It is reasonable to suggest that the high degree of similarity between the observed DHODH–HAQ interactions and those predicted for the enzyme-ubiquinone complex contributes to the success of these natural products in preventing binding of the native co-substrate, thereby inhibiting regeneration of oxidized FMN. Their effectiveness can be further rationalized through the observation that they engage all five subsites associated with inhibition of Class II enzymes, namely: (i) the hydrophobic tunnel, (ii) Arg102, (iii) His19, (iv) Leu100 and Leu109, which form a hydrophobic cap, and (v) residues, such as Tyr3, at the tunnel opening.20 The degree of inhibition of the human enzyme has been correlated with broader subsite engagement, perhaps providing the mechanism by which HMNQ and HQNO outcompete DCIP. The previously determined Ki values for HMNQ (51 nM) and HQNO (239 nM)11 with E. coli DHODH are generally consistent with this rationale.
Conclusions
Collectively, the results described herein confirm the hypothesis that competition with ubiquinone is the basis for the inhibitory activity of HAQs on DHODH. The presented structural models further provide important insights into binding interactions that may aid future development of HMNQ and HQNO-based therapeutics that target Gram-negative bacteria. The demonstrated similarities between binding modes of these HAQs and the native electron acceptor may even hint at expanded utility for the inhibition of other Class II enzymes. This stands in contrast to many inhibitors of the human enzyme, which do not function effectively in their bacterial counterparts.HAQ biosynthesis is widespread, especially in Burkholderia spp. and pseudomonads. One of the conserved features is the production of dozens of HAQ variants from a single biosynthetic pathway.10,29–32 Because quinone-utilizing enzymes are common, especially in the electron transport chain and energy production pathways, the varied HAQ congeners could target a broad range of quinone binding sites. In doing so, the structurally divergent HAQs may interfere with important enzymes of diverse origins, thereby providing a competitive advantage for producing organisms. Consistent with this notion are inhibitory activities that have been observed by HAQs against algal and fungal strains.33,34 In these cases, structures of the HAQs in complex with their molecular targets are not yet available, providing further avenues at the intersection of structural biology and natural product chemistry.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the National Institutes of Health (Pathway to Independence Award 4R00GM129460 to K. M. D. and R01GM140034 to M. R. S.), the Beckman Young Investigator Program (to K. M. D.), Emory University start-up funds, and by a JSPS Overseas Research Fellowship (grant 201960049 to S. H.). GM/CA@APS has been funded by the National Cancer Institute (ACB-12002) and the National Institute of General Medical Sciences (AGM-12006, P30GM138396). This research used resources of the Advanced Photon Source, a U.S. Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE-AC02-06CH11357. The Eiger 16M detector at GM/CA-XSD was funded by NIH grant S10 OD012289.Notes and references
- T. Rodrigues, D. Reker, P. Schneider and G. Schneider, Nat. Chem., 2016, 8, 531–541 CrossRef CAS PubMed.
- D. J. Newman and G. M. Cragg, J. Nat. Prod., 2020, 83, 770–803 CrossRef CAS PubMed.
- Z. Guo, Acta Pharm. Sin. B, 2017, 7, 119–136 CrossRef PubMed.
- S. E. Rossiter, M. H. Fletcher and W. M. Wuest, Chem. Rev., 2017, 117, 12415–12474 CrossRef CAS PubMed.
- S. Heeb, M. P. Fletcher, S. R. Chhabra, S. P. Diggle, P. Williams and M. Cámara, FEMS Microbiol. Rev., 2011, 35, 247–274 CrossRef CAS PubMed.
- L. M. Filkins, J. A. Graber, D. G. Olson, E. L. Dolben, L. R. Lynd, S. Bhuju and G. A. O'Toole, J. Bacteriol., 2015, 197, 2252–2264 CrossRef CAS PubMed.
- A. Fàbrega, S. Madurga, E. Giralt and J. Vila, Microb. Biotechnol., 2009, 2, 40–61 CrossRef PubMed.
- E. C. Pesci, J. B. J. Milbank, J. P. Pearson, S. McKnight, A. S. Kende, E. P. Greenberg and B. H. Iglewski, Proc. Natl. Acad. Sci. U. S. A., 1999, 96, 11229–11234 CrossRef CAS PubMed.
- M. Piochon, P. M. L. Coulon, A. Caulet, M.-C. Groleau, E. Déziel and C. Gauthier, J. Nat. Prod., 2020, 83, 2145–2154 CrossRef CAS PubMed.
- L. Vial, F. Lépine, S. Milot, M. C. Groleau, V. Dekimpe, D. E. Woods and E. Déziel, J. Bacteriol., 2008, 190, 5339–5352 CrossRef CAS PubMed.
- Y. Wu and M. R. Seyedsayamdost, Cell Chem. Biol., 2017, 24, 1437–1444 CrossRef CAS PubMed.
- B. Hacker, B. Barquera, A. R. Crofts and R. B. Gennis, Biochemistry, 1993, 32, 4403–4410 CrossRef CAS PubMed.
- N. Howell, J. Appel, J. P. Cook, B. Howell and W. W. Hauswirth, J. Biol. Chem., 1987, 262, 2411–2414 CrossRef CAS.
- R. L. Fagan and B. A. Palfey, Biochemistry, 2009, 48, 7169–7178 CrossRef CAS PubMed.
- R. L. Fagan, M. N. Nelson, P. M. Pagano and B. A. Palfey, Biochemistry, 2006, 45, 14926–14932 CrossRef CAS PubMed.
- B. A. Palfey, O. Björnberg and K. F. Jensen, Biochemistry, 2001, 40, 4381–4390 CrossRef CAS PubMed.
- S. Nørager, K. F. Jensen, O. Björnberg and S. Larsen, Structure, 2002, 10, 1211–1223 CrossRef.
- S. Liu, E. A. Neidhardt, T. H. Grossman, T. Ocain and J. Clardy, Structure, 2000, 8, 25–33 CrossRef CAS PubMed.
- Y. Zhou, L. Tao, X. Zhou, Z. Zuo, J. Gong, X. Liu, Y. Zhou, C. Liu, N. Sang, H. Liu, J. Zou, K. Gou, X. Yang and Y. Zhao, Cancer Metab., 2021, 9, 22 CrossRef PubMed.
- R. Baumgartner, M. Walloschek, M. Kralik, A. Gotschlich, S. Tasler, J. Mies and J. Leban, J. Med. Chem., 2006, 49, 1239–1247 CrossRef CAS PubMed.
- J. Zhu, L. Han, Y. Diao, X. Ren, M. Xu, L. Xu, S. Li, Q. Li, D. Dong, J. Huang, X. Liu, Z. Zhao, R. Wang, L. Zhu, Y. Xu, X. Qian and H. Li, J. Med. Chem., 2015, 58, 1123–1139 CrossRef CAS PubMed.
- D. E. Hurt, A. E. Sutton and J. Clardy, Bioorg. Med. Chem. Lett., 2006, 16, 1610–1615 CrossRef CAS PubMed.
- V. Patel, M. Booker, M. Kramer, L. Ross, C. A. Celatka, L. M. Kennedy, J. D. Dvorin, M. T. Duraisingh, P. Sliz, D. F. Wirth and J. Clardy, J. Biol. Chem., 2008, 283, 35078–35085 CrossRef CAS PubMed.
- M. Hansen, J. Le Nours, E. Johansson, T. Antal, A. Ullrich, M. Löffler and S. Larsen, Protein Sci., 2004, 13, 1031–1042 CrossRef CAS PubMed.
- J. P. Davis and R. A. Copeland, Biochem. Pharmacol., 1997, 54, 459–465 CrossRef CAS PubMed.
- A. Grosdidier, V. Zoete and O. Michielin, Nucleic Acids Res., 2011, 39, W270–W277 CrossRef CAS PubMed.
- J. Costeira-Paulo, J. Gault, G. Popova, M. J. G. W. Ladds, I. M. M. van Leeuwen, M. Sarr, A. Olsson, D. P. Lane, S. Laín, E. G. Marklund and M. Landreh, Cell Chem. Biol., 2018, 25, 309–317 CrossRef CAS PubMed.
- B. Walse, V. T. Dufe, B. Svensson, I. Fritzson, L. Dahlberg, A. Khairoullina, U. Wellmar and S. Al-Karadaghi, Biochemistry, 2008, 47, 8929–8936 CrossRef CAS PubMed.
- E. Déziel, F. Lépine, S. Milot, J. He, M. N. Mindrinos, R. G. Tompkins and L. G. Rahme, Proc. Natl. Acad. Sci. U. S. A., 2004, 101, 1339–1344 CrossRef PubMed.
- S. P. Diggle, P. Lumjiaktase, F. Dipilato, K. Winzer, M. Kunakorn, D. A. Barrett, S. R. Chhabra, M. Cámara and P. Williams, Chem. Biol., 2006, 13, 701–710 CrossRef CAS PubMed.
- J. R. Klaus, P. M. L. Coulon, P. Koirala, M. R. Seyedsayamdost, E. Déziel and J. R. Chandler, J. Ind. Microbiol. Biotechnol., 2020, 47, 877–887 CrossRef CAS PubMed.
- B. K. Okada, Y. Wu, D. Mao, L. B. Bushin and M. R. Seyedsayamdost, ACS Chem. Biol., 2016, 11, 2124–2130 CrossRef CAS PubMed.
- E. L. Harvey, R. W. Deering, D. C. Rowley, A. El Gamal, M. Schorn, B. S. Moore, M. D. Johnson, T. J. Mincer and K. E. Whalen, Front. Microbiol., 2016, 7 Search PubMed.
- O. Kilani-Feki, G. Culioli, A. Ortalo-Magné, N. Zouari, Y. Blache and S. Jaoua, Curr. Microbiol., 2011, 62, 1490–1495 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Materials & methods, Table S1, Fig. S1–S5 (PDF). See DOI: 10.1039/d1cb00255d |
| This journal is © The Royal Society of Chemistry 2022 |