Open Access Article
Open Access ArticleTrapping and structural characterisation of a covalent intermediate in vitamin B6 biosynthesis catalysed by the Pdx1 PLP synthase†
Matthew J.
Rodrigues
 ab,
Nitai
Giri
ab,
Nitai
Giri
 c,
Antoine
Royant
de,
Yang
Zhang
f,
Rachel
Bolton
ab,
Gwyndaf
Evans
b,
Steve E.
Ealick
c,
Antoine
Royant
de,
Yang
Zhang
f,
Rachel
Bolton
ab,
Gwyndaf
Evans
b,
Steve E.
Ealick
 f,
Tadhg
Begley
c and
Ivo
Tews
f,
Tadhg
Begley
c and
Ivo
Tews
 *a
*a
aBiological Sciences, Institute for Life Sciences, University of Southampton, Southampton, SO17 1BJ, UK. E-mail: ivo.tews@soton.ac.uk
bDiamond Light Source, Harwell Science and Innovation Campus, Didcot, OX11 0DE, UK
cDepartment of Chemistry, Texas A&M University, College Station, TX 77843, USA
dUniversité Grenoble Alpes, CNRS, CEA, Institut de Biologie Structurale (IBS), CS 10090, Grenoble Cedex 9 38044, France
eEuropean Synchrotron Radiation Facility, CS 40220, Grenoble Cedex 9 38043, France
fDepartment of Chemistry and Chemical Biology, Cornell University, Ithaca, NY 14853, USA
First published on 25th October 2021
Abstract
The Pdx1 enzyme catalyses condensation of two carbohydrates and ammonia to form pyridoxal 5-phosphate (PLP) via an imine relay mechanism of carbonyl intermediates. The I333 intermediate characterised here using structural, UV-vis absorption spectroscopy and mass spectrometry analyses rationalises stereoselective deprotonation and subsequent substrate assisted phosphate elimination, central to PLP biosynthesis.
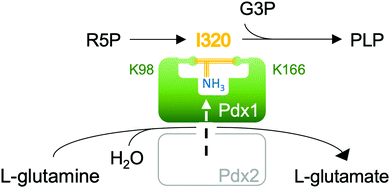
Pyridoxal 5-phosphate (10, PLP, vitamin B6) is an essential cofactor in all living systems where it plays a central role in the stabilisation of carbanions adjacent to amines primarily in amino acid metabolism.1 An enzyme complex consisting of Pdx1 and Pdx2 catalyses the conversion of glutamine, ribose 5-phosphate (R5P) and glyceraldehyde 3-phosphate (G3P) to PLP.2,3 Protein structures show two conserved phosphate binding sites in Pdx1, spaced 22 Å apart.4–6 During catalysis, the ribose phosphate group occupies the P1 phosphate binding site,5,7–9 while the product PLP phosphate group binds in P2.9,10 The Pdx1-associated glutaminase Pdx2 generates ammonia,2,3 which diffuses through an internal channel towards the Pdx1 P1 site4 where it then reacts with the covalent R5P adduct. This most intriguing catalytic step leads to formation of the double imine chromophoric intermediate I320 (Fig. 1).
In this communication we study the Arabidopsis Pdx1 enzyme; the plant has three pdx1 alleles,2,11 of which we studied Pdx1.3. Lys98 of Pdx1.3 was previously shown to form an imine with the R5P aldehyde,9,12 enabling a series of sugar modifications in the ribose tailoring site that culminate in the addition of Lys166 to generate I3209,12 (8). The Lys98/Lys166 crosslinking intermediate forms after phosphate elimination;13 however, the detailed mechanism is not understood. The vacated P1 site can then be occupied by the phosphate of the second substrate G3P, defining dual specificity for the Pdx1 active site.9 Incorporation of G3P leads to formation of PLP13–16 that is proposed to migrate to the PLP/P2 product site.9 Our previous study trapped an intermediate prior to phosphate elimination in the Lys166Arg exchange variant of the Pdx1.1 paralog (5LNT, see ESI†), suggested to form from R5P modification with ammonia.9 We now constructed the lysine/arginine exchange variant in the Pdx1.3 enzyme where structures of several other intermediates are available (reviewed in ESI†).9,12 We characterised the intermediate that formed after addition of R5P and ammonia in this variant. For its characteristic absorbance we named the adduct I333, determined its mass and demonstrated ammonia incorporation. The crystallographic structure suggests key catalytic residues for I333 formation, allowing us to propose a mechanism for deprotonation and phosphate elimination. The data rationalise how subsequent imine formation gives I320.
We used UV-vis absorption spectroscopy to characterise adducts formed by Pdx1.3 and its Pdx1.3K166R variant. R5P and (NH4)2SO4 were added to Pdx1.3 (10 mM and 200 mM final concentrations respectively) leading to accumulation of the chromophoric I320 intermediate over an hour at RT (8, λmax = 314 nm). Further addition of G3P (20 mM final concentration) leads to the formation of PLP in Pdx1.3 (λmax = 412 nm, Fig. 2). Pdx1.3K166R did not catalyse formation of the I320 intermediate (compare14); instead yielding a product with λmax = 333 nm (Fig. 2). Addition of G3P to the I333 complex did not lead to any significant changes in spectral signatures demonstrating that Pdx1.3K166R:I333 does not convert to PLP. We therefore conclude that Lys166 is required for I320 formation, consistent with recent structural proposals,9,12 and that I320 is a required intermediate for PLP biosynthesis.
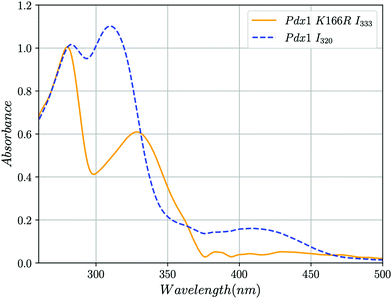 | ||
| Fig. 2 UV-vis absorption spectra showing the formation of the I320 and I333 intermediates in solution at RT. Blue dashed line, addition of R5P, ammonia and G3P to Pdx1.3 results in the formation of the chromophoric intermediate I320 (λmax = 315 nm) and PLP (λmax = 412 nm). Yellow line, addition of the same compounds to Pdx1.3K166R results in the formation of intermediate I333 (λmax = 333 nm) that does not convert to PLP after addition of G3P. The peak at λmax ∼ 280 nm represents protein. The discontinuity at 360 nm is caused by changeover of deuterium and halogen lamps. Spectra were smoothed and background correction for Rayleigh scattering was applied to account for protein aggregation as described in ESI.† | ||
To characterise intermediate formation by mass spectrometry, Pdx1.3 and Pdx1.3K166R were treated with substrates as indicated in Table 1. After tryptic digestion, the peptide mixtures were analysed by LC-ESI-TOF-MS. Comparison of the predicted and observed mass for the substrate free samples demonstrated that both Pdx1.3 and Pdx1.3K166R contained QAVTIPVMAK98, unmodified at lysine 98 ([M + H]+ = 1057.6 Da, Table 1 and Fig. S1, S2, ESI†). Addition of R5P to both samples leads to a mass difference of 212 Da, as seen in the samples of Pdx1.3 + R5P (Table 1 and Fig. S3, ESI†) and Pdx1.3K166R + R5P (Table 1 and Fig. S4, ESI†). For Pdx1.3K166R, the extra 16 Da on the Pdx1.3K166R peptide relative to Pdx1.3 is consistent with the oxidation of Met96 to its sulfoxide, which can occur during sample preparation.17 The observation of the 212 Da adduct (1) is consistent with earlier observations.3,15 From previous structural and MS analysis, it is known that I320 is formed after the addition of R5P and ammonia14,16 and crosslinks Lys98 and Lys166.9,12 In the present study, instead of trapping I320, treatment of Pdx1.3 with R5P and ammonia followed by trypsin gave unmodified QAVTIPVMAK and TK166GEAGTGNIIEAVR with a mass adduct of 96 Da (Table 1 and Fig. S5, ESI†), consistent with elimination of Lys98 from the I320 adduct to give (9). In the key experiment, treatment of Pdx1.3K166R with R5P and ammonia followed by trypsin gave the I333 modified QAVTIPVMAK98 peptide with a mass adduct of 194 Da (Table 1 and Fig. S6, ESI†). The Pdx1.3K166R TR166GEAGTGNIIEAVR peptide was not modified, demonstrating that it was not attached to I333. On the Bacillus subtilis Pdx1K149R variant (equivalent to Pdx1.3K166R) a 12-fold lower formation of I320 was reported,14 but the observation may indeed reflect formation of I333 in the bacterial enzyme.
| Subs | Pdx1.3 | Pdx1.3K166R |
|---|---|---|
| None | QAVTIPVMAK98 [M + H]+pred = 1057.6074 | QAVTIPVMAK98 [M + H]+pred = 1057.6074 |
| [M + H]+obs = 1057.6076 | [M + H]+obs = 1057.6085 | |
| +R5P | QAVTIPVMAK98AR [M + H]+pred = 1284.7457 | QAVTIPVM(O)AK98AR [M + H]+pred = 1300.7406 |
| [M + H]+obs = 1496.7546 | [M + H]+obs = 1512.7436 | |
| Modification (1) = 212.0089 | Modification (1) = 212.003 | |
| +R5P +NH3 | TK166GEAGTGNIIEAVR [M + H]+pred = 1515.8125 | QAVTIPVMAK98AR [M + H]+pred = 1284.7457 |
| [M + H]+obs = 1611.8326 | [M + H]+obs = 1478.7561 | |
| Modification (9) = 96.0201 | Modification (6) = 194.0104 |
The I333 adduct was further investigated by crystallographic analysis.‡ Equivalent to our earlier study of the I320 intermediate,9 we soaked Pdx1.3K166R crystals with R5P and (NH4)2SO4 where we used this protocol to generate the Pdx1.1K166R:preI320 complex (5LNT, see ESI†). UV-vis spectra were taken in crystallo18 to confirm formation of I333 (Fig. 3a) and corrected for scattering at the crystal-solvent/air interface (see ESI,†18,19). The observed absorbance at λmax = 333 nm was consistent with the in-solution data (Fig. 2). The crystal structure shows I333 as an adduct (6) between lysine 98 and C1 from R5P (Fig. 3b). Since the spectra indicated ammonia incorporation, the intermediate has been modelled with nitrogen in place of the oxygen at C2. This is consistent with previous NMR data showing that nitrogen incorporation precedes formation of the I320 intermediate,15 and is also consistent with the mass spectrometry analysis presented here (Table 1).
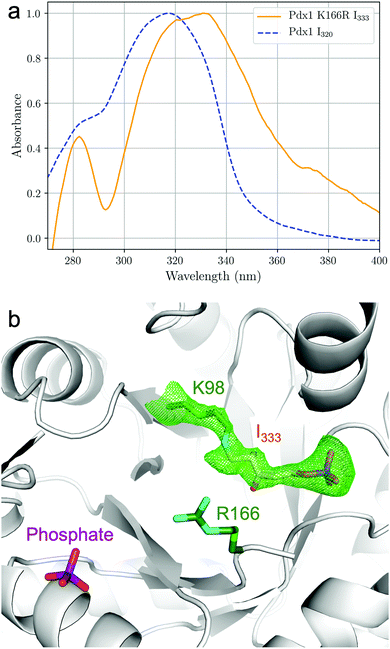 | ||
| Fig. 3 In crystallo 18 UV-vis absorption spectroscopy and structure of Pdx1.3K166R:I333. (a) UV-vis spectra normalised for the absorption maxima of the chromophores, collected from crystals of Pdx1.3:I320 (blue dashed line) and Pdx1.3K166R:I333 (orange line) after soaking with R5P and ammonia. Spectra were smoothed and background correction for light scattering at the crystal interface was applied as described in ESI.† (b) Structure of the ribose tailoring site of Pdx1.3K166R:I333 showing bound I333 (PDB:7NHE). The side chains of Lys98 and Arg166 are shown in stick format (carbon atoms green, nitrogen cyan). The I333 adduct is distinguished by color (carbon atoms orange, oxygen red, nitrogen blue, phosphorous purple). The electron density is contoured at 2.5σ and was calculated using a simulated annealing protocol with Lys98–I333 residues omitted from map calculation (see ESI†). A phosphate ion is seen in the PLP site. | ||
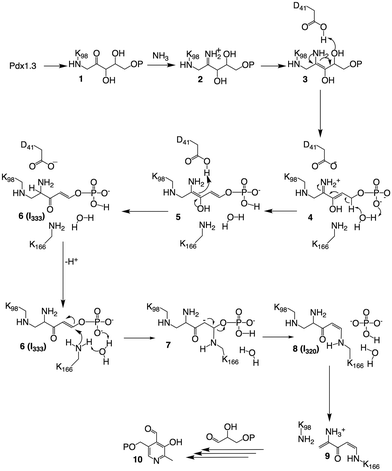
The novel crystal structure and mass-spectrometry data lead to a revised mechanistic proposal for the formation of I320 based on detection and characterisation of I333, as shown in Fig. 4. R5P forms an imine with Lys98, which then undergoes a tautomerisation to give 1. Imine formation with ammonia gives 2, which tautomerises to 3. Elimination of water, assisted by acid catalysis by Asp41 gives 4.
The crystal structure is particularly helpful for interpretation of catalytic steps required in I333 formation (Fig. S8, ESI†). The base catalyst for the tautomerisation of 4 to 5 is a conserved water molecule bound by hydrogen bonding to the amide NH of Gly230 and Val232 (β7α7 loop), the carbonyl oxygen of Phe250 (β8α8 loop) and the closest substrate phosphate oxygen, shown in Fig. 5. The active site bears a structure similar to a protease oxyanion hole.20 Interactions with the amide NH of Gly252, Ser253 (β8α8 loop), Gly231 (β7α7 loop) and Gly170 (β6α6 loop) stabilise the negatively charged R5P phosphate, which itself polarises a water molecule activating it as a base in substrate-assisted catalysis; phosphate groups are frequently observed to perform such function.21 The specific structure ensures stereospecificity of the C5 proS deprotonation reaction.13 Tautomerisation of 5 to 6 is assisted by proton transfer from Asp41.
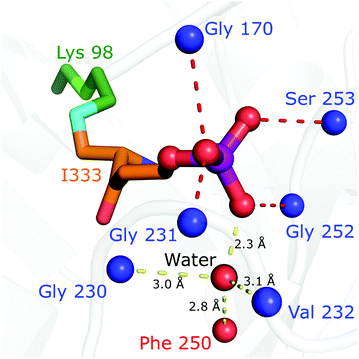 | ||
| Fig. 5 The structure of the Pdx1.3K166R:I333 complex (PDB:7NHE) argues for substrate assisted catalysis and provides an explanation for stereoselective deprotonation. A water, conserved in position across different PLP synthase structures and proposed to act in catalysis, is shown as red sphere. Positions of intermediate coordinating main chain nitrogen/oxygen atoms are shown as blue/red spheres. The covalent I333 adduct is shown in stick format (lysine carbon atoms green, I333 carbon atoms orange, nitrogen blue, oxygen red, phosphorous purple). | ||
In the native enzyme, a conjugate addition of Lys166 is proposed to 6 to give 7, where the substrate phosphate and the conserved water serve as catalytic base. Elimination of phosphate from carbanion 7 completes formation of I320, which is converted to 9 by elimination of Lys 98 in the presence of G3P to yield PLP, as previously described.9,15
Conclusions
Our data are consistent with I333 being on-pathway and serving a function in substrate-assisted catalysis during phosphate elimination. The active site structure allows rationalising stereoselective loss of the C5 hydrogen of R5P. The structure of I333 elucidates the catalytic mechanism for the formation of the novel lysine crosslinking intermediate I320 and is central to our understanding of the Pdx1-catalysed PLP biosynthesis.9,12Author contributions
MJR, NG and IT conducted research, RAB, AR, YZ, TB and IT analysed data, YZ, TB, GE and SE were involved in data curation, IT and TB wrote the manuscript; all authors have reviewed and edited the manuscript.§Conflicts of interest
There are no conflicts to declare.Acknowledgements
We thank S. Findlow and C. Holes at the Macromolecular Crystallisation Facility, Biological Sciences, University of Southampton for support with protein crystallisation. We thank the staff at beamline ID23-1 and at the icOS Lab of the European Synchrotron Radiation Facility for their support with collection of X-ray diffraction and UV-vis absorption spectroscopy data. We thank Nicolas Caramello at the ESRF for help with processing of data obtained at the icOS Lab.Notes and references
- H. C. Dunathan, Proc. Natl. Acad. Sci. U. S. A., 1966, 55, 712–716 CrossRef CAS.
- M. Tambasco-Studart, O. Titiz, T. Raschle, G. Förster, N. Amrhein and T. B. Fitzpatrick, Proc. Natl. Acad. Sci. U. S. A., 2005, 102, 13687–13692 CrossRef CAS.
- K. E. Burns, Y. Xiang, C. L. Kinsland, F. W. McLafferty and T. P. Begley, J. Am. Chem. Soc., 2005, 127, 3682–3683 CrossRef CAS PubMed.
- M. Strohmeier, T. Raschle, J. Mazurkiewicz, K. Rippe, I. Sinning, T. B. Fitzpatrick and I. Tews, Proc. Natl. Acad. Sci. U. S. A., 2006, 103, 19284–19289 CrossRef CAS.
- F. Zein, Y. Zhang, Y. N. Kang, K. Burns, T. P. Begley and S. E. Ealick, Biochemistry, 2006, 45, 14609–14620 CrossRef CAS.
- J. Zhu, J. W. Burgner, E. Harms, B. R. Belitsky and J. L. Smith, J. Biol. Chem., 2005, 280, 27914–27923 CrossRef CAS.
- G. Guédez, K. Hipp, V. Windeisen, B. Derrer, M. Gengenbacher, B. Böttcher, I. Sinning, B. Kappes and I. Tews, Structure, 2012, 20, 172–184 CrossRef.
- A. M. Smith, W. C. Brown, E. Harms and J. L. Smith, J. Biol. Chem., 2015, 290, 5226–5239 CrossRef CAS.
- M. J. Rodrigues, V. Windeisen, Y. Zhang, G. Guédez, S. Weber, M. Strohmeier, J. W. Hanes, A. Royant, G. Evans, I. Sinning, S. E. Ealick, T. P. Begley and I. Tews, Nat. Chem. Biol., 2017, 13, 290–294 CrossRef CAS.
- X. A. Zhang, Y. B. Teng, J. P. Liu, Y. X. He, K. Zhou, Y. X. Chen and C. Z. Zhou, Biochem. J., 2010, 432, 445–450 CrossRef CAS PubMed.
- G. C. Robinson, M. Kaufmann, C. Roux, J. Martinez-Font, M. Hothorn, S. Thore and T. B. Fitzpatrick, Acta Crystallogr D Struct Biol, 2019, 75, 400–415 CrossRef CAS PubMed.
- G. C. Robinson, M. Kaufmann, C. Roux and T. B. Fitzpatrick, Proc. Natl. Acad. Sci. U. S. A., 2016, 113, E5821–E5829 CrossRef CAS.
- J. W. Hanes, K. E. Burns, D. G. Hilmey, A. Chatterjee, P. C. Dorrestein and T. P. Begley, J. Am. Chem. Soc., 2008, 130, 3043–3052 CrossRef CAS PubMed.
- T. Raschle, D. Arigoni, R. Brunisholz, H. Rechsteiner, N. Amrhein and T. B. Fitzpatrick, J. Biol. Chem., 2007, 282, 6098–6105 CrossRef CAS.
- J. W. Hanes, I. Keresztes and T. P. Begley, Nat. Chem. Biol., 2008, 4, 425 CrossRef CAS PubMed.
- J. W. Hanes, I. Keresztes and T. P. Begley, Angew. Chem., Int. Ed., 2008, 47, 2102–2105 CrossRef CAS PubMed.
- H. Liu, G. Ponniah, A. Neill, R. Patel and B. Andrien, Anal. Chem., 2013, 85, 11705–11709 CrossRef CAS PubMed.
- D. von Stetten, T. Giraud, P. Carpentier, F. Sever, M. Terrien, F. Dobias, D. H. Juers, D. Flot, C. Mueller-Dieckmann, G. A. Leonard, D. de Sanctis and A. Royant, Acta Crystallogr D Biol Crystallogr, 2015, 71, 15–26 CrossRef CAS.
- F. S. Dworkowski, M. A. Hough, G. Pompidor and M. R. Fuchs, Acta Crystallogr D Biol Crystallogr, 2015, 71, 27–35 CrossRef CAS.
- R. Menard and A. C. Storer, Biol. Chem., 1992, 373, 393–400 CrossRef CAS PubMed.
- W. Dall'Acqua and P. Carter, Protein Sci., 2000, 9, 1–9 CrossRef.
Footnotes |
| † Electronic supplementary information (ESI) available: Experimental procedures and MS analysis (PDF). See DOI: 10.1039/d1cb00160d |
| ‡ Protein structures were deposited with the PDB under accession codes 7NHE for the Pdx1.3K166R:I333 complex and 7NHF for Pdx1.3K166R. |
| § MJR and RAB were supported by joint studentships between Diamond Light Source and the University of Southampton. This work used the icOS platform of the Grenoble Instruct-ERIC center (ISBG; UMS 3518 CNRS-CEA-UGA-EMBL) within the Grenoble Partnership for Structural Biology (PSB), supported by FRISBI (ANR-10-INBS-05-02) and GRAL, financed within the University Grenoble Alpes graduate school (Ecoles Universitaires de Recherche) CBH-EUR-GS (ANR-17-EURE-0003). NG and TPB were supported by the Robert A. Welch Foundation (A-0034). No competing financial interests have been declared. |
| This journal is © The Royal Society of Chemistry 2022 |