Open Access Article
Open Access ArticleA novel anti-tumor/anti-tumor-associated fibroblast/anti-mPEG tri-specific antibody to maximize the efficacy of mPEGylated nanomedicines against fibroblast-rich solid tumor†
Michael
Chen‡
 a,
Ming-Thau
Sheu‡
b,
Tian-Lu
Cheng
a,
Ming-Thau
Sheu‡
b,
Tian-Lu
Cheng
 cd,
Steve R.
Roffler
e,
Shyr-Yi
Lin
fgh,
Yi-Jou
Chen
a,
Yi-An
Cheng
cd,
Steve R.
Roffler
e,
Shyr-Yi
Lin
fgh,
Yi-Jou
Chen
a,
Yi-An
Cheng
 d,
Jing-Jy
Cheng
gi,
Hsin-Yu
Chang
a,
Tung-Yun
Wu
d,
Jing-Jy
Cheng
gi,
Hsin-Yu
Chang
a,
Tung-Yun
Wu
 j,
An-Pei
Kao
k,
Yuan-Soon
Ho
*hlp and
Kuo-Hsiang
Chuang
*acjmno
j,
An-Pei
Kao
k,
Yuan-Soon
Ho
*hlp and
Kuo-Hsiang
Chuang
*acjmno
aGraduate Institute of Pharmacognosy, Taipei Medical University, Taipei, Taiwan
bSchool of Pharmacy, College of Pharmacy, Taipei Medical University, Taipei, Taiwan
cDrug Development and Value Creation Research Center, Kaohsiung Medical University, Kaohsiung, Taiwan
dDepartment of Biomedical Science and Environmental Biology, Kaohsiung Medical University, Kaohsiung City, Taiwan
eInstitute of Biomedical Sciences, Academia Sinica, Taipei, Taiwan
fDepartment of Primary Care Medicine, Taipei Medical University Hospital, Taipei, Taiwan
gDepartment of General Medicine, School of Medicine, College of Medicine, Taipei Medical University, Taipei, Taiwan
hTMU Research Center of Cancer Translational Medicine, Taipei Medical University, Taipei, Taiwan
iNational Research Institute of Chinese Medicine, Ministry of Health and Welfare, Taipei, Taiwan
jPh.D. Program in Clinical Drug Development of Herbal Medicine, Taipei Medical University, Taipei, Taiwan
kStemforce Biotechnology Co., Ltd, Chiayi City, Taiwan
lSchool of Medical Laboratory Science and Biotechnology, College of Medical Science and Technology, Taipei Medical University, Taipei, Taiwan. E-mail: hoyuansn@tmu.edu.tw; Tel: +886-2-27361663-3327
mTraditional Herbal Medicine Research Center of Taipei Medical University Hospital, Taipei, Taiwan. E-mail: khchuang@tmu.edu.tw; Tel: +886-2-27361663-6163
nPh.D Program in Biotechnology Research and Development, Taipei Medical University, Taipei, Taiwan
oMaster Program for Clinical Pharmacogenomics and Pharmacoproteomics, Taipei Medical University, Taipei, Taiwan
pCancer Research Center, Taipei Medical University Hospital, Taipei, Taiwan
First published on 16th November 2021
Abstract
The therapeutic efficacy of methoxypolyethylene glycol (mPEG)-coated nanomedicines in solid tumor treatment is hindered by tumor-associated fibroblasts (TAFs), which promote tumor progression and form physical barriers. We developed an anti-HER2/anti-FAP/anti-mPEG tri-specific antibody (TsAb) for one-step conversion of mPEG-coated liposomal doxorubicin (Lipo-Dox) to immunoliposomes, which simultaneously target HER2+ breast cancer cells and FAP+ TAFs. The non-covalent modification did not adversely alter the physical characteristics and stability of Lipo-Dox. The TsAb-Lipo-Dox exhibited specific targeting and enhanced cytotoxicity against mono- and co-cultured HER2+ breast cancer cells and FAP+ TAFs, compared to bi-specific antibody (BsAb) modified or unmodified Lipo-Dox. An in vivo model of human breast tumor containing TAFs also revealed the improved tumor accumulation and therapeutic efficacy of TsAb-modified mPEGylated liposomes without signs of toxicity. Our data indicate that arming clinical mPEGylated nanomedicines with the TsAb is a feasible and applicable approach for overcoming the difficulties caused by TAFs in solid tumor treatment.
Introduction
Nanotechnology can provide benefits to traditional chemotherapy agents for solid tumor treatment. Cytotoxic compounds can be delivered using liposomes, micelles, or inorganic carriers at high loading amounts, and efficiently delivered to the tumor area through enhanced permeability and retention (EPR) effects.1–5 The surface modification of methoxypolyethylene glycol (mPEG) offers a shielding effect that prevents nanomedicines from non-specific interactions with the reticuloendothelial system6 and serum proteins,7 extends the in vivo half-life,8 reduces side effects,9 and enhances the therapeutic efficacy.10 Two mPEGylated liposomal drugs, liposomal doxorubicin11 and liposomal irinotecan,12 have been marketed for oncological therapy, and more liposomal formulations of chemotherapeutic agents are under development.13 However, problems using these mPEGylated formulations have become apparent, including lack of the necessary specificity to target tumor cells and that the shielding effect of mPEG hinders uptake by tumor cells.14,15One method that is commonly used to resolve the “PEG dilemma” is the conjugation of tumor-specific antibodies to PEG molecules to generate tumor-targeting drugs. Antibodies against tumor-specific antigens (TSAs) and tumor-associated antigens (TAAs)16 can offer nanomedicines with tumor-targeting abilities and enhanced the efficiency of endocytosis and fusion, thereby increasing the amount of the loaded agents entering cells.17–19 However, the process required for antibody conjugation includes the use of harsh conditions, such as antibody coupling via amines and thiols20–22 or PEG insertion,23 which can damage the stability and function of the antibodies. A non-covalent modification has previously been described using anti-tumor/anti-mPEG bi-specific antibodies (BsAbs) to arm mPEGylated drugs.24–27 BsAbs can be anchored to the mPEG surface and offer anti-tumor binding abilities and enhanced cellular uptake, improving the therapeutic efficacy of the modified agents. Compared with random chemical coupling, BsAb-based modification offers a feasible and time-saving procedure for the generation of antibody-modified liposomes or other nanomedicines, ensuring the outward orientation of functional anti-tumor antibodies on liposomes.24–26 The addition of a suitable anti-tumor moiety in the BsAbs allows for this approach to be adapted to various cancers associated with the overexpression of TSAs or TAAs on the cell surface, such as human epidermal growth factor receptor 2 (HER2), epidermal growth factor receptor (EGFR), prostate-specific membrane antigen (PSMA), and others.28,29
Another major problem associated with the treatment of solid tumors is the presence of tumor-associated fibroblasts (TAFs), which can prevent the entry of targeting nanomedicines, reducing therapeutic efficacy. TAFs are thought to derive from local fibroblasts30 under transforming growth factor β (TGF-β) stimulation conditions in the tumor stroma.31 TAFs contribute to tumor promotion, tissue remodeling, and metastasis32–34 and generate abnormal collagen and extracellular matrix, which can form physical barriers that result in the inefficient entry of therapeutic agents into tumor cells.35–37 High levels of TAFs in various types of solid tumors have been correlated with poor prognosis.38,39 Therapeutic strategies against TAFs can be achieved by targeting fibroblast activation protein (FAP), a membrane-bound serine protease expressed on activated fibroblasts, using monoclonal antibodies40,41 and small-molecule ligands. Although anti-FAP antibodies displayed only marginal clinical efficacy,42,43 previous studies have revealed that FAP is absent from normal human tissues,44,45 and the depletion of FAP expression in animal models did not cause systemic toxicity,46,47 indicating that FAP may represent an ideal target for cancer therapy. These findings indicated that FAP remains a valuable and safe drug target, suggesting that FAP-targeting drugs may require a combination with other more effective anti-cancer agents to achieve synergistic effects.
To simultaneously eliminate TAFs and enhance the therapeutic efficacy of liposomal drugs against tumor cells, we developed an anti-tumor/anti-FAP/anti-mPEG tri-specific antibody (TsAb) that enables liposomal drugs to target both cancer cells and FAP+ TAFs (Fig. 1A). An anti-HER2/anti-FAP/anti-mPEG TsAb was generated to treat HER2+ breast cancer, which is a TAA-expressing solid tumor. The non-covalent addition of TsAb to nanomedicines involved a simple, one-step process that did not require any changes to the nanomedicine formulation, retaining the original advantages of the nanomedicine, including stability and prolonged circulation. TsAb-armed nanomedicines selectively targeted HER2+ breast cancer cells and FAP+ fibroblasts and exhibited enhanced in vivo therapeutic efficacy against breast tumors surrounded by TAFs compared with non-targeting nanomedicines. The one-step TsAb modification represents a feasible method for generating tumor- and TAF-specific agents from well-prepared mPEGylated nanomedicines, enhancing the versatility of approved clinical drugs in cancer treatment.
Results
Design and production of functional TsAb and BsAbs
The anti-HER2/anti-FAP/anti-mPEG TsAb, anti-HER2/anti-mPEG BsAb, and anti-FAP/anti-mPEG BsAb were generated using genetic engineering in the following formats: scFv/scFv/Fab (TsAb) and scFv/Fab (BsAbs) (Fig. 1B). Light and heavy chains were assembled using interchain disulfide bonds in the anti-mPEG Fab. All production yields produced by Expi293 cells exceeded 0.1 mg mL−1. Both the heavy and light chains of the TsAb had estimated molecular weights of 55 kDa, and the heavy and light chains of the BsAbs had estimated molecular weights of 55 kDa and 25 kDa, respectively, which aligned with the results obtained from reducing protein electrophoresis (Fig. 2A). The successful formation of interchain disulfide bonds in the TsAb and BsAbs was demonstrated by the presence of single bands at 110 kDa and 80 kDa, respectively, in the non-reducing protein electrophoresis (Fig. 2A). Anti-mPEG ELISA indicated that the TsAb and BsAbs possessed comparable affinities for mPEG molecules (Fig. 2B and Table S1†). The anti-HER2 and anti-FAP functions of the TsAb and BsAbs were analyzed by flow cytometry using human breast cancer cells MCF-7/HER2 (HER2+/FAP−), human fibroblasts WS-1/FAP (HER2−/FAP+), and human keratinocytes HaCaT (HER2−/FAP−) (Fig. 2C). The TsAb bound to MCF-7/HER2 cells and WS-1/FAP cells, whereas the BsAbs only bound to those cell lines expressing the corresponding antigens. None of these antibodies demonstrated non-specific binding. These results supported the proper assembly of the light and heavy chains via disulfide bonds and indicated that the Fab and scFv moieties were functional.Characterization of TsAb-modified Lipo-Dox
The optimal TsAb modification ratios were determined by assessing the maximal cell targeting capacity and evaluating the minimal amount of unmodified TsAb. Liposomal doxorubicin (Lipo-Dox) was modified with the TsAb at various mPEG![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) TsAb molar ratios, ranging from 100
TsAb molar ratios, ranging from 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 to 2000
1 to 2000![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. The cell-targeting ability of TsAb-Lipo-Dox (also referred to as anti-HER2/anti-FAP-Lipo-Dox) against MCF-7/HER2 cells was detected by cell-based ELISA, which indicated that the binding capacity was dependent on the modification ratio (Fig. S1A†). Unmodified TsAb remaining in the TsAb-Lipo-Dox solution was detected by mPEG-coated ELISA, revealing that the unmodified amount was also dependent on the modification ratio (Fig. S1B†). An mPEG
1. The cell-targeting ability of TsAb-Lipo-Dox (also referred to as anti-HER2/anti-FAP-Lipo-Dox) against MCF-7/HER2 cells was detected by cell-based ELISA, which indicated that the binding capacity was dependent on the modification ratio (Fig. S1A†). Unmodified TsAb remaining in the TsAb-Lipo-Dox solution was detected by mPEG-coated ELISA, revealing that the unmodified amount was also dependent on the modification ratio (Fig. S1B†). An mPEG![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) TsAb (or BsAb) ratio of 200
TsAb (or BsAb) ratio of 200![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 was chosen for all further modifications using the TsAb and BsAbs. To investigate whether the antibody modifications affected the physical characteristics or stability of Lipo-Dox, the particle size, polydispersity index and zeta potential were analyzed (Table 1). Compared with the non-modified Lipo-Dox, the TsAb- and BsAb-modified Lipo-Dox presented with slightly increased particle sizes, up to 120.8–129.7 nm. TsAb-Lipo-Dox also has a larger PI value and a reduced absolute zeta potential than non-modified Lipo-Dox. No particle aggregation was observed after storage at 4 °C overnight. The influence of the antibody modifications on Lipo-Dox drug release was examined for 72 h (Fig. 3A). The drug release of all nanomedicines was less than 2%, and comparable levels of drug release were observed between the unmodified and modified Lipo-Dox samples (p = 0.9969 for Lipo-Dox vs. anti-HER2-Lipo-Dox; p = 0.8343 for Lipo-Dox vs. anti-FAP-Lipo-Dox; and p = 0.9989 for Lipo-Dox vs. anti-HER2/anti-FAP-Lipo-Dox). To measure the amounts of TsAb and BsAbs on the modified liposomes, the nanomedicines were assessed by sandwich ELISA using anti-PEG and anti-His antibodies (Fig. 3B). Similar amounts of TsAb and BsAbs were detected in the same concentrations of the modified Lipo-Dox. These results indicated that the TsAb and BsAbs did not damage the drug during the modification process and that level of antibodies found on the modified liposome surface was consistent.
1 was chosen for all further modifications using the TsAb and BsAbs. To investigate whether the antibody modifications affected the physical characteristics or stability of Lipo-Dox, the particle size, polydispersity index and zeta potential were analyzed (Table 1). Compared with the non-modified Lipo-Dox, the TsAb- and BsAb-modified Lipo-Dox presented with slightly increased particle sizes, up to 120.8–129.7 nm. TsAb-Lipo-Dox also has a larger PI value and a reduced absolute zeta potential than non-modified Lipo-Dox. No particle aggregation was observed after storage at 4 °C overnight. The influence of the antibody modifications on Lipo-Dox drug release was examined for 72 h (Fig. 3A). The drug release of all nanomedicines was less than 2%, and comparable levels of drug release were observed between the unmodified and modified Lipo-Dox samples (p = 0.9969 for Lipo-Dox vs. anti-HER2-Lipo-Dox; p = 0.8343 for Lipo-Dox vs. anti-FAP-Lipo-Dox; and p = 0.9989 for Lipo-Dox vs. anti-HER2/anti-FAP-Lipo-Dox). To measure the amounts of TsAb and BsAbs on the modified liposomes, the nanomedicines were assessed by sandwich ELISA using anti-PEG and anti-His antibodies (Fig. 3B). Similar amounts of TsAb and BsAbs were detected in the same concentrations of the modified Lipo-Dox. These results indicated that the TsAb and BsAbs did not damage the drug during the modification process and that level of antibodies found on the modified liposome surface was consistent.
| Nanomedicinesa | Particle sizeb [nm] | Polydispersity indexb | Zeta potentialb [mV] |
|---|---|---|---|
| a Nanomedicines were prepared by mixing Lipo-Dox with buffer, anti-HER2/anti-mPEG BsAb, anti-FAP/anti-mPEG BsAb, anti-HER2/anti-FAP/anti-mPEG TsAb. b Mean ± SD, n = 3. | |||
| Lipo-Dox | 118.8 ± 1.78 | 0.243 ± 0.044 | −29.3 ± 1.97 |
| Anti-HER2-Lipo-Dox | 122.1 ± 1.83 | 0.280 ± 0.035 | −23.5 ± 2.96 |
| Anti-FAP-Lipo-Dox | 120.8 ± 0.42 | 0.330 ± 0.020 | −26.9 ± 2.76 |
| Anti-HER2/anti-FAP-Lipo-Dox | 129.7 ± 3.21 | 0.356 ± 0.036 | −22.1 ± 2.97 |
HER2- and FAP-specific cellular targeting and enhanced cytotoxicity
The specific targeting abilities and enhanced cytotoxicity of Lipo-Dox modified with the TsAb or BsAbs were examined using MCF-7/HER2 (HER+/FAP−), WS-1/FAP (HER2−/FAP+), and HaCaT (HER−/FAP−) cells. Using cell-based ELISAs, the amount of liposome binding to cells was detected with an anti-PEG antibody. Unmodified Lipo-Dox was almost undetectable in all examined cell lines. The anti-HER2-Lipo-Dox and anti-FAP-Lipo-Dox bound to MCF-7/HER2 and WS-1/FAP cells, respectively, whereas the anti-HER2/anti-FAP-Lipo-Dox bound to both cell lines (Fig. 4A). Due to the autofluorescence of doxorubicin, the cellular uptake of liposomes could be directly observed by fluorescent microscopy (Fig. 4B). After incubation with liposomes, significant amounts of doxorubicin accumulated in the cell lines expressing the corresponding antigens (HER2 or FAP) targeted by the antibody modification used, which was consistent with the results of the cell-based ELISA. To investigate the enhancement of cell-specific cytotoxicity, MCF-7/HER2, WS-1/FAP, and HaCaT cells were separately treated with free doxorubicin, unmodified Lipo-Dox, and antibody-modified Lipo-Dox (Fig. 4C). The anti-HER2-Lipo-Dox exhibited enhanced cytotoxicity compared with unmodified Lipo-Dox in MCF-7/HER2 cells but not in WS-1/FAP cells. By contrast, the increased cytotoxicity of the anti-FAP-Lipo-Dox was observed in WS-1/FAP cells but not in MCF-7/HER2 cells. Only the anti-HER2/anti-FAP-Lipo-Dox was effectively cytotoxic for both cell lines. In HaCaT cells, no differences were observed for cell survival between the tested nanomedicines, demonstrating that the TsAb modification did not increase the off-targeting toxicity of Lipo-Dox. Therefore, the anti-HER2/anti-FAP/anti-mPEG TsAb was able to confer specificity against the overexpressed antigens on targeted cells, improving the cellular binding, drug accumulation, and cytotoxicity of the modified nanomedicines against tumor cells and TAFs.Cytotoxicity against co-culture of breast cancer cells and fibroblasts
Due to the high heterogeneity of cell populations and distributions in tumors, we investigated the cytotoxicity of the examined nanomedicines in environments featuring various ratios of breast cancer cells and fibroblasts. MCF-7/HER2 cells and WS-1/FAP cells were seeded together at ratios of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.2, 1
0.2, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, or 1
1, or 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5 using fixed total cell numbers. Anti-HER2-Lipo-Dox exhibited enhanced cytotoxicity against MCF-7/HER2 cell-rich wells (ratio 1
5 using fixed total cell numbers. Anti-HER2-Lipo-Dox exhibited enhanced cytotoxicity against MCF-7/HER2 cell-rich wells (ratio 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.2) but not against WS-1/FAP cell-rich wells (ratio 1
0.2) but not against WS-1/FAP cell-rich wells (ratio 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5). By contrast, anti-FAP-Lipo-Dox exhibited enhanced cytotoxicity against WS-1/FAP cell-rich wells (ratio 1
5). By contrast, anti-FAP-Lipo-Dox exhibited enhanced cytotoxicity against WS-1/FAP cell-rich wells (ratio 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5) but not against MCF-7/HER2 cell-rich wells (ratio 1
5) but not against MCF-7/HER2 cell-rich wells (ratio 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.2) (Fig. 4D and Table S2†). At all of the cell ratios tested (1
0.2) (Fig. 4D and Table S2†). At all of the cell ratios tested (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.2, 1
0.2, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, and 1
1, and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5), anti-HER2/anti-FAP-Lipo-Dox showed the most cytotoxic effects (Fig. 4D and Table S2†). The ability to simultaneously target HER2 and FAP allowed for the anti-HER2/anti-FAP-Lipo-Dox to induce enhanced cytotoxic effects regardless of whether the first-contacted cell was a breast cancer cell or a TAF, making this modified formulation suitable for the treatment of solid tumors, which are complex structures consisting of multiple cellular targets.
5), anti-HER2/anti-FAP-Lipo-Dox showed the most cytotoxic effects (Fig. 4D and Table S2†). The ability to simultaneously target HER2 and FAP allowed for the anti-HER2/anti-FAP-Lipo-Dox to induce enhanced cytotoxic effects regardless of whether the first-contacted cell was a breast cancer cell or a TAF, making this modified formulation suitable for the treatment of solid tumors, which are complex structures consisting of multiple cellular targets.
Pharmacokinetics and in vivo tumor-targeting of TsAb-modified nanomedicines
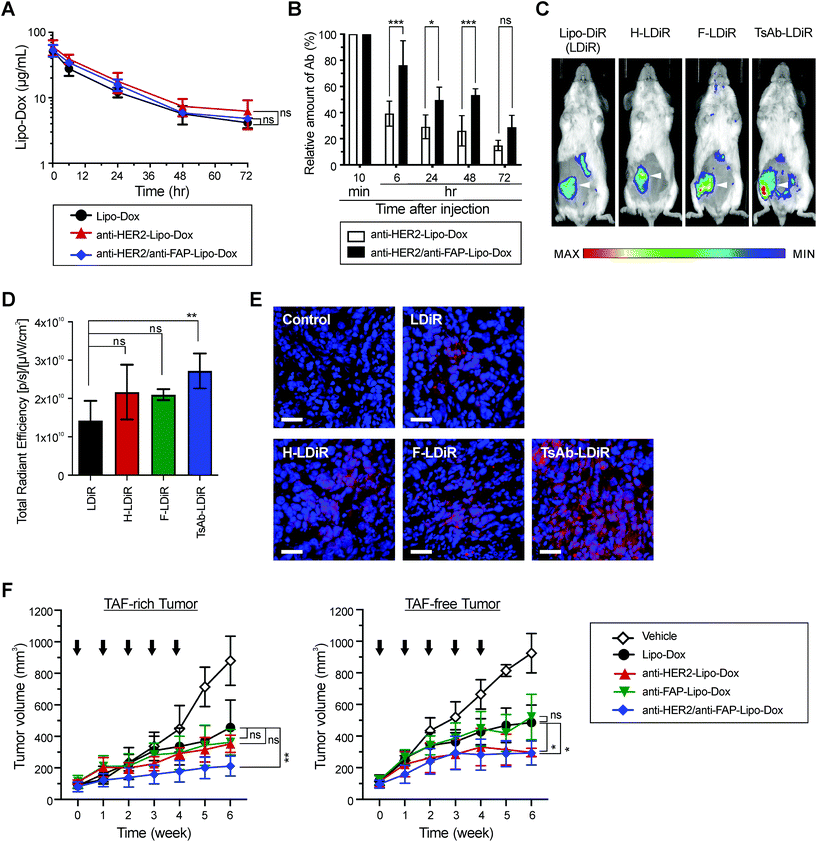
The influence of TsAb or BsAbs modifications on the in vivo pharmacokinetics of Lipo-Dox was investigated. The half-lives of drugs in plasma were not significantly different following antibody modifications (Fig. 5A). To investigate the retention of TsAb and BsAb in the in vivo environment, the antibodies on Lipo-Dox in mice plasma were measured using an anti-PEG/anti-IgG Fab sandwich ELISA after i.v. injection (Fig. 5B). These results indicated that 25% of the total TsAb quantity was retained on Lipo-Dox after 72 h, which was approximately 1.8-fold the quantity of retained anti-HER2/anti-mPEG BsAb, indicating that TsAb was able to remain attached to mPEGylated liposomes longer than BsAb. Additionally, the in vivo tumor targeting capabilities of the modified nanomedicines in a TAF-rich breast tumor animal model were assessed by using TsAb or BsAbs to modify Lipo-DiR, an mPEGylated near-infrared fluorescent liposome. Fig. 5C and D show that the fluorescent signal in the tumor region of anti-HER2/anti-FAP-Lipo-DiR-injected mice was 1.9-fold that of unmodified Lipo-DiR-injected mice. TsAb, which can simultaneously target tumor and TAF, allowed the armed Lipo-DiR homogeneously distributed in tumor section, while anti-HER2-Lipo-DiR and anti-FAP-Lipo-DiR only accumulated unevenly in a small area of the section (Fig. 5E). These results indicated that the TsAb-modified liposomes retained a prolonged circulation time and were able to specifically target TAF-rich breast tumors in the long term.Improved therapeutic efficacy of TsAb-Lipo-Dox against breast tumor surrounded by fibroblasts
To compare the in vivo therapeutic efficacy between TsAb- or BsAb-modified and non-modified Lipo-Dox, we established two xenograft models of human breast tumors: TAF-rich (MCF-7/HER2 cells and WS-1/FAP cells) and TAF-free (MCF-7/HER2 cells only). Anti-HER2/anti-FAP-Lipo-Dox significantly inhibited TAF-rich tumor growth, and the resulting tumor size was 53.6% smaller than that for the non-modified Lipo-dox group (Fig. 5F and Table S3†). Masson's trichrome staining also showed that anti-FAP-Lipo-Dox and TsAb-Lipo-Dox decreased collagen deposition in the tumor (Fig. S2†). By contrast, no significant improvement in tumor suppression was observed for the anti-HER2-Lipo-Dox and anti-FAP-Lipo-Dox groups. In the TAF-free tumor groups, the anti-HER2/anti-FAP-Lipo-Dox and anti-HER2-Lipo-Dox treatments but not the anti-FAP-Lipo-Dox treatment showed significant inhibitory effects against breast tumor compared with the unmodified Lipo-Dox group (Fig. 5F and Table S4†). These results indicated that TsAb-Lipo-Dox not only provides the same benefits as single-targeting Lipo-Dox in TAF-free tumors but also demonstrated a better treatment outcome than both single targeting and non-targeting Lipo-Dox in TAF-rich tumors. Additionally, none of the nanomedicines affected the body weights of the mice (Fig. S3†). No significant differences were observed in the populations of blood cells, liver function, or cardiotoxicity between the nanomedicine groups (Fig. S4†). Collectively, these data indicated that modification with anti-HER2/anti-FAP/anti-mPEG TsAb improved the anti-tumor efficacy of Lipo-Dox in breast tumors surrounded by fibroblasts and did not cause additional adverse effects.Specific targeting of TsAb to clinical breast cancer specimens
The clinical applicability of TsAb and BsAbs were evaluated by the immunohistochemistry staining of tumor sections from stage II and III HER2+ breast cancer patients (Fig. 6). The cell membranes of breast tumor cells were strongly stained with anti-HER2/anti-mPEG BsAb and anti-HER2/anti-FAP/anti-mPEG TsAb but not anti-FAP/anti-mPEG BsAb. Surrounding the tumor cells, spindle-shaped TAFs were stained with anti-FAP/anti-mPEG BsAb and anti-HER2/anti-FAP/anti-mPEG but not anti-HER2/anti-mPEG BsAb. These results demonstrated that the anti-HER2/anti-FAP/anti-mPEG TsAb was able to simultaneously target the FAP+ TAFs and HER2+ breast cancer cells in human clinical tumors, indicating the potential applicability of TsAb-modified nanomedicines for the future clinical treatment of solid tumors.Discussion
We investigated whether a TsAb specific to mPEG, HER2, and FAP can assist mPEGylated liposomal drugs in effectively eliminating breast cancer cells and TAFs and, thus, in treating breast solid tumors blended with fibroblasts. Our results show that the TsAb can easily be anchored on the methoxy terminus of mPEG in liposomes after a simple modification and that this did not affect the liposomal formation or drug release. The TsAb-Lipo-Dox exhibited specific cell targeting, as well as improved drug accumulation and cytotoxicity both to HER2+ breast cancer cells and FAP+ TAFs. Notably, the TsAb-Lipo-Dox inhibited the growth of tumors containing TAFs more efficiently than the anti-HER2-Lipo-Dox and anti-FAP-Lipo-Dox, and had a safety profile and pharmacokinetics similar to those of Lipo-Dox. By simultaneously targeting tumor cells and TAFs, the TsAb provides clinical mPEGylated nanomedicines with a promising solution for treating solid tumors while overcoming the obstacles caused by fibroblasts in cancer therapy.FAP has previously been considered a potential target of immunotherapy to deplete TAFs in solid tumors, but the single targeting of FAP has not been shown to be effective enough to inhibit tumor growth. Sibrotuzumab and other anti-FAP therapeutic antibodies require NK cells to trigger antibody-dependent cell-mediated cytotoxicity (ADCC), but NK cell depletion may occur due to the negative impact of the chemotherapy.48,49 Although more potent anti-FAP immunotherapies, such as antibody–drug conjugates (ADCs)50 and chimeric antigen receptor T cells.46 are being developed, targeting FAP alone also does not provide direct cytotoxic benefits against tumor cells which are usually FAP-negative. The conflicting roles of TAF were also discovered in pancreatic tumor models demonstrating that the depletion of stromal fibroblasts resulted in angiogenesis and suppressor T cell infiltration leads to a tumor-promoting environment.51,52 Hence, the delivery of cytotoxic agents by the dual targeting of FAP+ TAFs and TAA-expressing tumor cells is needed to address the complexity and heterogeneity of tumor. Liposomes chemically conjugated with anti-HER2 scFv and anti-FAP scFv have been found to demonstrate the feasibility of delivering florescent dye into tumor cells and TAFs.53 In this work, our non-chemical conjugate approach allowed clinical liposomal drugs to possess HER2- and FAP-targeting ability, and to delivery chemotherapeutic agents to tumor cells and TAFs (Fig. 4A and B). Unlike the anti-tumor/anti-mPEG BsAb-modified liposomal drugs only targeting tumor cells, the TsAb-modified liposomal drugs exhibited enhanced cytotoxicity against the targeted cells even when TAFs were more abundant than tumor cells (Fig. 4D), suggesting a significant therapeutic potential in complex environments of real solid tumors.
The non-covalently conjugation of TsAbs via the anti-mPEG moiety is expected to be conveniently applied to any mPEGylated nanomedicines. Traditional surface modification of antibodies or ligands is a time-consuming and laborious process and often leads the inconsistent direction or functional damage of modified proteins. By contrast, the anti-mPEG moiety of the TsAb developed in this study specifically binds to the methoxy terminus of mPEG, leading to the anchoring of the TsAb on the outer surfaces of mPEGylated nanomedicines. Meanwhile, the outward orientation ensured the functional binding ability of anti-HER scFv and anti-FAP scFv. Well-prepared liposomes were directly used for the modification with the TsAb or BsAbs, indicating that the manufacturing process of mPEGylated liposomes does not need to be changed for such modification. Notably, the TsAb-modification did not dramatically change the physical characteristics (Table 1 and Fig. 3A) and blood clearance rates (Fig. 5A), and nor did increase non-specific cytotoxicity (Fig. 4C). Additionally, the results of in vitro off-target toxicity and in vivo toxicological studies support the view that the non-covalent conjugation of TsAb does not alter the original safety of the liposomal drugs (Fig. 4C and Fig. S4†). For extended applications, the anti-mPEG moiety of the TsAb has shown a strong affinity to various mPEGylated molecules,24–26 so the TsAb can be used to modify a wide range of mPEGylated nanomedicines, including liposomes, micelles, and inorganic particles. Furthermore, the cancer indication can be expanded to other types of solid tumors, such as pancreatic cancer, by genetically engineering TsAbs with the appropriate anti-tumor moieties.
We further found in this study that modifying a TsAb is a better strategy than simultaneously using two types of BsAbs (e.g., in dual BsAbs-armed nanomedicines). First, pharmacology and pharmacokinetics studies become simpler when tracking only one type of antibody. Because two BsAbs share the same amino acid sequence as anti-mPEG Fab, it would be difficult to identify the contributions to the liposome targeting and cytotoxicity, and to investigate the blood clearance of each BsAb in in vivo studies. Second, the ratio of anti-HER2 and anti-FAP moieties on TsAb-armed nanomedicines is always 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, while the ratios of two BsAbs anchored on dual BsAbs-armed nanomedicine surfaces may be heterogeneous. Finally, the TsAb was retained on liposomes in vivo longer than were the BsAbs. One of possible explanations for this might be the cross-pairing of the TsAb's two scFv moieties (anti-HER2 scFv and anti-FAP scFv). The cross-pairing interaction of VH and VL of scFv moieties forms non-covalent scFv dimers, trimers, and tetramers54 and results in higher avidity,55 supporting the view that the scFv cross-pairing might provide advantages for the anchoring of TsAb on mPEGylated liposomes. By contrast, BsAbs only have one scFv moiety which has less opportunities to engage in cross-pairing. Taken together, we believed that these features indicate that using a TsAb, rather than two BsAbs, is more suitable for the one-step modification of mPEGylated nanomedicines.
1, while the ratios of two BsAbs anchored on dual BsAbs-armed nanomedicine surfaces may be heterogeneous. Finally, the TsAb was retained on liposomes in vivo longer than were the BsAbs. One of possible explanations for this might be the cross-pairing of the TsAb's two scFv moieties (anti-HER2 scFv and anti-FAP scFv). The cross-pairing interaction of VH and VL of scFv moieties forms non-covalent scFv dimers, trimers, and tetramers54 and results in higher avidity,55 supporting the view that the scFv cross-pairing might provide advantages for the anchoring of TsAb on mPEGylated liposomes. By contrast, BsAbs only have one scFv moiety which has less opportunities to engage in cross-pairing. Taken together, we believed that these features indicate that using a TsAb, rather than two BsAbs, is more suitable for the one-step modification of mPEGylated nanomedicines.
The optimization of antibody density on nanomedicine surfaces is critical for optimal drug stability, targeting, and killing ability. Increasing the amount of antibody on a nanomedicine may reduce the cellular uptake,56 or the steric stabilization effect of PEG that causes poor in vivo pharmacokinetics and targeting efficiency.57 Similarly, this study shows that higher amounts of TsAb or BsAb during modification resulted in enhanced specific targeting and cytotoxicity, but the excessive amount caused the presence of unbound antibody or negatively affected the stability of liposomal structure. The modification ratios ranging from 200![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 to 2000
1 to 2000![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 resulted in significant enhanced cellular targeting of modified Lipo-Dox (Fig. S1A†) without excessive unbound TsAb in solution (Fig. S1B†). Therefore, it is expected that the TsAb-Lipo-Dox maintained specific targeting even when only about 29% of the TsAb was retained on the nanomedicine surface at 72 h after its administration (Fig. 5B). Besides, our previous work on the modification of a mPEGylated lecithin-stabilized micellar drug delivery system (LsbMDDs) with anti-HER2/anti-mPEG BsAb showed that the mPEG
1 resulted in significant enhanced cellular targeting of modified Lipo-Dox (Fig. S1A†) without excessive unbound TsAb in solution (Fig. S1B†). Therefore, it is expected that the TsAb-Lipo-Dox maintained specific targeting even when only about 29% of the TsAb was retained on the nanomedicine surface at 72 h after its administration (Fig. 5B). Besides, our previous work on the modification of a mPEGylated lecithin-stabilized micellar drug delivery system (LsbMDDs) with anti-HER2/anti-mPEG BsAb showed that the mPEG![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) BsAb ratios of 100
BsAb ratios of 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 500
1 and 500![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 did not result in the presence of unbound BsAb,25 indicating that the modification ratio can be optimized for different mPEGylated nanomedicines.
1 did not result in the presence of unbound BsAb,25 indicating that the modification ratio can be optimized for different mPEGylated nanomedicines.
Experimental
Reagents
DMEM/F21 medium, MEM medium, Expi293 Expression System Kit, and bicinchoninic acid (BCA) Protein Assay Kit were purchased from Thermo Fisher Scientific (Waltham, MA, USA). FBS and HisTrap HP columns were purchased from GE Healthcare Life Sciences (Little Chalfont, UK). FITC-conjugated goat anti-human IgG F(ab′)2 and HRP-conjugated goat anti-human IgG F(ab′)2 were purchased from Jackson ImmunoResearch Laboratories (West Grove, PA, USA). Anti-PEG monoclonal antibody (clone AGP4) and biotin-conjugated AGP4 antibody (AGP4-biotin) were kindly provided by Dr Steve R. Roffler (Institute of Biomedical Science, Academia Sinica, Taiwan). Sesame oil, β-estradiol 17-valerate, and 2,2′-azino-bis(3-ethylbenzothiazoline-6-sulphonic acid) (ABTS) were purchased from Sigma-Aldrich (St Louis, MO, USA). Matrigel (high concentration) was purchased from Corning (Corning, NY, USA). ATPlite 1step luminescence assay kit was purchased from PerkinElmer (Waltham, MA, USA). Skim milk was purchased from BD Biosciences (San Jose, CA, USA). Lipo-Dox was purchased from TTY Biopharm Co. Ltd (Taipei, Taiwan).Cell lines
MCF-7/HER2 cells were human HER2-overexpresing breast cancer cells established via retroviral infection of MCF-7 cells,58 which were generously provided by Dr Mien-Chie Hung (Department of Molecular and Cellular Oncology, University of Texas, M. D. Anderson Cancer Center, Houston, TX, USA), and were maintained in DMEM/F12 medium with 10% (v/v) fetal bovine serum (FBS) and 1% (v/v) penicillin–streptomycin. WS-1/FAP cells were human fibroblasts WS-1 engineered to overexpress human FAP by lentiviral infection, and were maintained in MEM medium containing 10% FBS, 1% penicillin/streptomycin, 0.1 mM non-essential amino acid and 1 mM sodium pyruvate. HaCaT cells were HER2-/FAP-human keratinocytes used as negative control cell line in cellular experiments, and were maintained in DMEM medium containing 10% cosmic calf serum, and 1% penicillin/streptomycin. WS-1 cells and HaCaT cells were purchased from Bioresource Collection and Research Center (BCRC), Hsinchu, Taiwan.Animals
BALB/cByJNarl (BALB/c) mice and CB17/lcr-Prkdcscid/CrlNarl (SCID) mice were purchased from National Laboratory Animal Center, Taipei, Taiwan. All mice were kept in a specific-pathogen-free (SPF) facility, and the protocol of animal experiments performed (LAC-2014-0253) were approved by the Institutional Animal Care and Use Committee (IACUC) of Taipei Medical University (Taipei, Taiwan).Genetic construction of engineered antibodies
The anti-HER2/anti-mPEG anti-FAP/anti-mPEG BsAbs were constructed by genetic engineering as previously detailed24 with minor modifications. Briefly, the light chain and heavy chain of humanized anti-mPEG Fab (clone h15-2b) were genetically linked with an internal ribosome entry site (IRES)59 for co-expressing in a single plasmid. The Fv fragment of a humanized anti-HER2 (trastuzumab)60 or a humanized anti-FAP antibody (sibrotuzumab) was genetically linked after the heavy chain of humanized anti-mPEG Fab. To construct anti-HER2/anti-FAP/anti-mPEG TsAb, the anti-HER2 scFv and the anti-FAP scFv were respectively fused after the light chain and heavy chain of the anti-mPEG Fab. The genes of the engineered antibodies were added with a 6xHis tag for protein purification and subcloned into pLNCX vector for mammalian cell expression.Antibody production
TsAb and BsAbs were produced by transient transfection of pLNCX plasmid into Expi293 mammalian expression cells, according to the instructions of the Expi293 Expression System Kit. Cell supernatant was harvested at 5 days post-transfection for protein purification. Antibodies were purified using HisTrap HP columns, dialyzed in phosphate-buffered saline (PBS, pH 7.4, 10 mM), and sterilized by using 0.2 μm filter discs. Antibody concentrations were determined by BCA assay.SDS-PAGE analysis
To assess the molecular weight and antibody folding, purified TsAb and BsAbs were separated by electrophoresis in 10% (w/v) reducing or non-reducing SDS-PAGE gels and stained with EBL Easy Blue-Plus CBB Stain Reagent (EBL Biotechnology, New Taipei City, Taiwan). In reduced condition, the light chain and heavy chain of the BsAbs were expected to have molecular weights of approximately 25 kDa and 55 kDa, respectively, and both chains of TsAb were expected to have molecular weights of approximately 55 kDa. In non-reduced condition, the intact BsAbs and TsAb with the correct formations of inter-chain disulfides were expected to have molecular weights of 80 kDa and 110 kDa, respectively.Anti-mPEG ELISA
ELISA plates were coated with mPEG5000-NH2 (20 μg per well) diluted in coating buffer (0.1 M NaHCO3, pH 9.0) at 37 °C for 2 h, and then blocked with 5% (w/v) skim milk (dissolved in PBS, pH 7.4) at 4 °C overnight. TsAb and BsAbs were serially diluted with 2% (w/v) skim milk (dissolved in PBS, pH 7.4) to the same molar concentrations, and then incubated in the wells (50 μL per well). After being washed with PBS, the wells were sequentially incubated with 50 μL of HRP-conjugated goat anti-human IgG F(ab′)2 and ABTS substrate. Color development was measured at 405 nm with an Epoch microplate reader (BioTek Instruments, Winooski, VT, USA).Flow cytometry
MCF-7/HER2 cells (HER2+/FAP−), WS-1/FAP cells (HER2−/FAP+) and HaCaT cells (HER2−/FAP−) were used to evaluate the anti-HER2 and anti-FAP function of TsAb and BsAbs. Cells were harvested and suspended in polystyrene tubes (2 × 105 cells per tube) with PBS/BSA buffer (0.05% (w/v) BSA dissolved in cold PBS, pH 7.4), and stained with 20 μg mL−1 of TsAb and BsAbs on ice for 1 h. After being washed with PBS/BSA buffer, cells were sequentially stained with FITC-conjugated goat anti-human IgG F(ab′)2 on ice for 1 h and counterstained with propidium iodide. The FITC signal of viable cells was detected by a FACSCalibur flow cytometer (BD Biosciences, San Jose, CA, USA) and analyzed using the CellQuest software (BD Biosciences, San Jose, CA, USA).Non-covalent modification of Lipo-Dox with TsAb or BsAbs
Modified Lipo-Dox was freshly prepared for each experiment. Lipo-Dox was incubated with TsAb or BsAbs in serum-free medium or PBS buffer containing 0.05% of BSA (DMEM/F12 + BSA or PBS + BSA) at RT for 1 h. The DSPE-mPEG2000 of Lipo-Dox![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) antibody molar ratio was 2000
antibody molar ratio was 2000![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 1000
1, 1000![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 500
1, 500![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 200
1, 200![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 or 100
1 or 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. In most of the experiments, the ratio of 200
1. In most of the experiments, the ratio of 200![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 was chosen for the balance between the maximal cellular targeting and the minimal free antibody amount determined by cell-based ELISA and mPEG-coated ELISA, respectively (Fig. S1†). Lipo-Dox contains 2 mg mL−1 (3.68 mM) doxorubicin HCl and 4.2 mg mL−1 (1.497 mM) DSPE-mPEG2000 according to the compositions provided by the manufacturers.61 The number of doxorubicin HCl per nanoparticle was estimated at about 13
1 was chosen for the balance between the maximal cellular targeting and the minimal free antibody amount determined by cell-based ELISA and mPEG-coated ELISA, respectively (Fig. S1†). Lipo-Dox contains 2 mg mL−1 (3.68 mM) doxorubicin HCl and 4.2 mg mL−1 (1.497 mM) DSPE-mPEG2000 according to the compositions provided by the manufacturers.61 The number of doxorubicin HCl per nanoparticle was estimated at about 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 doxorubicin molecules,62 so we inferred that there are approximately 5289 mPEG and 26 TsAb (or BsAb) molecules on each modified Lipo-Dox nanoparticle with the mPEG
000 doxorubicin molecules,62 so we inferred that there are approximately 5289 mPEG and 26 TsAb (or BsAb) molecules on each modified Lipo-Dox nanoparticle with the mPEG![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) TsAb molar ratio of 200
TsAb molar ratio of 200![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.
1.
Drug release
The release rates of doxorubicin from the nanomedicines (Lipo-Dox, anti-HER2-Lipo-Dox, anti-FAP-Lipo-Dox, and anti-HER2/anti-FAP-Lipo-Dox) were measured with a previously described dialysis method.63 One ml of free doxorubicin and the nanomedicines (equivalent to 0.1 mg mL−1 of doxorubicin) was placed into a dialysis bag of MWCO 6000 (Cellu-Sep V T1, Orange Scientific, Seguin, TX) and dialyzed against 20 mL of pre-warmed 50 mM PBS (containing 0.1% (v/v) Tween 80, pH 7.4) at 37 °C with shaking at a speed of 100 rpm. At the indicated time points, released doxorubicin in the dialysis buffer was quantified by high-performance liquid chromatography (HPLC) assay, and the dialysis buffer was replaced with fresh buffer to maintain the sink condition. The HPLC conditions were as follows: a Discovery® C18 column (250 × 4.6 mm, 5 μm; SUPELCO, Bellefonte, PA, USA) maintained at 40 °C; the composition of the mobile phase was water![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) acetonitrile
acetonitrile![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) methanol
methanol![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) acetic acid (650
acetic acid (650![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 250
250![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100
100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2, v/v/v/v) adjusted to pH 3.6 by using 2 N NaOH; the flow rate was 1 mL min−1; and the fluorescence detector was set at λex 480 nm/λem 550 nm. All measurements were performed in triplicate.
2, v/v/v/v) adjusted to pH 3.6 by using 2 N NaOH; the flow rate was 1 mL min−1; and the fluorescence detector was set at λex 480 nm/λem 550 nm. All measurements were performed in triplicate.
Physical analysis of TsAb-Lipo-Dox
To measure the average particle size and polydispersity index, the nanomedicines were diluted with DDW and then analyzed by an N5 submicron particle size analyzer (Beckman Coulter, Brea, CA, USA). Each measurement was performed at 25 °C with a detection angle of 90°. To measure zeta potentials, the nanomedicines were diluted with DDW and then analyzed with a Zetasizer (Nano ZS) (Malvern Instrument, Worcestershire, UK).Sandwich ELISA
To measure the amount of TsAb or BsAbs on liposome surface, anti-PEGbackbond antibody (clone AGP3) (5 μg per well) was coated in ELISA plates with coating buffer (0.1 M NaHCO3, pH 9) at 37 °C for 4 h and then at 4 °C overnight. The plates were blocked with 5% skim milk (dissolved in PBS, pH 7.4) at 37 °C for 3 h. The nanomedicines were serially diluted with 2% (w/v) skim milk (dissolved in PBS, pH 7.4) to 300, 100, and 33.3 μg mL−1, and then incubated in the wells for 1 h (50 μL per well). After being washed with PBS, the wells were sequentially incubated with 50 μL of mouse anti-His antibody and HRP-conjugated goat anti-mouse IgG Fc and ABTS substrate. Color development was measured at 405 nm with the microplate reader.Cell-based ELISA
MCF-7/HER2, WS-1/FAP, or HaCaT cells were seeded in 96-well plates (2 × 105 cells per well). On the next day, the cells were treated with 50 μL of the nanomedicines (serially diluted with serum-free medium containing 0.05% BSA) at 37 °C for 1 h. After being washed with serum-free medium, the wells were sequentially incubated with 50 μL of anti-PEG mAb (AGP4-biotin, 5 μg mL−1) HRP-conjugated streptavidin (1 μg mL−1) and ABTS substrate. Color development was measured at 405 nm with the microplate reader.Deconvolution microscopy of cellular uptake
MCF-7/HER2 cells or WS-1/FAP cells were seeded in 35 mm glass bottom dishes. On the next day, the cells were treated with phenol red-free medium containing 0.05% BSA and 20 μg mL−1 of the nanomedicines at 37 °C. After 10 min, the cells were washed with phenol red-free medium and analyzed with a DeltaVision Elite microscope (GE Healthcare, USA). The fluorescence of doxorubicin was imaged with a filter set of λex 475 nm/λem 597 nm.In vitro cytotoxicity
MCF-7/HER2, WS-1/FAP, or HaCaT cells were seeded in 96-well plates at 4 × 103 cells per well. On the next day, the cells were treated with 50 μL of the nanomedicines (serially diluted with serum-free medium containing 0.05% BSA) at 37 °C for 1 h. After the removal of the drugs, the cells were further cultured in growth medium at 37 °C for 72 h. Cell survival was determined with ATPlite assay. To investigate the cytotoxicity against tumor cell/fibroblast co-culture, MCF-7/HER2 cells and WS-1/FAP cells were mixed with at the indicated cell ratios (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.2, 1
0.2, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, and 1
1, and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5) and seeded into 96-well plates (4000 cells per well). On the next day, the cells were treated with the nanomedicines, and cell survival was determined as described above.
5) and seeded into 96-well plates (4000 cells per well). On the next day, the cells were treated with the nanomedicines, and cell survival was determined as described above.
Pharmacokinetics of TsAb-Lipo-Dox
TsAb-Lipo-Dox and anti-HER2-Lipo-Dox were i.v. injected into 5-week-old female BALB/c mice. At the indicated timepoint (10 min and 6, 24, 48, and 72 h), blood from the tail vein was collected in a heparinized microhematocrit tube and centrifuged to obtain plasma. The plasma concentration of the nanomedicines was measured with a commercial anti-PEG sandwich ELISA. The plasma samples were diluted with 2% skim milk and incubated in ELISA plates coated with an anti-PEG capture antibody (clone AGP4). After being washed with PBS, the plates were sequentially incubated with an anti-PEG detection antibody (clone 6-3), HRP-conjugated goat anti-mouse IgG Fc, and ABTS substrate. Color development was measured at 405 nm with the microplate reader.To measure the resident TsAb and BsAbs bound on Lipo-Dox after i.v. injection, the plasma samples were diluted with 2% skim milk at an equivalent concentration of 0.5 μg mL−1, and the resident TsAb and BsAbs were measured with a sandwich ELISA. Anti-PEG antibody AGP4 and HRP-conjugated goat anti-human IgG F(ab′)2 were used as a capture/detection antibody pairing. The freshly prepared nanomedicines were serially diluted and used as standards to calculate the amounts of the resident TsAb and BsAbs on Lipo-Dox in the plasma samples.
In vivo imaging of TsAb- or BsAb-modified Lipo-DiR
Female 5-week-old SCID mice were weekly s.c. injected with β-estradiol 17-valerate (25 μg per mice, dissolved in sesame oil). MCF-7/HER2 cells and WS-1/FAP cells (3 × 106 cells for each cell line) were mixed together with Matrigel and s.c. injected into the right abdominal mammary fat pads of the mice. After the tumor size reached 100 mm3, Lipo-DiR (F60203F-DR, FormuMax Scientific Inc., Sunnyvale, CA, USA) modified with TsAb or BsAbs was i.v. injected (1 μg per mice). After 24 h, non-invasive imaging was performed by IVIS Lumina XRMS (PerkinElmer, Waltham, Massachusetts, USA) with a filter set of λex 740 nm/λem 790 nm. Tumor tissues were embedded in OCT compound and cut at 10 μm. The slides were fixed in 4% paraformaldehyde, stained with DAPI, and then imaged with fluorescent microscopy.In vivo treatment against fibroblast-containing breast tumor
SCID mice bearing TAF-rich tumors consisting of mixes of MCF-7/HER2 cells and WS-1/FAP cells were established as above. After the tumor size reached 100 mm3, Lipo-Dox modified with TsAb or BsAbs was i.v. injected into the mice weekly at a dose of 2 mg kg−1 5 times. Tumor size and body weight were measured once weekly. Two weeks after the last dose, collagen amount in tumor sections was evaluated with Masson's trichrome stain. To evaluate the efficacy of TsAb- or BsAb-Lipo-Dox in the absence of human TAFs (TAF-free tumor), SCID mice received a s.c. injection of MCF-7/HER2 cells alone and the same therapeutic regiment.Immunohistochemistry
Frozen breast tumor sections of anonymous patients were obtained from the biobanks of the Taipei Medical University (Taipei, Taiwan), and were examined according to a protocol approved by the Taipei Medical University-Joint Institutional Review Board (TMU-JIRB N 201501029). The sections were fixed with 4% (v/v) formaldehyde for 15 min, and were immersed in 3% (v/v) hydrogen peroxide (H2O2) for 5 min to block endogenous peroxidase activity. Then, the slides were incubated in 0.1% BSA and gelatin blocking buffer for 15 min. The TsAb or BsAbs were incubated with the slides in a humidified chamber at 4 °C overnight. The staining then was developed according to Dako REAL EnVision detection system (Glostrup, Denmark). The sections were also stained with hematoxylin and eosin (HE).Toxicological evaluation of TsAb- and BsAb-Lipo-Dox
Healthy BALB/c mice were i.v. injected weekly with Lipo-Dox, anti-HER2-Lipo-Dox, or anti-HER2/anti-FAP-Lipo-Dox (6 mg kg−1) three times for a total cumulative dose of 18 mg kg−1, which is a known dosage of free doxorubicin to cause cardiomyopathy in mice.64 Whole blood via submandibular bleeding was collected in EDTA-coated microcollection tubes prior to the administration and 2 weeks after the last dose for subsequent measurements. Red blood cells (RBC), hemoglobin level, white blood cells (WBC), neutrophils, and lymphocytes for myelotoxicity were measured by a ProCyte Dx Hematology Analyzer (IDEXX, Westbrook, ME, USA). Serum aspartate aminotransferase (AST) and alanine aminotransferase (ALT) levels for liver toxicity were measured by VetTest Chemistry Analyzer (IDEXX, Westbrook, ME, USA). Serum troponin I level after the administration for cardiac toxicity was measured by an ELISA kit (E-EL-M0086, Elabscience, Wuhan, PRC).Statistical analysis
Data was analyzed by using GraphPad Software Prism (San Diego, CA, USA). Comparisons were determined using one-way or two-way analysis of variance (ANOVA) with Bonferroni's test for two groups or with Dunnett's test for three or more groups. Tumor sizes and relative body weights were compared at the end of the 6th week. A significant difference was defined as a p value <0.05.Conclusions
According to the results in this study, we developed an engineered antibody binding to three targets, mPEGylated nanomedicines, breast tumor cells, and TAFs, to provide clinical nanomedicines with a dual targeting ability to treat TAF-rich solid tumors. The rapid and non-covalent conjugation of TsAb on mPEGylated nanomedicines constitutes a one-step modification without damaging the stability of the drugs. The TsAb-armed nanomedicines exhibited enhanced cellular specificity and simultaneously destroyed HER2+ breast tumor containing heterogeneously distributed FAP+ TAFs. Since TAFs hinder the therapeutic effects of clinical mPEGylated nanomedicines in diverse types of solid tumors, the modification with TsAb could be a feasible approach for enabling such drugs to eliminate TAFs while also treating tumors.Author contributions
M. Chen and M. T. Sheu contributed equally to this work. K. H. Cuang, M. Chen, and M. T. Sheu designed the TsAb-armed nanomedicine platform technology. T. L. Cheng, S. R. Roffler, and Y. A. Cheng provided anti-PEG antibodies and the gene of anti-mPEG Fab. M. Chen, Y. J. Chen, and H. Y. Chang performed the experiments whose results are shown in Fig. 2, 4, S1–S4 and Tables S1–S4.† M. T. Sheu performed the experiments whose results are shown in Table 1 and Fig. 3. M. Chen and T. Y. Wu performed, and J. J. Cheng guided the animal experiments shown in Fig. 5. Y. S. Ho performed the experiments whose results are shown in Fig. 6. A. P. Kao assisted in the data analysis and drew Fig. 1. K. H. Chuang, M. Chen, M. T. Sheu, and S. Y. Lin composed the manuscript and figures.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by grants from the Ministry of Science and Technology, Taiwan [MOST 110-2320-B-038-030, 110-2628-B-038-011, and MOST 106-2632-B-038-001]; the National Health Research Institutes, Taiwan [NHRI-EX110-10935SI]; the Jin-lung-Yuan Foundation [2020–2021]; the Ministry of Education [RSC 110-5804-002-400] and the Health and Welfare Surcharge of Tobacco Products, Taiwan [MOHW110-TDU-B-212-144014].Notes and references
- J. O. Eloy, M. Claro de Souza, R. Petrilli, J. P. Barcellos, R. J. Lee and J. M. Marchetti, Colloids Surf., B, 2014, 123, 345–363 CrossRef CAS PubMed.
- M. Coimbra, B. Isacchi, L. van Bloois, J. S. Torano, A. Ket, X. Wu, F. Broere, J. M. Metselaar, C. J. Rijcken, G. Storm, R. Bilia and R. M. Schiffelers, Int. J. Pharm., 2011, 416, 433–442 CrossRef CAS PubMed.
- F. Wang, Y. C. Wang, S. Dou, M. H. Xiong, T. M. Sun and J. Wang, ACS Nano, 2011, 5, 3679–3692 CrossRef CAS.
- G. Gaucher, M. H. Dufresne, V. P. Sant, N. Kang, D. Maysinger and J. C. Leroux, J. Controlled Release, 2005, 109, 169–188 CrossRef CAS PubMed.
- J. Fang, H. Nakamura and H. Maeda, Adv. Drug Delivery Rev., 2011, 63, 136–151 CrossRef CAS.
- K. Maruyama, T. Yuda, A. Okamoto, S. Kojima, A. Suginaka and M. Iwatsuru, Biochim. Biophys. Acta, 1992, 1128, 44–49 CrossRef CAS.
- J. S. Suk, Q. Xu, N. Kim, J. Hanes and L. M. Ensign, Adv. Drug Delivery Rev., 2016, 99, 28–51 CrossRef CAS PubMed.
- F. Danhier, O. Feron and V. Preat, J. Controlled Release, 2010, 148, 135–146 CrossRef CAS.
- A. Fukuda, K. Tahara, Y. Hane, T. Matsui, S. Sasaoka, H. Hatahira, Y. Motooka, S. Hasegawa, M. Naganuma, J. Abe, S. Nakao, H. Takeuchi and M. Nakamura, PLoS One, 2017, 12, e0185654 CrossRef.
- M. Xing, F. Yan, S. Yu and P. Shen, PLoS One, 2015, 10, e0133569 CrossRef.
- Y. Barenholz, J. Controlled Release, 2012, 160, 117–134 CrossRef CAS.
- A. H. Ko, Int. J. Nanomed., 2016, 11, 1225–1235 CrossRef CAS.
- U. Bulbake, S. Doppalapudi, N. Kommineni and W. Khan, Pharmaceutics, 2017, 9, 12 CrossRef.
- H. Hatakeyama, H. Akita and H. Harashima, Biol. Pharm. Bull., 2013, 36, 892–899 CrossRef CAS.
- L. Y. Song, Q. F. Ahkong, Q. Rong, Z. Wang, S. Ansell, M. J. Hope and B. Mui, Biochim. Biophys. Acta, 2002, 1558, 1–13 CrossRef CAS.
- N. E. Papaioannou, O. V. Beniata, P. Vitsos, O. Tsitsilonis and P. Samara, Ann. Transl. Med., 2016, 4, 261 CrossRef.
- J. O. Eloy, R. Petrilli, L. N. F. Trevizan and M. Chorilli, Colloids Surf., B, 2017, 159, 454–467 CrossRef CAS.
- K. B. Johnsen, A. Burkhart, F. Melander, P. J. Kempen, J. B. Vejlebo, P. Siupka, M. S. Nielsen, T. L. Andresen and T. Moos, Sci. Rep., 2017, 7, 10396 CrossRef PubMed.
- K. W. Peng, K. A. Donovan, U. Schneider, R. Cattaneo, J. A. Lust and S. J. Russell, Blood, 2003, 101, 2557–2562 CrossRef CAS.
- P. Marques-Gallego and A. I. de Kroon, BioMed Res. Int., 2014, 2014, 129458 Search PubMed.
- A. S. Manjappa, K. R. Chaudhari, M. P. Venkataraju, P. Dantuluri, B. Nanda, C. Sidda, K. K. Sawant and R. S. Murthy, J. Controlled Release, 2011, 150, 2–22 CrossRef CAS.
- H. Xu, X. Zhao, C. Grant, J. R. Lu, D. E. Williams and J. Penfold, Langmuir, 2006, 22, 6313–6320 CrossRef CAS.
- M. L. Geddie, N. Kohli, D. B. Kirpotin, M. Razlog, Y. Jiao, T. Kornaga, R. Rennard, L. Xu, B. Schoerberl, J. D. Marks, D. C. Drummond and A. A. Lugovskoy, mAbs, 2017, 9, 58–67 CrossRef CAS.
- C. H. Kao, J. Y. Wang, K. H. Chuang, C. H. Chuang, T. C. Cheng, Y. C. Hsieh, Y. L. Tseng, B. M. Chen, S. R. Roffler and T. L. Cheng, Biomaterials, 2014, 35, 9930–9940 CrossRef CAS PubMed.
- C. Y. Su, M. Chen, L. C. Chen, Y. S. Ho, H. O. Ho, S. Y. Lin, K. H. Chuang and M. T. Sheu, Drug Delivery, 2018, 25, 1066–1079 CrossRef CAS PubMed.
- Y. A. Cheng, I. J. Chen, Y. C. Su, K. W. Cheng, Y. C. Lu, W. W. Lin, Y. C. Hsieh, C. H. Kao, F. M. Chen, S. R. Roffler and T. L. Cheng, Biomater. Sci., 2019, 7, 3404–3417 RSC.
- Y. A. Cheng, T. H. Wu, Y. M. Wang, T. L. Cheng, I. J. Chen, Y. C. Lu, K. H. Chuang, C. K. Wang, C. Y. Chen, R. A. Lin, H. J. Chen, T. Y. Liao, E. S. Liu and F. M. Chen, J. Nanobiotechnol., 2020, 18, 118 CrossRef CAS PubMed.
- S. Yu, A. Li, Q. Liu, T. Li, X. Yuan, X. Han and K. Wu, J. Hematol. Oncol., 2017, 10, 78 CrossRef.
- S. M. Chiavenna, J. P. Jaworski and A. Vendrell, J. Biomed. Sci., 2017, 24, 15 CrossRef CAS PubMed.
- A. Arina, C. Idel, E. M. Hyjek, M. L. Alegre, Y. Wang, V. P. Bindokas, R. R. Weichselbaum and H. Schreiber, Proc. Natl. Acad. Sci. U. S. A., 2016, 113, 7551–7556 CrossRef CAS.
- Y. Kojima, A. Acar, E. N. Eaton, K. T. Mellody, C. Scheel, I. Ben-Porath, T. T. Onder, Z. C. Wang, A. L. Richardson, R. A. Weinberg and A. Orimo, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 20009–20014 CrossRef CAS PubMed.
- B. Erdogan and D. J. Webb, Biochem. Soc. Trans., 2017, 45, 229–236 CrossRef CAS.
- Y. Yu, C. H. Xiao, L. D. Tan, Q. S. Wang, X. Q. Li and Y. M. Feng, Br. J. Cancer, 2014, 110, 724–732 CrossRef CAS.
- J. T. O'Connell, H. Sugimoto, V. G. Cooke, B. A. MacDonald, A. I. Mehta, V. S. LeBleu, R. Dewar, R. M. Rocha, R. R. Brentani, M. B. Resnick, E. G. Neilson, M. Zeisberg and R. Kalluri, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 16002–16007 CrossRef PubMed.
- S. Torosean, B. Flynn, J. Axelsson, J. Gunn, K. S. Samkoe, T. Hasan, M. M. Doyley and B. W. Pogue, Nanomedicine, 2013, 9, 151–158 CrossRef CAS PubMed.
- L. Eikenes, O. S. Bruland, C. Brekken and L. Davies Cde, Cancer Res., 2004, 64, 4768–4773 CrossRef CAS.
- C. H. Heldin, K. Rubin, K. Pietras and A. Ostman, Nat. Rev. Cancer, 2004, 4, 806–813 CrossRef CAS PubMed.
- L. Tao, G. Huang, R. Wang, Y. Pan, Z. He, X. Chu, H. Song and L. Chen, Sci. Rep., 2016, 6, 38408 CrossRef CAS.
- A. Marusyk, D. P. Tabassum, M. Janiszewska, A. E. Place, A. Trinh, A. I. Rozhok, S. Pyne, J. L. Guerriero, S. Shu, M. Ekram, A. Ishkin, D. P. Cahill, Y. Nikolsky, T. A. Chan, M. F. Rimawi, S. Hilsenbeck, R. Schiff, K. C. Osborne, A. Letai and K. Polyak, Cancer Res., 2016, 76, 6495–6506 CrossRef CAS PubMed.
- E. Ostermann, P. Garin-Chesa, K. H. Heider, M. Kalat, H. Lamche, C. Puri, D. Kerjaschki, W. J. Rettig and G. R. Adolf, Clin. Cancer Res., 2008, 14, 4584–4592 CrossRef CAS.
- B. Brocks, P. Garin-Chesa, E. Behrle, J. E. Park, W. J. Rettig, K. Pfizenmaier and D. Moosmayer, Mol. Med., 2001, 7, 461–469 CAS.
- W. J. Rettig, S. L. Su, S. R. Fortunato, M. J. Scanlan, B. K. Raj, P. Garin-Chesa, J. H. Healey and L. J. Old, Int. J. Cancer, 1994, 58, 385–392 CrossRef CAS PubMed.
- P. Garin-Chesa, L. J. Old and W. J. Rettig, Proc. Natl. Acad. Sci. U. S. A., 1990, 87, 7235–7239 CrossRef CAS PubMed.
- R. D. Hofheinz, S. E. Al-Batran, F. Hartmann, G. Hartung, D. Jager, C. Renner, P. Tanswell, U. Kunz, A. Amelsberg, H. Kuthan and G. Stehle, Onkologie, 2003, 26, 44–48 CAS.
- A. M. Scott, G. Wiseman, S. Welt, A. Adjei, F. T. Lee, W. Hopkins, C. R. Divgi, L. H. Hanson, P. Mitchell, D. N. Gansen, S. M. Larson, J. N. Ingle, E. W. Hoffman, P. Tanswell, G. Ritter, L. S. Cohen, P. Bette, L. Arvay, A. Amelsberg, D. Vlock, W. J. Rettig and L. J. Old, Clin. Cancer Res., 2003, 9, 1639–1647 CAS.
- L. C. Wang, A. Lo, J. Scholler, J. Sun, R. S. Majumdar, V. Kapoor, M. Antzis, C. E. Cotner, L. A. Johnson, A. C. Durham, C. C. Solomides, C. H. June, E. Pure and S. M. Albelda, Cancer Immunol. Res., 2014, 2, 154–166 CrossRef CAS PubMed.
- J. Niedermeyer, M. Kriz, F. Hilberg, P. Garin-Chesa, U. Bamberger, M. C. Lenter, J. Park, B. Viertel, H. Puschner, M. Mauz, W. J. Rettig and A. Schnapp, Mol. Cell. Biol., 2000, 20, 1089–1094 CrossRef CAS PubMed.
- G. P. Adams and L. M. Weiner, Nat. Biotechnol., 2005, 23, 1147–1157 CrossRef CAS.
- R. Verma, R. E. Foster, K. Horgan, K. Mounsey, H. Nixon, N. Smalle, T. A. Hughes and C. R. Carter, Breast Cancer Res., 2016, 18, 10 CrossRef PubMed.
- M. Fabre, C. Ferrer, S. Dominguez-Hormaetxe, B. Bockorny, L. Murias, O. Seifert, S. A. Eisler, R. E. Kontermann, K. Pfizenmaier, S. Y. Lee, M. D. Vivanco, P. P. Lopez-Casas, S. Perea, M. Abbas, W. Richter, L. Simon and M. Hidalgo, Clin. Cancer Res., 2020, 26, 3420–3430 CrossRef CAS PubMed.
- B. C. Ozdemir, T. Pentcheva-Hoang, J. L. Carstens, X. Zheng, C. C. Wu, T. R. Simpson, H. Laklai, H. Sugimoto, C. Kahlert, S. V. Novitskiy, A. De Jesus-Acosta, P. Sharma, P. Heidari, U. Mahmood, L. Chin, H. L. Moses, V. M. Weaver, A. Maitra, J. P. Allison, V. S. LeBleu and R. Kalluri, Cancer Cell, 2014, 25, 719–734 CrossRef CAS PubMed.
- A. D. Rhim, P. E. Oberstein, D. H. Thomas, E. T. Mirek, C. F. Palermo, S. A. Sastra, E. N. Dekleva, T. Saunders, C. P. Becerra, I. W. Tattersall, C. B. Westphalen, J. Kitajewski, M. G. Fernandez-Barrena, M. E. Fernandez-Zapico, C. Iacobuzio-Donahue, K. P. Olive and B. Z. Stanger, Cancer Cell, 2014, 25, 735–747 CrossRef CAS.
- F. L. Tansi, R. Ruger, C. Bohm, F. Steiniger, R. E. Kontermann, U. K. Teichgraeber, A. Fahr and I. Hilger, Acta Biomater., 2017, 54, 281–293 CrossRef CAS PubMed.
- F. Viti, L. Tarli, L. Giovannoni, L. Zardi and D. Neri, Cancer Res., 1999, 59, 347–352 CAS.
- C. R. MacKenzie, T. Hirama, S. J. Deng, D. R. Bundle, S. A. Narang and N. M. Young, J. Biol. Chem., 1996, 271, 1527–1533 CrossRef CAS.
- F. L. Tansi, R. Ruger, A. M. Kollmeier, M. Rabenhold, F. Steiniger, R. E. Kontermann, U. K. Teichgraber, A. Fahr and I. Hilger, Pharmaceutics, 2020, 12, 370 CrossRef CAS PubMed.
- A. Schnyder and J. Huwyler, NeuroRx, 2005, 2, 99–107 CrossRef.
- C. C. Benz, G. K. Scott, J. C. Sarup, R. M. Johnson, D. Tripathy, E. Coronado, H. M. Shepard and C. K. Osborne, Breast Cancer Res. Treat., 1992, 24, 85–95 CrossRef CAS.
- J. Li, C. Menzel, D. Meier, C. Zhang, S. Dubel and T. Jostock, J. Immunol. Methods, 2007, 318, 113–124 CrossRef CAS.
- P. Carter, L. Presta, C. M. Gorman, J. B. Ridgway, D. Henner, W. L. Wong, A. M. Rowland, C. Kotts, M. E. Carver and H. M. Shepard, Proc. Natl. Acad. Sci. U. S. A., 1992, 89, 4285–4289 CrossRef CAS PubMed.
- TTY Biopharm Company Limited, Taiwan Pat., TWI270379B, 2007 Search PubMed.
- H. Benasutti, G. Wang, V. P. Vu, R. Scheinman, E. Groman, L. Saba and D. Simberg, Bioconjugate Chem., 2017, 28, 2747–2755 CrossRef CAS.
- M. T. Sheu, H. J. Jhan, C. Y. Su, L. C. Chen, C. E. Chang, D. Z. Liu and H. O. Ho, Colloids Surf., B, 2016, 143, 260–270 CrossRef CAS.
- J. Park, P. M. Fong, J. Lu, K. S. Russell, C. J. Booth, W. M. Saltzman and T. M. Fahmy, Nanomedicine, 2009, 5, 410–418 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: Supplementary Tables S1–S4 and Fig. S1–S4. See DOI: 10.1039/d1bm01218e |
| ‡ Michael Chen and Ming-Thau Sheu contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2022 |