Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Evaluating commercial thermoplastic materials in fused deposition modeling 3D printing for their compatibility with DNA storage and analysis by quantitative polymerase chain reaction†
Derek R.
Eitzmann
and
Jared L.
Anderson
 *
*
Department of Chemistry, Iowa State University, Ames, Iowa 50011, USA. E-mail: andersoj@iastate.edu
First published on 20th June 2022
Abstract
Nucleic acids are ubiquitous in biological samples and can be sensitively detected using nucleic acid amplification assays. To achieve highly accurate and reliable results, nucleic acid isolation and purification is often required and can limit the accessibility of these assays. Encapsulation of these workflows onto a single device may be achieved through fabrication methodologies featuring commercial three-dimensional (3D) printers. This study aims to characterize fused deposition modeling (FDM) filaments based on their compatibility with nucleic acid storage using quantitative polymerase chain reaction (qPCR). To study the adsorption of nucleic acids, storage vessels were fabricated using six common thermoplastics including: polylactic acid (PLA), nylon, acrylonitrile butadiene styrene (ABS), co-polyester (CPE), polycarbonate (PC), and polypropylene (PP). DNA adsorption of a short 98 base pair and a longer 830 base pair fragment to the walls of the vessel was shown to vary significantly among the polymer materials as well as the color varieties of the same polymer. PLA storage vessels were found to adsorb the least amount of the 98 base pair DNA after 12 hours of storage in 2.5 M NaCl TE buffer whereas the ABS and PC vessels adsorbed up to 97.2 ± 0.2% and 97.5 ± 0.2%. DNA adsorption could be reduced by decreasing the layer height of the 3D printed object, thereby increasing the functionality of the ABS storage vessel. Nylon was found to desorb qPCR inhibiting components into the stored solution which led to erroneous DNA quantification data from qPCR analysis.
Introduction
The molecular detection of nucleic acids is a crucial tool used to gain insight into genetic predespositions,1 disease identification,2 and disease prognosis.3 Nucleic acids found within biological samples co-exist with other components such as proteins, salts, and organic compounds which can significantly degrade the quality of downstream nucleic acid analysis.4,5 Purification and preconcentration approaches are often confined to laboratories equipped with significant resources and skilled technicians which decreases the accessibility of high quality nucleic acid analysis. The field of microfluidics aims to increase accessibility, reduce reagent costs, and decrease the need for highly skilled labor by shrinking entire laboratory workflows onto a single enclosed device. DNA analysis techniques such as electrophoresis,6 sequencing,7 and amplification-based assays8,9 have been successfully employed in the study of nucleic acids on microfluidic devices. However, without implementation of nucleic acid purification within the microfluidic device, its applicability to point-of-care (POC) diagnostics is often diminished. Custom-designed devices can accomplish these tasks with channels, reaction chambers, and flow modulation with syringe pumps, and are typically constructed using soft lithography to produce feature dimensions on the order of tens of micrometers; however, this can require significant effort and must be performed in a semi-clean room.10 The resulting cost and complexity of construction has been pointed out as a stumbling block for device commercialization and wider implementation.11–13Three-dimensional (3D) printing is an alternative fabrication method for microfluidic devices and is rising in prominence as it enables rapid transformation of digital 3D models into objects, while circumventing the preconditions of soft lithography.14–16 Furthermore, a wide variety of printing methods for the creation of microfluidic devices have been reported for various applications, including stereolithography (SLA), inkjet, fused deposition modeling (FDM), and many others. SLA printers use light to photopolymerize a liquid resin in a layer-by-layer fashion and have been extensively used to construct microfluidic devices.17–19 This approach is popular for microfluidic devices due to the optical clarity of resins and the minimum size of channel dimensions (approximately 250 μm and 18 μm have been achieved using commercial and customized printers, respectively).18,20 However, the use of a liquid resin as a printing medium leads to the capture of semi-polymerized resin within device channels and requires the device to be subjected to significant post-processing washes with organic solvents.17 Additionally, it has been shown that SLA-printed devices may be subject to channel blockages17,18 following post-processing as well as the questionable biocompatibility of acrylate-based resins due to the toxicity of residual unreacted resin.21
FDM is the most common 3D printing technique due to its simple mechanism of heating, liquification, precise deposition in a layer-by-layer fashion and cooling of thermoplastic materials. FDM is an attractive alternative as it is relatively inexpensive, requires no post-processing, produces channel widths as low as 260 μm,22 and offers a wide variety of compatible thermoplastic polymers. Furthermore, FDM 3D printers have been used to construct reactionware for organic23 and inorganic synthesis,22 as well as bioreactors for DNA amplification.24 Additionally, FDM printers are compatible with a plethora of thermoplastic polymers25 which can be doped with metals,26 pharmaceuticals,27 or conductive components.28,29 Furthermore, modern 3D printers have been developed with additional extrusion nozzles enabling the construction of devices with multiple materials, such as internal or external coatings, to impart additional functionalization.
Despite the advantages of deploying task-specific thermoplastic polymers, there is a current dearth of literature focused on material selection.30 One broad study examined 25 FDM materials in the fabrication of bioreactors for colorimetric loop-mediated isothermal amplification (LAMP) and found that several materials produced inaccurate results derived from the vessels leaching components leading to disruption of the biochemical reaction.24 Microfluidic devices featuring separate stages for sample purification are greatly preferred to increase the accuracy and reliability for downstream assays. It has been previously reported that the nature of materials comprising DNA storage vessels can drastically impact the extent of surface adsorption.31 In another study, it was demonstrated that vessels fabricated with FDM polymers can adsorb DNA in high ionic strength solutions leading to lower amounts of DNA measured downstream.32
Herein, we perform a systematic examination of six common FDM 3D printing filaments and study their compatibility with DNA storage and downstream amplification assays. Two DNA fragments were studied to mimic short, heavily fragmented DNA samples (98 base pair) and longer genes or plasmids (830 base pair) which are commonly stored by many users. A standard DNA solution at high and low salt concentrations was stored in FDM fabricated vessels and the amount of DNA in solution was monitored using a time-course. The vessels were screened for the leaching of inhibitory components into the contained solution that can affect the accuracy of downstream analysis. Color varieties of the acrylonitrile butadiene styrene (ABS) polymer, which are produced through the addition of different additives in the formulation process, exhibited significantly different DNA adsorption behavior. Lastly, it is shown that the adsorption of DNA can be partially mitigated using printing parameters that decrease surface roughness and increase vessel functionality.
Experimental
Reagents
All primers used in this study (Table S1 of the ESI†) were purchased from Integrated DNA Technologies (Coralville, IA, USA) and purified with standard desalting. Deionized water (18.2 MΩ cm) used for all experiments was obtained from a Millipore Milli-Q purification system (Bedford, MA, USA). Buffers were prepared using Tris(hydroxymethyl)aminomethane (Tris) (98%) obtained from P212121 (Ypsilanti, MI, USA), sodium chloride (≥99.0%) from Fisher Scientific (Hampton, NH, USA), ethylenediaminetetraacetic acid dipotassium salt dihydrate (99%) from Acros Organics (Geel, Belgium) and pH adjusted with hydrochloric acid (Fisher Scientific).FDM printer and filaments
All filaments (2.85 mm) used in the study were acquired from Dynamism (Chicago, IL, USA) and printed using an Ultimaker S5 printer (Utrecht, The Netherlands). For creation of DNA adsorption-time profiles, Ultimaker white acrylonitrile butadiene styrene (ABS), Ultimaker transparent polylactic acid (PLA), Ultimaker natural polypropylene (PP), Ultimaker transparent co-polyester (CPE), Ultimaker transparent nylon, and Ultimaker transparent polycarbonate (PC) were employed. For subsequent testing of additional ABS filament replicates, Ultimaker red, grey, and Dynamism white were used.Preparation of DNA templates
Two model DNA templates, found in Table S1,† were deployed in this study to compare the adsorption behavior of small and larger fragments. The smaller 98 bp fragment was generated as a PCR amplicon from a plasmid containing a 210 bp BRAF gene insert using the primers and a standard qPCR protocol. The amplicon was separated on a 2.0% agarose gel using a H4 gel bed (Bethesda Research Laboratories, Gaithersburg, MD) coupled to a Neo/Sci (Rochester, NY) power supply. Following separation, the amplicon bands were excised and purified by QIA quick gel extraction kit (Qiagen, Hilden, Germany). The larger 830 bp dsDNA fragment was synthesized by Integrated DNA Technologies and contains the same internal 98 bp sequence of the small fragment. Both DNA samples were quantified by the high sensitivity dsDNA assay using a Invitrogen 2.0 Qubit Fluorometer (Fisher Scientific) prior to study.qPCR amplification protocol
Amplification of both DNA sequences was carried out on a Bio-Rad CFX96 Touch real-time PCR system (Hercules, CA, USA). Each 20.0 μL reaction contained 10.0 μL of SsoAdvanced Universal SYBR green supermix (Bio-Rad), 1.0 μL of 10 μM forward and reverse primers, 8.0 μL of water, and 1.0 μL of 200 mM NaCl DNA containing solution. The reactions were heated to 95.0 °C for 2 minutes, followed by 40 cycles of 95.0 °C for 5 seconds and 60.0 °C for 30 seconds followed by a plate read. Cycle of quantification (Cq) values were determined by manually setting a threshold of 500 RFU for all reactions.DNA adsorption time-course
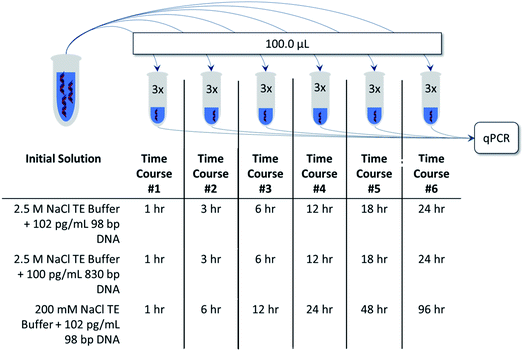
For this work, a 10.0 mM Tris–HCl (pH 7.50) and 1.0 mM EDTA (TE) buffer was prepared to control DNA solubility and prevent enzymatic degradation. The TE buffer also included 2.5 M NaCl to facilitate DNA adsorption. To represent more modest salt concentrations, the previous TE buffer was diluted to a final concentration of 200 mM NaCl. These solutions were either spiked with DNA to a concentration of 100 pg mL−1 for the 830 bp long fragment or 102 pg mL−1 for the 98 bp short fragment. A 100.0 μL portion of the spiked TE buffer was added in triplicate to vessels, covered, and allowed to sit for a time-course. Following the end of the time-course, a portion of the solution was removed from the vessel and subjected to qPCR analysis. A schematic of this procedure is shown in Fig. 1 along with the corresponding time-courses used for each experiment.Water contact angle measurements
Sessile drop contact angle measurements were performed to determine the correlation of surface wettability and DNA adsorption of selected 3D printed materials. A rectangular, L-shaped 3D model (Fig. S1(A)†) was generated so that two large surfaces featuring significantly different morphologies are produced. The upright wall surface (Fig. S1(B)†) features a ridged morphology derived from the individually deposited layers which mimics the inside of the storage vessel. The horizontal face represents a flat surface that is formed using a single layer of extruded filament and provides a simpler surface for contact angle measurements. A video-based OCA 15Pro instrument (Charlotte, NC, USA) was used to determine contact angles for both surfaces following water droplet equilibration.Results and discussion
Development of 3D printed storage vessels
Commonly, DNA samples are stored in commercial microcentrifuge tubes. These vessels were found to be an ideal benchmark for the FDM materials, and subsequently a 3D model matching the internal geometry of microcentrifuge tubes featuring a volume of 1.5 mL was generated using Autodesk's Inventor software (San Rafael, CA). Vessels were printed using the following Ultimaker filaments: acrylonitrile butadiene styrene (ABS), polylactic acid (PLA), polypropylene (PP), co-polyester (CPE), nylon, and polycarbonate (PC). Images of the printed vessels are shown in Fig. S2† and were printed using optimized settings shown in Table S2.†Investigating DNA adsorption within 3D printed storage vessels
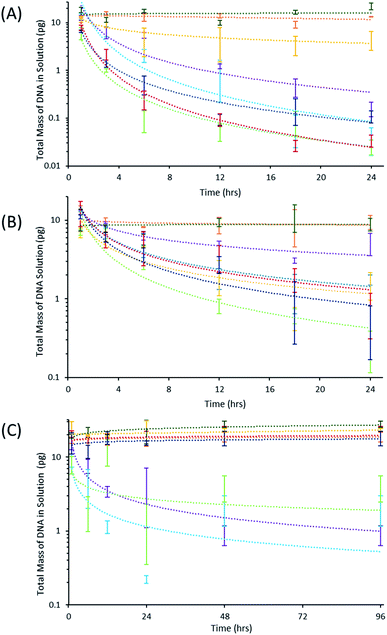
Quantitative polymerase chain reaction (qPCR) is the gold standard analysis technique for measuring small amounts of DNA and employs a thermally-regulated biochemical reaction to exponentially amplify DNA present in a sample. By monitoring the amplification in real-time, the time required for amplification is inversely related to the initial concentration of DNA allowing for the original template DNA mass to be determined in an optimized assay. In this study, standard curves for a short 98 bp and longer 830 bp BRAF DNA template were constructed and are shown in Fig. S3.†To study DNA adsorption characteristics, the 2.5 M NaCl TE buffer was spiked with 102 pg mL−1 of the small DNA fragment, aliquoted into each vessel, and stored for a time-course. The concentration of DNA was monitored with qPCR and the results are shown in Fig. 2A. The choice of polymer material significantly impacts the efficacy of the vessels for DNA storage. For example, nylon and PLA materials exhibited little decrease in the DNA mass over a period of 24 hours; however, a measurable decrease in DNA mass was observed for all other polymers. Varying adsorption rates were observed for the polymer materials, including CPE, which showed little adsorption followed by a leveling off at longer time points (Fig. 2A). From this data, the percent of DNA adsorbed was calculated by comparing the DNA remaining in solution to the initial DNA standard and is shown in Table S3A.† Interestingly, vessels constructed from the nylon filament yielded higher amounts of DNA than the actual initial DNA spike and produced large negative (>−20.0%) adsorption values. However, it is important to note that inhibitory molecules present in the DNA sample interfere with the enzymatic reaction leading to inaccurate DNA quantification.4 This result may be due to the nylon material leeching components into the stored solution which leads to inaccurate quantification of DNA within the storage vessel, thereby making it impossible to compare to the other vessels. At the end of the time-course, it was determined that PLA adsorbed the least DNA followed by CPE, then PP and the DNA LoBind® vessels, while the PC, ABS, and the Fisherbrand™ vessels adsorbed the most.
The 830 bp DNA sequence was employed with the 2.5 M NaCl TE buffer to study the effect of larger sequences on adsorption. Once again, nylon and PLA did not produce a significant decrease in the DNA mass, as shown in Fig. 2B. A decreased rate of DNA adsorption was observed, and is likely due to the smaller diffusion coefficient of the larger DNA fragment (Fig. 2B). DNA LoBind® and Fisherbrand™ vessels were both observed to adsorb greater than 96% of the short DNA template at the end of the time-course (Table S3A†), but the DNA LoBind® vessels adsorbed a significantly smaller fraction of the longer 830 base pair fragment, approximately 59.3% and 86.3%, respectively (Table S3B†). PLA adsorbed the least amount of the 830 bp DNA fragment, followed by the DNA LoBind® vessels at the conclusion of the time-course. The Fisherbrand™ and CPE vessels showed intermediate adsorption results compared to high DNA adsorption of the ABS, PP, and PC vessels.
DNA storage conditions involving high concentrations of NaCl may not reflect realistic working conditions; therefore, a TE buffer containing 200 mM NaCl was examined. It is important to note that the decreased salt concentration led to decreased DNA adsorption and required the use of a longer time-course. The results shown in Fig. 2C demonstrate that most FDM materials did not adsorb a measurable amount of DNA over the course of the study. Two of these materials, nylon and CPE, intermittently produced negative percent adsorption values, but it is only with nylon that these values are statistically significant for experiments carried out at the 48 and 96 hour time-courses (Table S3C†). The concentration of DNA was only observed to decrease in the commercial DNA LoBind®, Fisherbrand™, and Ultimaker PC vessels (Fig. 2C). High variation in the measured DNA mass for these vessels indicates DNA adsorption may be facilitated by impurities which vary from vessel-to-vessel at low salt concentrations.
Survey of 3D printed storage vessels for leaching of qPCR inhibitors
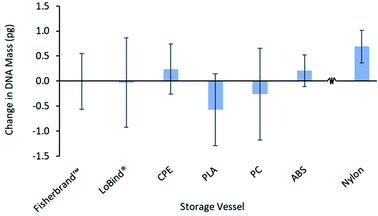
For FDM materials to be compatible in the analysis of DNA, the vessels should not desorb inhibitory molecules into the solution which may negatively influence the accuracy and reliability of results from downstream amplification assays.5 To thoroughly examine this phenomenon for the studied vessels, they were used to store the 200 mM NaCl TE buffer for 24 hours enabling components of the vessels to leach into the solution. Following this time-course, a portion of the solution was removed and a known amount of DNA standard was added prior to qPCR. As a control, the same amount of DNA standard was added to fresh solution and immediately subjected to qPCR analysis allowing for a direct comparison of the stored versus fresh solution. The results shown in Fig. 3 indicate that all storage vessels were compatible with qPCR, except for nylon, as the experimental data revealed no deviation from the control. Through this experiment, it can be concluded that the nylon material likely desorbs components into the solution leading to contamination of the qPCR reaction and inaccurate quantification.Effect of filament color varieties and printing conditions on DNA adsorption behavior
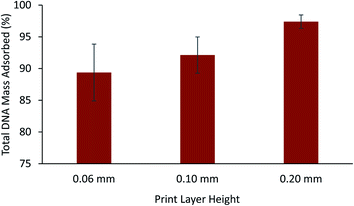
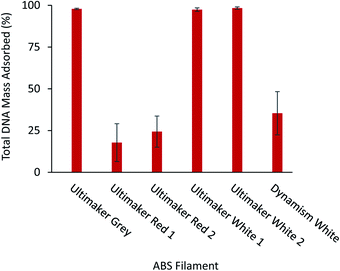
Prior to the printing process, slicing software such as Cura (Ultimaker, The Netherlands) was used to convert models into horizontal slices providing a path for the printer nozzle to travel for fabrication of the object. A fundamental parameter in the printing process is the vertical height of each horizontal slice which determines the surface quality, surface roughness, and overall print time. To determine the effect of the layer height on the functionality of the printed vessel for DNA storage, three sets of vessels were created featuring decreasing layer heights using the white Ultimaker ABS filament. The amount of DNA remaining in solution following a time-course is shown for each vessel in Fig. 4. The vessel featuring the largest layer height (0.20 mm) was shown to adsorb approximately 98.4% of DNA from the solution compared to an approximate 89.4% using vessels printed with the smallest layer height of 0.06 mm. It has been shown previously that printing with smaller layer heights reduces surface roughness33,34 leading to decreased vessel surface area and DNA adsorption.For any given thermoplastic material, a choice of color varieties is often available and are created through the addition of color additives. To study the effect that color additives may play in DNA adsorption, the ABS filament was chosen since it was observed to adsorb a significant amount of DNA (Fig. 2A–C) and many colors are readily available for purchase. A total of 6 various filament rolls were used to compare the adsorption of short DNA fragments for a time-course of 6 hours using the 2.5 M NaCl TE buffer. As shown in Fig. 5, vessels fabricated with the grey and white filaments were observed to adsorb 98.8% and 98.4% of the total DNA from solution, respectively. Significantly less total DNA (approximately 24.4%) was adsorbed in vessels fabricated with the red ABS filament. This finding was also reproduced using replicate rolls of the white and red ABS filament (labeled as Ultimaker White 2 and Ultimaker Red 2 in Fig. 5) and no statistically significant difference in the adsorption of DNA was found. Furthermore, a roll of white ABS filament acquired from a separate manufacturer, Dynamism, was found to adsorb approximately 35.4% of the total DNA compared to the same color of Ultimaker filament (98.4% and 97.4%, Fig. 5). To compare the surface wettability of the white ABS filaments originating from separate manufacturers, sessile drop contact angle measurements were performed. As shown in Table S4,† it was found that similar water contact angles were obtained for the Ultimaker and Dynamism white ABS filaments with both the wall and flat surfaces (see Fig. S1†) indicating that both of these surfaces have similar hydrophilicity and does not completely explain the differences in interfacial interactions or DNA adsorption.
Conclusions
Since microfluidic devices utilize small volumes and channel dimensions, interactions between analytes and the material comprising the device must be considered to avoid analyte loss and inaccurate analysis. In this study, several FDM thermoplastic filaments were examined to assess their compatibility with DNA storage and downstream analysis. DNA adsorption within 3D printed vessels was monitored by qPCR analysis for two DNA sequences of varied size and buffers containing 2.5 M or 200 mM NaCl. PLA storage vessels were shown to be very compatible with DNA storage as less than 20% of the initial DNA mass of either DNA fragment was adsorbed after 24 hours in 2.5 M NaCl TE buffer. Less compatible materials such as PP, ABS, and PC adsorbed approximately 98% of the initial DNA in the same period of time and salt concentration. To decrease DNA adsorption, it was shown that reducing the layer height from 0.2 mm to 0.06 mm and reducing the surface of the walls led to a decrease in adsorption from approximately 97.4% to 89.4%. The nylon filament was determined to be incompatible with qPCR analysis due to leaching of inhibiting components, leading to inaccurate quantification. Results from this study will assist users in identifying suitable materials for 3D printed devices that minimize DNA loss and/or inhibition of downstream amplification assays. Conversely, the results may prove useful to other users who seek to use thermoplastic materials as a simple approach to remove small amounts of DNA from solution.Conflicts of interest
The authors have no conflicts of interest to declare.Acknowledgements
The authors acknowledge financial support from the Chemical Measurement and Imaging Program at the National Science Foundation (Grant No. CHE-1709372). J. L. A. and D. R. E. thank the Alice Hudson Professorship for generous support of this work. Shu-An Hsieh is thanked for assisting in designing the 3D model used in this study.References
- G. Kurzawski, D. Dymerska, P. Serrano-Fernández, J. Trubicka, B. Masojć, A. Jakubowska and R. J. Scott, Hered. Cancer Clin. Pract., 2012, 10, 17 CrossRef PubMed.
- J. Löffler, H. Hebart, U. Schumacher, H. Reitze and H. Einsele, J. Clin. Microbiol., 1997, 35, 3311–3312 CrossRef PubMed.
- M. Xing, W. H. Westra, R. P. Tufano, Y. Cohen, E. Rosenbaum, K. J. Rhoden, K. A. Carson, V. Vasko, A. Larin, G. Tallini, S. Tolaney, E. H. Holt, P. Hui, C. B. Umbricht, S. Basaria, M. Ewertz, A. P. Tufaro, J. A. Califano, M. D. Ringel, M. A. Zeiger, D. Sidransky and P. W. Ladenson, J. Clin. Endocrinol. Metab., 2005, 90, 6373–6379 CrossRef CAS PubMed.
- M. Emaus, M. Varona, D. Eitzmann, S.-A. Hsieh, V. Zeger and J. Anderson, TrAC, Trends Anal. Chem., 2020, 130, 115985 CrossRef CAS.
- C. Schrader, A. Schielke, L. Ellerbroek and R. Johne, J. Appl. Microbiol., 2012, 113, 1014–1026 CrossRef CAS PubMed.
- D. Wu, J. Qin and B. Lin, J. Chromatogr. A, 2008, 1184, 542–559 CrossRef CAS PubMed.
- F. Lan, B. Demaree, N. Ahmed and A. R. Abate, Nat. Biotechnol., 2017, 35, 640–646 CrossRef CAS PubMed.
- A. K. White, M. VanInsberghe, O. I. Petriv, M. Hamidi, D. Sikorski, M. A. Marra, J. Piret, S. Aparicio and C. L. Hansen, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 13999 CrossRef CAS PubMed.
- H. Zhang, Y. Xu, Z. Fohlerova, H. Chang, C. Iliescu and P. Neuzil, TrAC, Trends Anal. Chem., 2019, 113, 44–53 CrossRef CAS PubMed.
- S. K. Tiwari, S. Bhat and K. K. Mahato, Sci. Rep., 2020, 10, 9215 CrossRef CAS PubMed.
- N. Convery and N. Gadegaard, Micro Nano Eng., 2019, 2, 76–91 CrossRef.
- P. Cui and S. Wang, J. Pharm. Anal., 2019, 9, 238–247 CrossRef PubMed.
- V. Ortseifen, M. Viefhues, L. Wobbe and A. Grünberger, Front. Bioeng. Biotechnol., 2020, 8 DOI:10.3389/fbioe.2020.589074.
- C. Chen, B. T. Mehl, A. S. Munshi, A. D. Townsend, D. M. Spence and R. S. Martin, Anal. Methods, 2016, 8, 6005–6012 RSC.
- B. C. Gross, J. L. Erkal, S. Y. Lockwood, C. Chen and D. M. Spence, Anal. Chem., 2014, 86, 3240–3253 CrossRef CAS PubMed.
- V. Mehta and S. N. Rath, Bio-Des. Manuf., 2021, 4, 311–343 CrossRef.
- A. I. Shallan, P. Smejkal, M. Corban, R. M. Guijt and M. C. Breadmore, Anal. Chem., 2014, 86, 3124–3130 CrossRef CAS PubMed.
- A. K. Au, W. Lee and A. Folch, Lab Chip, 2014, 14, 1294–1301 RSC.
- Z. Chen, J. Y. Han, L. Shumate, R. Fedak and D. L. DeVoe, Adv. Mater. Technol., 2019, 4, 1800511 CrossRef.
- H. Gong, B. P. Bickham, A. T. Woolley and G. P. Nordin, Lab Chip, 2017, 17, 2899–2909 RSC.
- S. M. Oskui, G. Diamante, C. Liao, W. Shi, J. Gan, D. Schlenk and W. H. Grover, Environ. Sci. Technol. Lett., 2016, 3, 1–6 CrossRef CAS.
- L. P. Bressan, T. M. Lima, G. D. da Silveira and J. A. F. da Silva, SN Appl. Sci., 2020, 2, 984 CrossRef CAS.
- P. J. Kitson, M. H. Rosnes, V. Sans, V. Dragone and L. Cronin, Lab Chip, 2012, 12, 3267–3271 RSC.
- A. K. Pantazis, G. Papadakis, K. Parasyris, A. Stavrinidis and E. Gizeli, Sens. Actuators, B, 2020, 319, 128161 CrossRef CAS.
- M. D. Symes, P. J. Kitson, J. Yan, C. J. Richmond, G. J. T. Cooper, R. W. Bowman, T. Vilbrandt and L. Cronin, Nat. Chem., 2012, 4, 349–354 CrossRef CAS PubMed.
- Z. Liu, Q. Lei and S. Xing, J. Mater. Res. Technol., 2019, 8, 3741–3751 CrossRef CAS.
- J. A. Weisman, U. Jammalamadaka, K. Tappa and D. K. Mills, Bioeng, 2017, 4, 96 Search PubMed.
- K. Gnanasekaran, T. Heijmans, S. van Bennekom, H. Woldhuis, S. Wijnia, G. de With and H. Friedrich, Appl. Mater. Today, 2017, 9, 21–28 CrossRef.
- R. H. Sanatgar, A. Cayla, C. Campagne and V. Nierstrasz, J. Appl. Polym. Sci., 2019, 136, 47040 CrossRef.
- D. Pranzo, P. Larizza, D. Filippini and G. Percoco, Micromachines, 2018, 9, 374 CrossRef PubMed.
- C. Gaillard and F. Strauss, Tech. Tips Online, 1998, 3, 63–65 CrossRef.
- D. R. Eitzmann, M. Varona and J. L. Anderson, Anal. Chem., 2022, 94, 3677–3684 CrossRef CAS PubMed.
- R. Anitha, S. Arunachalam and P. Radhakrishnan, J. Mater. Process. Technol., 2001, 118, 385–388 CrossRef.
- P. J. Nuñez, A. Rivas, E. García-Plaza, E. Beamud and A. Sanz-Lobera, Procedia Eng., 2015, 132, 856–863 CrossRef.
Footnote |
| † Electronic supplementary information (ESI) available. See https://doi.org/10.1039/d2ay00772j |
| This journal is © The Royal Society of Chemistry 2022 |