Multi-element stable isotope Raman microspectroscopy of bacterial carotenoids unravels rare signal shift patterns and single-cell phenotypic heterogeneity†
Julian
Weng
 ,
Kara
Müller
,
Kara
Müller
 ,
Oleksii
Morgaienko
,
Martin
Elsner
,
Oleksii
Morgaienko
,
Martin
Elsner
 and
Natalia P.
Ivleva
and
Natalia P.
Ivleva
 *
*
Technical University of Munich, Institute of Water Chemistry, Chair for Analytical Chemistry and Water Chemistry, Lichtenbergstr. 4, 85748 Garching, Germany. E-mail: natalia.ivleva@tum.de
First published on 29th November 2022
Abstract
The combination of single-cell Raman microspectroscopy (SCRM) and stable isotope probing (SIP) enables in situ tracking of carbon or hydrogen fluxes into microorganisms at the single-cell level. Therefore, it has high potential for the analysis of metabolic processes and biogeochemical cycles. However, especially for high throughput applications such as imaging or cell sorting, it is hampered by low Raman scattering intensities (and therefore long acquisition times). In order to overcome these limitations, this study brings forward a systematic investigation of Resonance Raman (RR) enhanced SCRM for SIP of bacterial carotenoids. Dynamic carbon uptake from 13C-glucose was successfully monitored and quantified utilizing 13C stable isotope-induced red-shifts of RR signals. High single-cell phenotypic heterogeneity was revealed in terms of carbon uptake and, unlike in previous studies, clear evidence for de novo synthesis of carotenoids was found. For the first time, hydrogen uptake into carotenoids was systematically investigated by deuterium labeling (providing a direct probe for metabolic activity of single cells). In carotenoid single-cell Resonance Raman (SCRR) spectra, a unique pattern of signal red-shifts and apparent blue-shifts was observed and quantitatively evaluated. Finally, a novel combined approach for simultaneous monitoring of carbon and hydrogen uptake revealed complementary effects in carotenoid SCRR spectra that can be analyzed in parallel. Overall, it was shown that the high RR intensity, simplicity of spectral features and straightforward signal processing make microbial carotenoids an ideal target for quantitative multi-element SIP, with great potential for high throughput applications.
1. Introduction
Microorganisms play essential roles in biogeochemical cycles as they are responsible for the conversion of nutrients from abiotic to biotic compartments.1,2 For a better understanding of biogeochemical processes, it is crucial to identify active microbes as well as to clarify their ecological functions and metabolic pathways.3–6 In this context, the involved element fluxes (e.g. hydrogen and carbon) have to be monitored which can conclusively be achieved by stable isotope probing (SIP).7–9 However, several challenges for the analysis are associated with this approach. Microbes are commonly isolated and grown in pure culture before investigation. Yet, a vast majority of species remains unculturable10–12 or behaves differently in the absence of their natural habitat which necessitates an in situ approach.13–15 Additionally, to differentiate between functions of community members, to investigate cell–cell-interactions and to unravel phenotypical diversity within single species, individual cells need to be probed.14,16–18The combination of Raman microspectroscopy (RM) with SIP provides an ideal platform for this.19,20 It has proven to be useful for in situ studies of microorganisms in complex environments like seawater,6,9,21,22 soil5 or biological samples.3,4 Moreover, by achieving the high spatial resolution of a confocal optical microscope (i.e. in the low micrometer range), it allows to acquire Raman spectra of single bacteria cells (denoted as single-cell Raman microspectroscopy – SCRM).
Cellular biomolecules like proteins, nucleic acids, lipids or chromophoric pigments are Raman active and show vibrational fingerprint spectra.20,23,24 Thus, SCRM provides whole-organism fingerprints for the characterization and identification of different biological systems. SCRM-SIP is based on the dependency of vibrational frequencies on the reduced mass of involved elements.15 Isotopologues with heavier isotopes (e.g. D or 13C) are characterized by bands shifted towards lower wavenumbers, so called red-shifted bands.
Compared to alternative single-cell SIP approaches like nanoscale secondary ion mass spectrometry (nano-SIMS),5,25 SCRM requires only little sample preparation, is non-destructive and provides additional structural chemical information. The relatively rapid and easy measurements allow high throughput screening, a prerequisite for reliable statistic evaluation of single-cell data, especially for complex biological consortia.26,27 In addition, high throughput analysis opens new horizons like Raman imaging for spatial distributions21,28 or Raman-based cell counting29 and sorting.28,30–33
However, since these applications rely on the rapid measurement and detection of sharp Raman bands, they are limited by the low quantum efficiency of the Raman effect. One option to overcome low Raman intensities is Resonance Raman (RR), i.e. when the excitation wavelength of the laser is close to an electronic transition of the compound leading to coupled enhancement of vibrational and electronic states.20,34 Consequently, single-cell Resonance Raman (SCRR) requires only low laser powers at the sample (preventing photodegradation) and short acquisition times (enabling rapid spectra acquisition for high throughput applications).32 Common targets for SCRR are cellular pigments like carotenoids or cytochromes.35 Carotenoids are widespread in photoautotrophic microorganisms, where they are either used for light-harvesting or as protection against reactive singlet oxygen species.9,35,36
In previous studies, Li et al. were able to track carbon dioxide fixation of single photosynthetic bacteria cells by combination of 13C-SIP and SCRR of carotenoids.21 Linear relationships of 13C-content and gradual signal red-shifts were identified and the high potential for rapid Raman imaging was demonstrated. Taylor et al. evaluated 13C-induced red-shifts of carotenoid signals quantitatively in order to obtain single cell growth rates.9
Despite these promising results, the application of SCRR for carotenoid SIP is still subject to several limitations. (i) The dynamics of 13C-uptake were up to now solely investigated for labeled bicarbonate carbon sources and isotope mixtures.9,21 While this is excellent to probe for carbon dioxide fixation, it does not represent more nutrient-rich substances. In this study, it is shown that dynamic signal shifts considerably deviate for fast-growing bacteria in rich media. (ii) Previous SCRR carotenoid SIP approaches are constrained to the exclusive tracking of 13C-flows which has several pitfalls.3 Many compounds are very expensive or not at all commercially available in a 13C-labeled form. Furthermore, the composition of suitable substances is not known for the majority of ecosystems. In many cases, interrupting and biasing of microbial communities by adding external organic carbon sources might not be wanted. Therefore, in this study the scope of SCRR carotenoid SIP is extended to D-labeling with heavy water (D2O) which was introduced by Berry et al. as a more general marker for tracking metabolic activity in SCRM.3 It is systematically analyzed how the microbial uptake of D is quantitatively reflected in SCRR spectra of carotenoid forming bacteria (also in comparison with Raman spectra of other cellular biomass). Additionally, a multiple stable isotope approach (13C and D in parallel) is explored, combining the benefits and orthogonal information of both labeling schemes.
2. Experimental
2.1. Growth conditions and stable isotope labeling
Unless otherwise stated, medium constituents and materials were purchased from Carl Roth (Karlsruhe, Germany) and Sigma-Aldrich (Steinheim, Germany).The Sphingomonas sp. strain was isolated from aged suspensions of polylactide (PLA) microparticles in water prepared according to the procedure of von der Esch et al.37 After several weeks of storage in glass vials at ambient temperature, microbial growth was observed in scanning electron microscopy (SEM) images of the sample. Isolation of bacteria was carried out from single colonies of a spread plate (NZCYM agar) using an inoculation loop and subsequent pure culture in NZCYM medium. Orange pigmented bacteria were identified as Sphingomonas sp. by matrix-assisted laser desorption/ionization time of flight mass spectrometry (MALDI-TOF-MS) and 16S rRNA sequencing (based on SepsiTest Blast, Molzym, Bremen, Germany).
In this study, Sphingomonas sp. was cultivated in minimal M9 medium (optimized composition according to Weiss et al.38) supplemented with trace elements (Trace Metal Mix A5, Merck, Darmstadt, Germany). The exact M9 medium composition is presented in ESI Table 1.† As the sole carbon source, 4 g L−1 unlabeled D-glucose, 13C6-labeled D-glucose (99 atom% 13C) or their corresponding mixtures were applied. Media were prepared with ultrapure water (Milli-Q™, Merck, Darmstadt, Germany). For deuteration experiments, mixtures of ultrapure water and D2O (99.9 atom% D) were used. Cultivation was carried out in reaction vials (1.5 mL, PP) in an incubator shaker at 37 °C and 100 rpm. Experiments were performed in triplicates with a negative control (M9 medium without bacteria inoculation).
2.2. Sample preparation and single-cell Raman microspectroscopy
Bacteria were harvested in stationary growth phase after at least three days of incubation (with the exception of the 13C-labeling dynamics time series). After centrifugation (6000 rpm, 3 min), the cell pellet was washed three times by resuspension and centrifugation with ultrapure water (with twice the volume of the initial sample) and finally resuspended in ultrapure water. The samples harvested in stationary phase were additionally 1/100 diluted with ultrapure water to avoid cell aggregation during deposition on the sample carrier. 1 μL of cell suspension was pipetted on an Al-coated glass slide (EMF Dynasil, USA) and dried in a laminar flow box (ENVIAR eco air, Germany).SCRM was conducted with a apyron confocal Raman microscope (WITec, Germany) equipped with a power-adjustable frequency-doubled Nd:YAG laser (532 nm) and a 300 g mm−1 diffraction grating. Al-coated glass slides with dried sample spots were placed on a motorized microscope stage. For each sample spot, a defined number of random coordinates was determined for the measurements (cf. ESI† for detailed procedure). A 100× objective (Zeiss EC Epiplan-Neofluar, NA = 0.9, WD = 1 mm, Zeiss, Germany) was used for single-cell measurements. SCRR spectra were recorded with an integration time of 2 s and a laser power of 1 mW at the sample (leading to Raman fingerprints dominated by bacterial carotenoids). These parameters were adjusted to 20 s and 10 mW for regular SCRM spectra, respectively (leading to Raman fingerprints dominated by cellular biomolecules other than carotenoids).
2.3. Spectra processing and fitting procedures
SCRM spectra were processed and analyzed with an in-house script in R (R-4.0.3, R Core Team, Vienna, Austria). A detailed description of the automated processing steps (including rolling ball background-correction, min–max scaling, averaging, peak picking, principal component analysis) is given in the ESI.† For determination of the exact signal positions and signal full width at half maximum (FWHM) in SCRR spectra, a peak fitting algorithm based on a Gaussian density function was applied on the individual spectra. Likewise, the ratio of signal integrals I12C/(I12C + I13C) was calculated based on the integrals of Gaussian fits. A detailed explanation of the fitting algorithm and its modification for each case can be found in the ESI.†3. Results and discussion
3.1. Red-shifts of carotenoid Resonance Raman signals quantitatively reflect 13C-uptake of Sphingomonas sp. cells
The subject of this study is Sphingomonas sp., a bacterial strain isolated from aged suspensions of PLA microparticles. Upon examination by SCRM, RR signals were observed (ESI Fig. 1A†) that can be assigned to bacterial carotenoids.9,20,35 The three predominant signals in the spectrum – designated as ν1, ν2 and ν3 – represent C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C stretchings, C–C stretchings and methyl rocking vibrations (cf. carotenoid scheme in ESI Fig. 1B†).35
C stretchings, C–C stretchings and methyl rocking vibrations (cf. carotenoid scheme in ESI Fig. 1B†).35
A time series of consecutive SCRM spectra of one single Sphingomonas sp. cell is shown in ESI Fig. 2.† Due to their strongly enhanced Raman intensity, carotenoid signals dominate the integrated SCRM spectra for short integration times (cf. orange spectrum of ESI Fig. 2†). However, they are rapidly photobleached by the irradiated laser and therefore signals representing other cellular biomolecules (i.e. microbial biomass) become prevalent (cf. blue spectrum of ESI Fig. 2†).
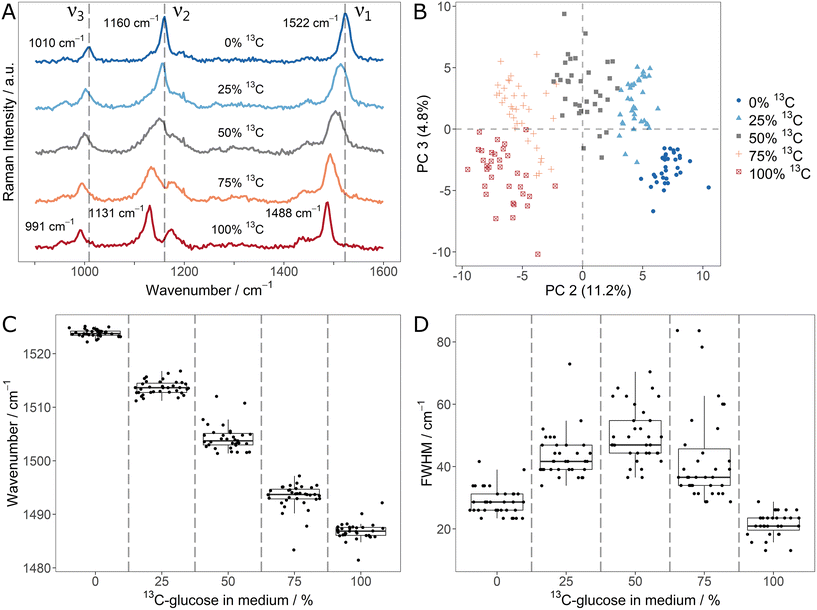
In order to explore the changes of carotenoid SCRR signals upon incorporation of different stable isotopes, Sphingomonas sp. was used as a model microorganism. First, it was cultivated with mixtures of unlabeled glucose and 13C6-glucose. The average SCRR spectra (n = 35) of five different isotope contents are shown in Fig. 1A. For comparison, the normal SCRM analogues (representing other cellular biomass) are shown in ESI Fig. 3† (average spectra, n = 30–40). As reported earlier by Li et al., the three dominant RR signals in carotenoid spectra are gradually red-shifted with increasing 13C-content.21 The individual SCRR spectra of each 12C/13C-mixture are clearly distinguished for each condition as reflected in the separate clusters of the PCA depicted in Fig. 1B.
The fitted ν1-signal positions in Fig. 1C suggest a linear correlation of red-shift and isotope content as proposed by Li et al.21 (examples of Gaussian fits of individual carotenoid ν1-signals and their residuals are shown in ESI Fig. 4 and 5†). Remarkably, the cellular isotope content is also clearly reflected in the FWHM of the carotenoid signals, as quantified for the ν1-signal in Fig. 1D. The lowest FWHMs are observed for pure 12C or 13C media. For mixtures, the FWHMs as well as the intra-population variability (i.e. interquartile range – IQR – of the boxplots) increase gradually. In average, the FWHM reaches a maximum at equal isotope content.
Both, the gradual shift and the increased FWHM for isotope mixtures can be explained by the presence and overlap of signals corresponding to different isotopologues/isotopomers. Taylor et al. suggested a superposition of three signal positions related to unlabeled (12C–12C), partially labeled (12C–13C) and fully labeled (13C–13C) vibrations.9 While this is plausible for the ν1- and ν2-signal, the ν3-signal represents methyl rocking vibrations. Based on density functional theory (DFT) calculations by Tschirner et al., only one carbon atom is directly associated to them.35 Nonetheless, it can be assumed that also adjacent labeled atoms in the carotenoid chain contribute indirectly to the vibration modes and therefore to red-shifts of the vibrational signals. This would explain the gradual character of the signal shift as well as the increased FWHM.
As reported earlier by Li et al., the linearity of 12C/13C stable isotope content and signal shifts may not only allow identification of isotope-labeled cells but also quantification of 13C-incorporation.21 The present study shows that the signal FWHM can be used as an additional independent indicator. Apart from the considerably higher Raman intensity of SCRR carotenoid spectra, the simple quantification of the stable isotope content is an advantage over normal SCRM spectra representing cellular biomass. As shown in the corresponding average SCRM spectra (ESI Fig. 3†), not all signals are red-shifted (e.g. Amide 3 vibrations). Furthermore, instead of gradual shifts, signal separation may be observed for isotopologues, hampering direct analysis (e.g. phenylalanine as reported by Kubryk et al.34).
3.2. Monitoring of 13C-uptake dynamics with SCRR reveals high phenotypic heterogeneity on a single-cell level
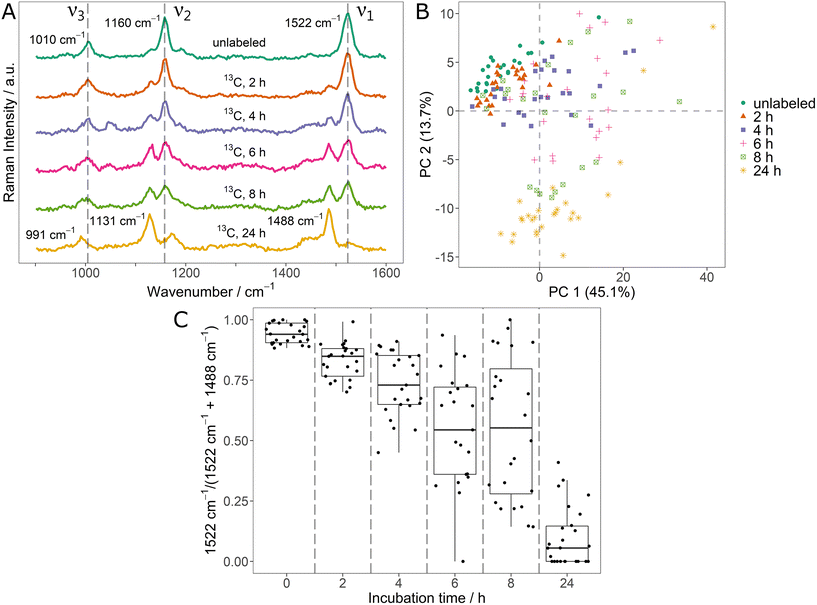
In the context of monitoring stable isotope uptake, it is highly relevant to dynamically track the element fluxes as they determine incorporation rates and metabolism kinetics.17,18 For this, the potential of SCRR carotenoid spectra was tested on initially unlabeled Sphingomonas sp. cells that were incubated in medium containing 100% 13C6-glucose. The averaged SCRR spectra (n = 25) of randomly picked cells after different incubation times are shown in Fig. 2A.Remarkably, all signals are clearly separated into only two distinct peaks, representing unlabeled and fully 13C-labeled carotenoids (cf. signal positions in Fig. 1A). No considerable signals originating from mixed isotopologues are observed. This suggest that upon incubation with the easily accessible 13C-glucose, carotenoid biosynthesis exclusively takes place as de novo synthesis.
In the SCRR average spectra (Fig. 2A), the dynamic uptake of 13C-glucose is reflected by a gradual increase of the according 13C related signals. After 24 h, these 13C-signals are observed almost exclusively.
The single-cell resolution allows a more detailed analysis of these findings. Fig. 2B shows the PCA of individual spectra contributing to the average spectra of Fig. 2A. Clearly, the spectra of initially unlabeled cells (green dots) and fully 13C-labeled cells (after 24 h, yellow asterisks) constitute two separate clusters. However, data points corresponding to spectra taken during the dynamic phase after 2 h to 8 h are scattered in between the clusters. This indicates that both, fully unlabeled and fully 13C-labeled carotenoids are present simultaneously in single cells following the transition to a new 13C-source. The spectra of selected individual cells corresponding to data points of the 6 h sample confirm this observation (ESI Fig. 6†).
In order to quantify this relation, the two ν1-signals (12C and 13C) were fitted and integrated for the individual spectra. The ratio of integrals I12C – 1522 cm−1/(I12C – 1522 cm−1 + I13C – 1488 cm−1) is shown in Fig. 2C. In the course of the 13C-uptake, the ratio shifts from 1 (only unlabeled carotenoid signal) to 0 (only 13C-labeled carotenoid signal). However, during the dynamic phase, a high variety of ratios is observed for individual cells (broad IQR of the boxplots). For example, even for the 4 h and 6 h sample, some cells show ratios close to 1 indicating no substantial 13C-uptake (e.g. due to metabolic inactivity). Therefore, the ratios of Fig. 2C constitute a direct measure for the phenotypic heterogeneity within the bacteria population in terms of 13C-incorporation dynamics. These results also reflect the inadequacy of bulk analysis and average spectra to describe dynamics of a population of individual cells (highlighting the importance of single-cell approaches).
3.3. D-Uptake of Sphingomonas sp. leads to unique pattern of red-shifts and apparent blue-shifts in carotenoid SCRR spectra
While 13C stable isotope labeling is a powerful concept for monitoring carbon fluxes, in many cases it may be hampered by the unavailability of (appropriate) labeled compounds.3 Moreover, it does not indicate microorganisms that are metabolically active but do not grow (i.e. do not take up carbon in a considerable amount). Therefore, D-labeling with easily accessible D2O can be applied in SIP as a more general marker for metabolic activity.3,14,26 To the best of our knowledge, no systematic investigation was performed yet for cellular carotenoid SCRR spectra in the context of in vivo D-labeling.In order to close this gap, Sphingomonas sp. was cultivated in media with different contents of D2O and analyzed by SCRM and SCRR. It is well known that high ratios of D in the medium can lead to growth inhibition for both, mammalian and bacterial cells.3,39 Also in our study, reduced growth rates were observed for bacteria grown in media with 50% and 90% D2O (cf. growth curves in ESI Fig. 7†). It cannot be excluded that this affects physiological processes in the cells while incorporating D. Nevertheless, provided that longer incubation times are applied, the equilibrated isotope contents in the stationary growth phase bacteria are shown to relate closely to the media D-contents. This was quantified by monitoring of D-incorporation in regular biomass SCRM spectra (ESI Fig. 8A and B†). Apart from a range of signals in the fingerprint region (e.g. ν(Amide 3)), a distinct red-shift of the CH/CD-stretch signal is observed. The ratio of the fitted integrals I12C – 2938 cm−1/(I12C – 2938 cm−1 + I13C – 2152 cm−1) shown in ESI Fig. 8C† points out that the actual D-content in the cells ranges slightly below the D2O-contents in the media. This is plausible since both, H2O/D2O and the unlabeled glucose in the medium, contribute as H/D-sources. Besides, a kinetic isotope effect may be at work that would discriminate against D-incorporation into biomass and therefore lead to lower D-contents in the cells. In contrast to Berry et al., large phenotypic heterogeneity between different individual cells with respect to the amount of D-incorporation could not be observed, even at high D concentrations (cf. narrow distributions in boxplots of ESI Fig. 8C†).3 Therefore, we assume for this system that also at high D levels spectral artifacts due to growth inhibition do not substantially hinder the tracking of hydrogen fluxes by RM. Moreover, the high D-contents allow a systematical analysis of signal shift patterns for carotenoids.
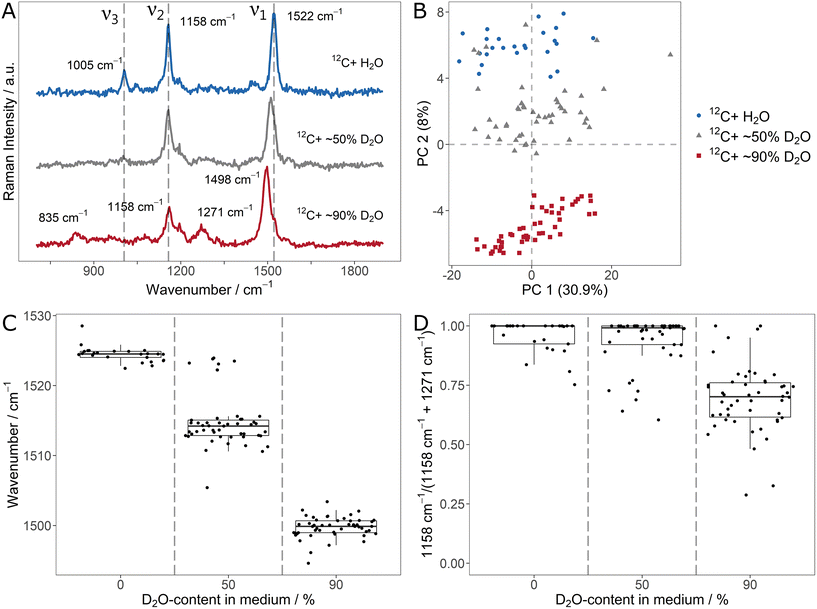
The average (n = 25/50) carotenoid SCRR spectra for the different D2O-contents are shown in Fig. 3A. Similar to the 13C-labeling, the ν1-signal is gradually red-shifted for increasing D-content. The ν2- and ν3-signals however, behave differently. While the initial ν3-signal is already imperceptible at 50% D2O, the initial ν2-signal at 1158 cm−1 does not alter its position but gradually decreases in intensity compared to ν1. At the same time, for the 90% D2O sample, new signals of lower intensity evolve at 835 cm−1 and 1271 cm−1. We note that although associated with C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C- and C–C-stretching vibrations (i.e. without direct involvement of H-atoms), both, ν1-and ν2, are directly affected by D-incorporation. This highlights once more that adjacent atoms exert influence on the vibration modes in the carotenoid molecules.
C- and C–C-stretching vibrations (i.e. without direct involvement of H-atoms), both, ν1-and ν2, are directly affected by D-incorporation. This highlights once more that adjacent atoms exert influence on the vibration modes in the carotenoid molecules.
It can be assumed that the signal at 835 cm−1 is a red-shifted D-equivalent to the vibrations of the ν3-signal. This assumption is supported by the fact that no other band is observed at lower wavenumbers that might be associated to ν3 (cf. the SCRR spectra with extended spectral range in ESI Fig. 9†). Furthermore, Eyring et al. reported a red-shift of similar deuterated methyl rocking vibrations (also in a diene system of a biopigment) from 1038 cm−1 to 874 cm−1 which supports our assignment.40
However, the signal at 1271 cm−1 is blue-shifted compared to the initial ν2-signal. Nevertheless, in previous studies comprising isolated fully D-labeled carotenoids, Nagae et al. confirmed by computational assignments that the signal at 1271 cm−1 is the predominant band for D-labeled C–C-stretchings (just as the initial ν2-signal is for unlabeled carotenoids).41 The fact that no H-atom is directly involved in ν2-vibrations (just as in ν1) suggest an underlying effect other than the simple mass dependency of a single vibration mode. According to Nagae et al., the origin of the apparent blue-shift lies in a coupling of C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C- and C–C-stretchings (i.e. the ν1- and ν2-signals) with less intensive C–H-bendings (within the range of 1200 cm−1 to 1300 cm−1) in the unlabeled carotenoids.41 Upon deuteration, these bendings are strongly red-shifted and therefore decoupled from the stretching vibrations. This results in the apparent red-shift of ν1 and blue-shift of ν2. Compared to the unlabeled ν2-signal, the D-labeled counterpart is less intensive (and therefore not clearly observed in the 50% D2O sample).
C- and C–C-stretchings (i.e. the ν1- and ν2-signals) with less intensive C–H-bendings (within the range of 1200 cm−1 to 1300 cm−1) in the unlabeled carotenoids.41 Upon deuteration, these bendings are strongly red-shifted and therefore decoupled from the stretching vibrations. This results in the apparent red-shift of ν1 and blue-shift of ν2. Compared to the unlabeled ν2-signal, the D-labeled counterpart is less intensive (and therefore not clearly observed in the 50% D2O sample).
Fig. 3B shows the PCA of the individual spectra of the D-labeling series. For each condition, the spectra form distinct clusters. Therefore, it is easily resolved that a few spectra of the 50% D2O sample lie in the area of the unlabeled equivalents. This indicates that these individual cells did not take up D, i.e. remained metabolically inactive.
In order to quantify the D-uptake into carotenoids, the characteristic signal shifts allow different approaches. Similar as for the 13C-labeling, the gradual red-shift of the ν1-signal can be correlated with the D-content (Fig. 3C). Once more, inactive cells that don't take up D are clearly identified by the unchanged signal position. Another approach is to monitor the signal intensity of the initial ν2-signal compared to its D-counterpart at 1271 cm−1. For this, both signals were fitted and integrated. The integral ratio IH – 1158 cm−1/(IH – 1158 cm−1 + ID – 1271 cm−1) is shown in Fig. 3D. Due to the relatively low intensity of the deuterated ν2-signal, a substantial effect on the ratio is only observed for the 90% D2O sample, limiting the applicability of this approach to high D-contents.
3.4. Simultaneous 13C- and D-labeling of Sphingomonas sp. leads to complementary effects in carotenoid SCRR spectra
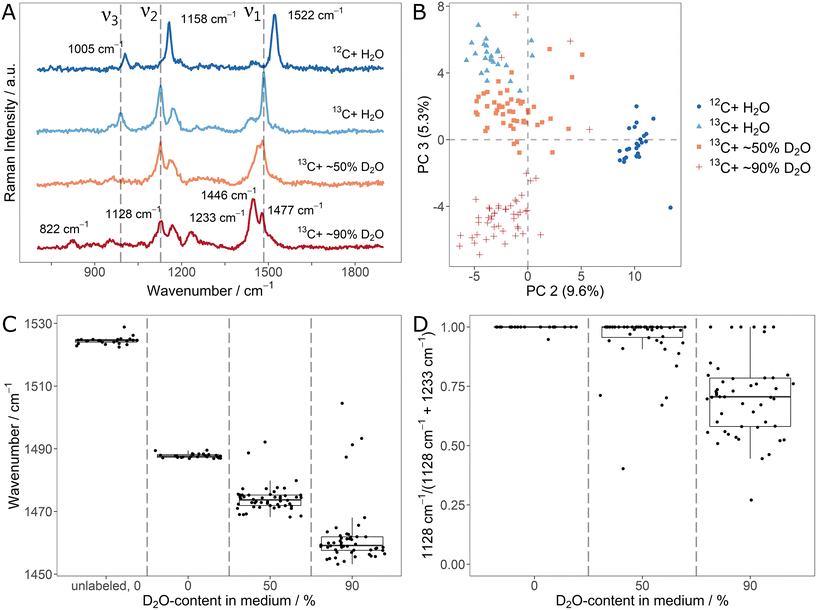
Both, 13C- and D-stable isotope fluxes, are characteristically reflected in SCRR spectra of cellular carotenoids. Knowing that, there is a high motivation for monitoring them simultaneously in order to exploit their orthogonal information. Therefore, we explored the characteristics of a parallel monitoring scheme. For this, fully 13C-labeled Sphingomonas sp. were incubated in medium containing 13C6-glucose and different ratios of H2O/D2O. Successful labeling was achieved as shown by the red-shifted signals in the regular biomass SCRM spectra (ESI Fig. 10A and B†). Once more, D-incorporation ranges at levels slightly below the D2O-content in the media (cf. signal integral ratios in ESI Fig. 10C†).The average (n = 25/50) carotenoid SCRR spectra for the different conditions (unlabeled, 13C-labeled and [1H/D,13C]-labeled) are shown in Fig. 4A. With respect to the signal shifts, the effects of the two labeling schemes complement each other. The D-labeling characteristics are mainly preserved (e.g. red-shift of ν1, apparent blue-shift of ν2). However, due to the 13C-labeling, all signals are additionally red-shifted compared to the mere D-labeling (cf.Fig. 3A). At high D-contents, a separation of the ν1-signal into two shoulders is observed. The less intensive right shoulder remains at the initial position, corresponding to [1H,13C]-labeled carotenoids. Therefore, it presumably represents a fraction of carotenoids in the cells that had (not yet) incorporated D. This might be due to a reduced growth rate of the cells in a medium comprising both, 13C- and D-isotopes (cf. growth curves in ESI Fig. 7†).
In the PCA (Fig. 4B), the individual spectra cluster according to their growth condition where outliers of the 50% and 90% D2O-samples confirm the potential of SCRR to identify metabolically inactive cells. As in the case of separate labeling schemes, the spectral characteristics can be used to simultaneously quantify 13C- and D-incorporation. As shown in the fitted signal positions of Fig. 4C, both, 13C- and D-uptake, contribute to the shift of the ν1-signal. Hence, it cannot be applied as a measure to quantify isotope contents as it was done for the separate labeling approaches. However, with respect to the 13C-content in cellular carotenoids, the signal position of the initial H-ν2-band is an unambiguous quantitative marker. While it is gradually red-shifted for 13C-incorporation, the position remains unchanged for D-incorporation. On the other hand, the ν2-signal intensity exclusively decreases with D-incorporation (as reflected for the simultaneous labeling scheme in the integral ratios of Fig. 4D). For high D-contents, this might be used as a marker (due to the low intensity of the D-ν2-band, it is not applicable for low D-contents). As an alternative, an unambiguous quantitative measure for the D-content can be found in the integral ratio of the CH/CD-stretch signal in the regular biomass SCRM spectra.
4. Conclusion
These results reveal that SCRR carotenoid spectra are a powerful marker to quantitatively track carbon as well as hydrogen stable isotope fluxes into microbial cells, and also both at the same time. To the best of our knowledge, this is the first systematic investigation of this matter in situ on a single-cell level and for diverging isotope profiles. The single-cell resolution and comprehensive response for simultaneous stable isotope labeling allow an in-depth analysis regarding phenotypic heterogeneities of bacteria populations. Major advantages of the carotenoid SCRR approach over regular cellular biomass SCRM spectra lie not only in the simplicity of spectral features, in easily quantifiable correlations and in a high capability for automated spectra processing. The high signal intensities at low measurement times also make cellular carotenoid SCRR spectra an ideal marker for tracking element fluxes in high throughput applications such as cell sorting, counting and imaging. In this context, the simultaneous mode of monitoring carbon and hydrogen fluxes might also allow the differentiation of metabolically active cells that grow – as indicated by 13C- and D-uptake – metabolically active cells that do not grow – indicated by D-uptake only – and metabolically inactive cells – indicated by no 13C- or D-uptake – in future studies.Author contributions
JW, ME and NI designed the experiments. JW carried out lab work and Raman microspectroscopy studies. Data analysis and programming of automated evaluation scripts was done by JW with contribution by KM. Growth curves were recorded and process by KM. OM contributed to microbiological lab work. All authors discussed the results, the manuscript was written by JW and KM under supervision of NI, and corrected by ME and NI. All authors have given approval to the final version of the manuscript.Data availability statement
The code for processing and evaluation of SCRM and SCRR spectra can be found at GitHub and Zenodo with https://doi.org/10.5281/zenodo.7343989. Data for this paper, including SCRM and SCRR raw spectra are available at Zenodo with https://doi.org/10.5281/zenodo.7343786.Conflicts of interest
There are no conflicts to declare.Acknowledgements
Financial funding by the German Research Foundation (Deutsche Forschungsgemeinschaft, DFG) Project IV 110/2-2 is gratefully acknowledged. We thank Lisa Göpfert, Elisabeth von der Esch and Christian Schwaferts for their efforts in isolating the Sphingomonas sp. strain.References
- P. G. Falkowski, T. Fenchel and E. F. Delong, Science, 2008, 320, 1034–1039 CrossRef CAS PubMed.
- E. L. Madsen, Curr. Opin. Biotechnol., 2011, 22, 456–464 CrossRef CAS PubMed.
- D. Berry, E. Mader, T. K. Lee, D. Woebken, Y. Wang, Di Zhu, M. Palatinszky, A. Schintlmeister, M. C. Schmid, B. T. Hanson, N. Shterzer, I. Mizrahi, I. Rauch, T. Decker, T. Bocklitz, J. Popp, C. M. Gibson, P. W. Fowler, W. E. Huang and M. Wagner, Proc. Natl. Acad. Sci. U. S. A., 2015, 112, E194–E203 CAS.
- M. Li, W. E. Huang, C. M. Gibson, P. W. Fowler and A. Jousset, Anal. Chem., 2013, 85, 1642–1649 CrossRef CAS PubMed.
- S. A. Eichorst, F. Strasser, T. Woyke, A. Schintlmeister, M. Wagner and D. Woebken, FEMS Microbiol. Ecol., 2015, 91, 1–14 CrossRef PubMed.
- X. Jing, H. Gou, Y. Gong, X. Su, La Xu, Y. Ji, Y. Song, I. P. Thompson, J. Xu and W. E. Huang, Environ. Microbiol., 2018, 20, 2241–2255 CrossRef CAS PubMed.
- S. Radajewski, P. Ineson, N. R. Parekh and J. C. Murrell, Nature, 2000, 403, 646–649 CrossRef CAS PubMed.
- M. T. Zumstein, R. Narayan, H.-P. E. Kohler, K. McNeill and M. Sander, Environ. Sci. Technol., 2019, 53, 9967–9969 CrossRef CAS PubMed.
- G. T. Taylor, E. A. Suter, Z. Q. Li, S. Chow, D. Stinton, T. Zaliznyak and S. R. Beaupré, Front. Microbiol., 2017, 8, 1449 CrossRef PubMed.
- W. E. Huang, Y. Song and J. Xu, Microb. Biotechnol., 2015, 8, 15–16 CrossRef PubMed.
- B. P. Hedlund, J. A. Dodsworth, S. K. Murugapiran, C. Rinke and T. Woyke, Extremophiles, 2014, 18, 865–875 CrossRef CAS PubMed.
- Y. Song, L. Cui, J. Á. S. López, J. Xu, Y.-G. Zhu, I. P. Thompson and W. E. Huang, Sci. Rep., 2017, 7, 16648 CrossRef PubMed.
- W. E. Huang, K. Stoecker, R. Griffiths, L. Newbold, H. Daims, A. S. Whiteley and M. Wagner, Environ. Microbiol., 2007, 9, 1878–1889 CrossRef CAS PubMed.
- Y. Wang, Y. Song, Y. Tao, H. Muhamadali, R. Goodacre, N.-Y. Zhou, G. M. Preston, J. Xu and W. E. Huang, Anal. Chem., 2016, 88, 9443–9450 CrossRef CAS PubMed.
- Y. Wang, W. E. Huang, L. Cui and M. Wagner, Curr. Opin. Biotechnol., 2016, 41, 34–42 CrossRef CAS PubMed.
- J.-K. Hong, S. B. Kim, E. S. Lyou and T. K. Lee, J. Microbiol., 2021, 59, 249–258 CrossRef CAS PubMed.
- G. Azemtsop Matanfack, M. Taubert, S. Guo, T. Bocklitz, K. Küsel, P. Rösch and J. Popp, Anal. Chem., 2021, 93, 7714–7723 CrossRef CAS PubMed.
- M. Chisanga, H. Muhamadali, D. McDougall, Y. Xu, N. Lockyer and R. Goodacre, Analyst, 2021, 146, 1734–1746 RSC.
- N. P. Ivleva, P. Kubryk and R. Niessner, Anal. Bioanal. Chem., 2017, 409, 4353–4375 CrossRef CAS PubMed.
- K. S. Lee, Z. Landry, F. C. Pereira, M. Wagner, D. Berry, W. E. Huang, G. T. Taylor, J. Kneipp, J. Popp, M. Zhang, J.-X. Cheng and R. Stocker, Nat. Rev. Methods Primers, 2021, 1, 1–25 CrossRef.
- M. Li, D. P. Canniffe, P. J. Jackson, P. A. Davison, S. FitzGerald, M. J. Dickman, J. G. Burgess, C. N. Hunter and W. E. Huang, ISME J., 2012, 6, 875–885 CrossRef CAS PubMed.
- M. Taubert, W. A. Overholt, B. M. Heinze, G. A. Matanfack, R. Houhou, N. Jehmlich, M. von Bergen, P. Rösch, J. Popp and K. Küsel, ISME J., 2021, 16, 1153–1162 CrossRef PubMed.
- W. E. Huang, R. I. Griffiths, I. P. Thompson, M. J. Bailey and A. S. Whiteley, Anal. Chem., 2004, 76, 4452–4458 CrossRef CAS PubMed.
- N. P. Ivleva, M. Wagner, H. Horn, R. Niessner and C. Haisch, Anal. Bioanal. Chem., 2009, 393, 197–206 CrossRef CAS PubMed.
- M. T. Zumstein, A. Schintlmeister, T. F. Nelson, R. Baumgartner, D. Woebken, M. Wagner, H.-P. E. Kohler, K. McNeill and M. Sander, Sci. Adv., 2018, 4, eaas9024 CrossRef CAS PubMed.
- G. A. Matanfack, M. Taubert, S. Guo, R. Houhou, T. Bocklitz, K. Küsel, P. Rösch and J. Popp, Anal. Chem., 2020, 92, 11429–11437 CrossRef CAS PubMed.
- G. Azemtsop Matanfack, A. Pistiki, P. Rösch and J. Popp, J. Biophotonics, 2021, 14, e202100013 CrossRef CAS PubMed.
- M. Li, J. Xu, M. Romero-Gonzalez, S. A. Banwart and W. E. Huang, Curr. Opin. Biotechnol., 2012, 23, 56–63 CrossRef CAS PubMed.
- M. Li, P. C. Ashok, K. Dholakia and W. E. Huang, J. Phys. Chem. A, 2012, 116, 6560–6563 CrossRef CAS PubMed.
- K. S. Lee, M. Palatinszky, F. C. Pereira, J. Nguyen, V. I. Fernandez, A. J. Mueller, F. Menolascina, H. Daims, D. Berry, M. Wagner and R. Stocker, Nat. Microbiol., 2019, 4, 1035–1048 CrossRef CAS PubMed.
- Y. Wang, Y. Ji, E. S. Wharfe, R. S. Meadows, P. March, R. Goodacre, J. Xu and W. E. Huang, Anal. Chem., 2013, 85, 10697–10701 CrossRef CAS PubMed.
- Q. Zhang, P. Zhang, H. Gou, C. Mou, W. E. Huang, M. Yang, J. Xu and B. Ma, Analyst, 2015, 140, 6163–6174 RSC.
- P. Zhang, L. Ren, X. Zhang, Y. Shan, Y. Wang, Y. Ji, H. Yin, W. E. Huang, J. Xu and B. Ma, Anal. Chem., 2015, 87, 2282–2289 CrossRef CAS PubMed.
- P. Kubryk, J. S. Kölschbach, S. Marozava, T. Lueders, R. U. Meckenstock, R. Niessner and N. P. Ivleva, Anal. Chem., 2015, 87, 6622–6630 CrossRef CAS PubMed.
- N. Tschirner, M. Schenderlein, K. Brose, E. Schlodder, M. A. Mroginski, C. Thomsen and P. Hildebrandt, PCCP Phys. Chem. Chem. Phys., 2009, 11, 11471–11478 RSC.
- E. A. Johnson and W. A. Schroeder, Adv. Biochem. Eng./Biotechnol., 1996, 53, 119–178 CrossRef CAS PubMed.
- E. von der Esch, M. Lanzinger, A. J. Kohles, C. Schwaferts, J. Weisser, T. Hofmann, K. Glas, M. Elsner and N. P. Ivleva, Front. Chem., 2020, 8, 169 CrossRef CAS PubMed.
- R. Weiss, M. Palatinszky, M. Wagner, R. Niessner, M. Elsner, M. Seidel and N. P. Ivleva, Analyst, 2019, 144, 943–953 RSC.
- J. F. Thomson, Ann. N. Y. Acad. Sci., 1960, 84, 736–744 CrossRef CAS PubMed.
- G. Eyring, B. Curry, R. Mathies, R. Fransen, I. Palings and J. Lugtenburg, Biochemistry, 1980, 19, 2410–2418 CrossRef CAS PubMed.
- H. Nagae, M. Kuki, J.-P. Zhang, T. Sashima, Y. Mukai and Y. Koyama, J. Phys. Chem. A, 2000, 104, 4155–4166 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2an01603f |
| This journal is © The Royal Society of Chemistry 2023 |