A near-infrared optical nanosensor for measuring aerobic respiration in microbial systems†
Samuel C.
Saccomano
 a and
Kevin J.
Cash
a and
Kevin J.
Cash
 *ab
*ab
aChemical and Biological Engineering Department, Colorado School of Mines, Golden, CO, USA. E-mail: kcash@mines.edu
bQuantitative Biosciences and Engineering, Colorado School of Mines, Golden, CO, USA
First published on 25th November 2021
Abstract
We developed a ratiometric oxygen-sensitive nanosensor and demonstrated application in monitoring metabolic oxygen consumption in microbial samples over time. Based on a near-infrared (NIR) emitting oxygen-quenched luminophore, platinum(II) octaethylporphine ketone (PtOEPK), along with a stable dioctadecyl dicarbocyanine reference dye (DiD), this nanosensor system provides an advantageous approach for overcoming imaging issues in biological systems, such as autofluorescence and optical scattering in the visible wavelength region. The dyes are encapsulated within a polymer-based nanoparticle matrix to maintain them at a constant ratio in biological samples, precluding the need for complex synthetic approaches. With this constant ratio of the two dyes, the nanosensor response can be measured as a ratio of their two signals, accounting for nanosensor concentration artifacts in measurements. The nanosensors are reversible, which enabled us to temporally monitor systems in which dissolved oxygen concentrations both increase and decrease. These sensors were applied for the monitoring of oxygen in samples of Saccharomyces cerevisiae (brewing yeast) in a 96-well optical fluorescence plate reader format over 60 h. By mixing the nanosensors directly into the sample well with the yeast, we were able to dynamically track metabolic activity changes over time due to varying cell concentration and exposure to an antimicrobial agent. This system could be a potential platform for high-throughput screening of various species or variants of microbes with unknown metabolic rates in response to external stimuli (antimicrobials, metabolites, etc.).
Introduction
Oxygen is an essential component in many biological systems as an electron acceptor in cellular respiration. Thus, oxygen measurements are a common way of characterizing metabolic activity in a variety of clinical and environmental settings.1,2 We can learn much about the function of these systems through determining key parameters such as biochemical oxygen demand (BOD),3 oxygen diffusion rates4 and dissolved oxygen concentration gradients.5 One common approach to measure dissolved oxygen is through electrochemical measurements, which involve reading the potential generated from the chemical reduction of oxygen at an electrode or microelectrode.6–8 While this method is the most well studied, it has some distinct drawbacks that include consumption of the analyte, physical disruption of the system under investigation, and a limited ability to measure temporal and 3-dimensional spatial dynamics.9 An orthogonal approach is based on optical measurements, which involve the use of a luminescent dye which is quenched in the presence of oxygen.9–11 This can be done through a planar optode approach which measures oxygen along a 2-dimensional surface,12 a fiber optic oxygen sensor13 or a micro/nanoparticle approach, which we use in this work. While, this paper will primarily focus on sensing oxygen for determining cell metabolic activity, Braissant et al. provides a broader review of methods for measuring metabolic rates.14Chemically responsive nanoparticle-based sensors (nanosensors) are an emerging technology which aims to overcome many of the common challenges in making biological measurements. These nanoparticles consist of a highly plasticized polymer matrix which encapsulates luminescent indicator dyes and is permeable to molecular oxygen.15 Many hydrophobic dyes are known to have poor solubility in aqueous environments and can be difficult to apply in a biological settings.16 By encapsulating these dyes in a polymer matrix, we can better control the sensor response which allows for sensing in a wider variety of samples.17 Additionally, these nanoparticles are stable enough to measure temporal dynamics over several hours to several days and can be evenly dispersed throughout the sample for measuring 3-dimensional gradients18 when paired with the necessary optical instrumentation. Applications which have used oxygen-sensitive nanosensors include, intracellular oxygen measurements in cancer cells,19–22 confocal imaging of various cell types,23–25 oxygen dynamics in biofilms,18,26,51 oxygen sensing in microfluidics27 and in vivo monitoring.28
Many optical probes still face the challenge of overcoming the background signal produced from biomolecules present in biological tissues.29 In thin films and surface tissues the background is relatively low, however in deeper tissue samples there can be significant absorbance or emission signals in the visible light range (400–700 nm). Probes that emit photons in the near-infrared (NIR) wavelength range (700 nm–3000 nm) can be much easier to detect for these types of biological measurements.30 NIR probes have been used for measuring many analytes in different applications. For example, Jo et al. used a fluorescent NIR tracer to measure glucose uptake in human cervical cancer cells.31 Barone et al. demonstrated the applicability of carbon nanotubes as near-infrared sensors which respond to changes in dielectric properties of the material in the presence of glucose.32 Cash et al. used fluorescent nanosensors for in vivo imaging and measurement of lithium in mice.33 Hirayama et al. measured copper ion regulation in mice using NIR copper sensors due to its link to progression of degenerative diseases such as Alzheimer's and Wilson's disease.34 A more extensive review of near-IR probes for bioimaging applications was written by Li et al.30
Platinum(II) octaethylporphine ketone (PtOEPK) is a luminescent dye which emits in the NIR range at approximately 760 nm when excited, but is reversibly quenched in the presence of oxygen, thus making it a good candidate for optical measurements of oxygen in biological samples.35 PtOEPK is a metalloporphyrin dye which has a longer emission wavelength and phosphorescent lifetime compared to many other oxygen-sensitive dyes.
Mechanistically, molecular oxygen quenches PtOEPK luminescence through a process called collisional quenching.36 As the luminophore is excited, the triplet state oxygen collides with the chelated platinum in the center of the molecule quenching the excited energy state of the dye and dispersing the additional energy through non-radiative decay.11
Emission wavelengths for metalloporphyrins start at around 650 nm and range up to as high as 950 nm (PtOEPK ∼760 nm) with large Stokes’ shifts (PtOEPK ∼360 nm) and relatively high quantum yields (PtOEPK ∼12%).16 Additionally, metalloporphyrin phosphorescence lifetimes are very long ranging from 10 microseconds to 1 millisecond16 (PtOEPK ∼60 μs (ref. 35)). Other factors such as brightness and photostability can vary greatly depending on the exact dye that is used.37
In this study, we developed a ratiometric nanosensor to measure oxygen. We used PtOEPK as the oxygen-sensitive component and 1,1′-dioctadecyl-3,3,3′,3′-tetramethylindodicarbocyanine (DiD) as a reference dye which does not respond to oxygen. The two dyes are both in the same nanoparticle matrix upon fabrication meaning that the ratio of the dye signals can be used to measure oxygen even when there are variations in signal due to nanoparticle concentration or some changing optical conditions of the samples.38 A diagram of the nanosensor and the oxygen quenching mechanism can be seen in Fig. S1.† In this work we apply these oxygen sensors to a high-throughput assay for measuring oxygen in biological systems, demonstrated here with oxygen consumption in different strains of brewing yeast.
Experimental
Materials
Polyvinyl chloride (PVC), bis ethylhexyl sebacate (BEHS), tetrahydrofuran (THF), dichloromethane (DCM), Dubelco's phosphate buffered saline (PBS), 0.22 μm sterile vacuum filter, 15-guage needles and 18-guage needles were all purchased from Sigma-Aldrich (St Louis, MI, USA). Platinum(II) octaethylphorphine ketone was purchased from Frontier Scientific (Logan, UT, USA). 1,1′-Dioctadecyl-3,3,3′,3′-tetramethylindodicarbocyanine, 4-chlorobenzenesulfonate salt (DiD; Invitrogen), potassium phosphate dibasic, potassium phosphate monobasic and 96-well black-walled optical bottom plates were purchased from Thermo Fisher Scientific (Waltham, MA, USA). 1,2-Dipalmitoyl-sn-glycero-3-phosphoethanolamine-N-[methoxy(polyethylene glycol)-750] ammonium salt (PEG-750) was purchased from Avanti Polar Lipids (Alabaster, AL, USA). 0.8 μm polyethersulfone membrane filters were purchased from Pall Corporation (New York, NY, USA). Ultrahigh purity nitrogen gas and compressed air were purchased from Matheson (Denver, CO, USA). 10 mm pathlength quartz cuvette with rubber septa seal cap was purchased from Starna Cells (Atascadero, CA, USA). Campden tablets (potassium metabisulfite, PMB) were obtained from Crosby and Baker (Westport, MA, USA).Methods
Fabrication of nanosensors
Prior to nanosensor fabrication, we fabricated an optode formulation with the sensor components. 15 mg of polyvinyl chloride (PVC) was weighed out into a 2 mL glass vial and combined with 33 μL of BEHS and vortexed. 2.5 mg of platinum(II) octaethylporphyrin ketone (PtOEPK) and 0.2 mg of DID were dissolved in 250 μL of tetrahydrofuran (THF) and transferred to the PVC slurry and mixed until all PVC was dissolved. 375 μL dichloromethane (DCM) was added to the vial containing the PVC-dye mixture. The optode was then capped and stored at 4 °C. To make the nanoparticles, 80 μL of 25 mg mL−1 PEG-750 in chloroform was added to an 8 mL scintillation vial. An air stream was used to dry off the chloroform and the PEG-750 was then brought back up in 5 mL of phosphate buffered saline (PBS) and sonicated with a probe tip sonicator (Branson Ultrasonics, Brookfield, CT, USA) for 30 s at 20% power. 125 μL of the optode was added and simultaneously sonicated for another 3 minutes at 20% intensity. Once done, the nanosensor mixture was filtered through a 0.8 μm polyethersulfone membrane into an 8 mL glass vial and stored at room temperature away from light. To measure potential losses of sensor within this process we analysed samples without any filtering, samples which had been centrifuged for 2 min at 2000 rpm to remove precipitated polymer, and samples which had been filtered by the PES membrane to compare to the raw sonicated sensor. Absorbance scans and fluorescence scans (590 exc.) were taken in triplicate for each sample (see Fig. S2†).Gas flow and oxygen sensor characterization
Calibration of the oxygen sensors was done using a gas flow system of mixed ultrapure nitrogen and air streams. 2 mL of nanosensor solution was transferred to a quartz cuvette with 1 cm pathlength and rubber septa seal cap. Luminescence measurements were taken with an Avantes Avaspec2408L spectrometer with excitation from a 532 nm fiber coupled LED (Thor Labs, Newton, NJ, USA). Nitrogen and air gas flow rates were controlled by two mass flow controllers. The nitrogen controller was obtained from Aalborg (Orangeburg, NY, USA) and the air controller was obtained from Alicat Scientific (Tuscon, AZ). The gas streams were then mixed in a 50 mL stainless-steel gas mixing chamber (see Fig. S3†). The gas mixture was bubbled directly into the cuvette through the septa seal using a 15-gauge needle attached to the end of the gas line. An additional needle was inserted through the septa to allow excess gas to vent from the cuvette during bubbling. The total gas flow rate was kept constant at 20 mL min−1 with the ratio of nitrogen and air being used to adjust the oxygen concentration between 0% and 21%. Measurements were taken at 0%, 5%, 10%, 15% and 21% oxygen in the gas phase, which was equivalent to 0, 1.58, 3.17, 4.75 and 6.65 mg L−1 in solution respectively. The initial equilibration time for 0% O2 stream was 40 min and the subsequent equilibration steps were 20 min.A calibration curve was generated by measuring the ratio of luminescence at 757 nm (oxygen-sensitive dye) to 675 nm (reference dye) at each of the oxygen concentrations. A pseudo-Stern–Volmer equation is used to plot these values as a ratio of the intensity in the absence of the quenching species (0 mg L−1 O2) to the intensity at another fixed concentration. The Stern–Volmer equation is described by eqn (1) in the Results section. The pseudo-Stern–Volmer constant, or KpSV was found with a linear regression of the plot.
Reversibility was tested by cycling between pure nitrogen (deoxygenation) and air (21% oxygenation) to see if the signal would change as a function of cycle number. Each bubbling step was run for 40 min between measurements, and they were tested for 5 cycles.
Temperature response was tested by performing nitrogen and air bubbling steps while the cuvette was partially submerged in a heated water bath (Fisher Scientific, Waltham, MA, USA) followed by a rapid measurement with the system outlined above before being returned to the bath. pH response was tested by carefully diluting the sensors 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 in a mixture of potassium phosphate dibasic and potassium phosphate monobasic buffers. Nanosensor concentration was tested by diluting the stock concentration in PBS to either 50%, 20%, 10% or 5% of its original concentration.
1 in a mixture of potassium phosphate dibasic and potassium phosphate monobasic buffers. Nanosensor concentration was tested by diluting the stock concentration in PBS to either 50%, 20%, 10% or 5% of its original concentration.
The response time of the nanosensor was estimated using oxygen purging by glucose and glucose oxidase (GOx) to artificially drive the oxygen concentration down in a well plate. 160 μL of sensor and 20 μL of 100 mM glucose were placed into 3 wells of a 96-well optical bottom plate. The wells received either 20 μL of PBS, 20 μL of 20 units per mL GOx, or 20 μL of 100 units per mL GOx. Immediately after addition of the final reagents the plate was loaded into a microplate reader and fluorescence was measured every 5 seconds at 590 nm excitation with 675 nm and 760 nm emissions for the two dyes. The delay between reagent addition and the first read was measured at 15 s. Size and charge of the nanoparticles were measured using a Malvern ZEN360 ZetaSizer (Southborough, MA, USA).
Brewing yeast growth
Two strains of brewing yeast were tested from Propagate Labs (Golden, CO, USA). Strains MIP-510 (Kolsch I) and MIP-354 (Kveik:Oslo) were used. Yeast packets contained 200 billion cells per mL in roughly 200 mL of media. Fast Pitch canned wort was obtained from Northern Brewer (Roseville, MN, USA). The wort was prepared by diluting 1 can (16 oz.) with 16 oz. of ultrapure grade water. The diluted wort was then filter sterilized through a 0.22 μm sterile vacuum filter. Yeast stocks were prepared by diluting 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100, 1
100, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 30, 1
30, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 in filter sterilized PBS. Yeast cells were resuspended and mixed in solution to ensure accuracy of pipetting. Campden tablets (also known as potassium metabisulfite (PMB)) were prepared based on recommended concentration of 1 tablet per 1 gallon of wort (∼3785 mL). A 100× concentrated solution was prepared by dissolving the tablet into 37.85 mL of ultrapure MilliQ water. Two stocks were then prepared from the 100× stock. In one stock, 77 μL of the 100× solution was diluted in 4.923 mL of wort. In the other stock 100 μL of the 100× solution was diluted in 900 μL of PBS. These values were determined so that the final concentration of PMB in each well was equivalent to 1 tablet in 3785 mL of volume.
10 in filter sterilized PBS. Yeast cells were resuspended and mixed in solution to ensure accuracy of pipetting. Campden tablets (also known as potassium metabisulfite (PMB)) were prepared based on recommended concentration of 1 tablet per 1 gallon of wort (∼3785 mL). A 100× concentrated solution was prepared by dissolving the tablet into 37.85 mL of ultrapure MilliQ water. Two stocks were then prepared from the 100× stock. In one stock, 77 μL of the 100× solution was diluted in 4.923 mL of wort. In the other stock 100 μL of the 100× solution was diluted in 900 μL of PBS. These values were determined so that the final concentration of PMB in each well was equivalent to 1 tablet in 3785 mL of volume.
Plate setup
A diagram of the plate layout and the well contents can be seen in Fig. S4.† Samples were run in a 96-well black-walled non-treated optical bottom plate. The plate was setup so that each combination of variables (3 yeast dilutions, 2 yeast strains, 3 PMB conditions, 18 total combinations) would be tested in quadruplicate. The 3 different PMB conditions were as follows: no PMB, PMB added at t = 0 h and PMB added at t = 42 h. Additionally, 4 total control experiments were included: (1, 2) 4 replicates of each strain in the absence of nanosensor, (3) 8 replicates with no yeast in wort, and (4) 8 replicates with no yeast in wort with PMB added. First the wort was added to each well. For wells where the PMB was added at t = 0 h, they received 130 μL of the PMB-wort stock solution. The control wells containing nanosensors only with PMB also received 130 μL of the PMB-wort stock. The remainder of the wells received 130 μL of the normal wort stock solution. Next, the nanosensors (fabricated as described above) were added to each well. Every well except the yeast only no nanosensor controls received 50 μL of the nanosensor stock. In the control wells, the nanosensors were replaced with 50 μL of PBS. Finally, each well received 20 μL from one of the 6 different yeast strain/dilution stocks. For wells that did not have yeast, this addition was replaced with 20 μL of PBS. Prior to starting the run, each well was mixed by gently pipetting the solution several times. The plate was then sealed with an aluminum foil adhesive cover and running a finger over each well to ensure proper seal. A hole was poked in each well's cover by gently pressing an 18 gauge needle through the foil in the center of the well. All yeast operations were performed in a biosafety cabinet for optimal sample sterility.Plate reader setup
The assay was run on a Synergy H1 microplate reader (Biotek, Winooski, VT, USA). Prior to any reads the plate reader was pre-heated to 30 °C. Optical reads were taken every 15 minutes with no additional exposure to light in between luminescent measurements. The well was subject to orbital shaking, on low speed at 693 rpm for 5 min prior to each read. Three different optical settings were read at each 15 min interval. This included the OD600, luminescence at 675 nm emission with 590 nm excitation (DiD channel), luminescence at 760 nm emission with 590 nm excitation (PtOEPK channel). Both luminescence measurements were read from the bottom of the plate with a gain setting of 100. The first part of the plate reader run was set for 42 h. At this point the plate was briefly removed from the plate reader. The wells intended to receive PMB at the 42 h mark got 20 μL of the 10× PMB stock in PBS by pipetting through the small hole in the aluminum foil covering each well. All other wells received 20 μL of PBS. The plate was placed back in the plate reader and allowed to run for an additional 18 hours under the same temperature, shaking and readout settings.Yeast counting and cytotoxicity
Yeast cell counts were confirmed via haemocytometer. The original cell stock used was diluted in PBS 1000-fold prior to cell counting. 15 μL of the diluted cells were pipetted onto the gridded haemocytometer plate (Hausser Scientfic, Horsham, PA, USA). The haemocytometer was placed on to a microscope at 10× objective magnification. Cells were counted across 6 different 1 mm × 1 mm grids and averaged together. The total cell concentration of the original stock was calculated by multiplying the cell count by the dilution factor (1000×) and the volume factor (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000×) to achieve the number of cells per mL. S. cerevisiae cultures for determining nanosensor cytotoxicity were grown up in wort from frozen stocks obtained from ATCC (Manassas, Virginia, USA) for 24 h at 30° C while shaking at 150 rpm. 150 μL of yeast stock was mixed with 50 μL of either nanosensor or PBS (3 wells each) and allowed to sit at room temperature away from light for 48 h. 25 μL from each well was mixed with 75 μL of PBS in a separate well. 10 μL of CCK-8 solution was added to each well along with 3 more wells containing 100 μL of PBS. An initial absorbance scan was taken from 350 nm to 650 nm on the Synergy H1 microplate reader. After 24 h the same measurements were taken again. The net change in absorbance was determined by subtracting the initial measurement from the final measurement at each wavelength on the spectra. A larger number of remaining live cells was indicated by a greater increase in absorbance at 460 nm due to greater production of NADPH by the cells.
000×) to achieve the number of cells per mL. S. cerevisiae cultures for determining nanosensor cytotoxicity were grown up in wort from frozen stocks obtained from ATCC (Manassas, Virginia, USA) for 24 h at 30° C while shaking at 150 rpm. 150 μL of yeast stock was mixed with 50 μL of either nanosensor or PBS (3 wells each) and allowed to sit at room temperature away from light for 48 h. 25 μL from each well was mixed with 75 μL of PBS in a separate well. 10 μL of CCK-8 solution was added to each well along with 3 more wells containing 100 μL of PBS. An initial absorbance scan was taken from 350 nm to 650 nm on the Synergy H1 microplate reader. After 24 h the same measurements were taken again. The net change in absorbance was determined by subtracting the initial measurement from the final measurement at each wavelength on the spectra. A larger number of remaining live cells was indicated by a greater increase in absorbance at 460 nm due to greater production of NADPH by the cells.
Results
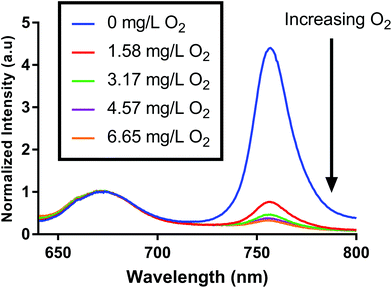
We fabricated oxygen-sensitive nanosensors by combining two dyes (PtOEPK, DiD) in our nanoparticle matrix. These dyes were chosen primarily based on their optical properties. DiD and PtOEPK both emit at relatively long wavelengths (675 nm and 757 nm respectively) but are far enough apart to show sufficient separation of the two peaks. Additionally, PtOEPK can be excited simultaneously with DiD using an excitation wavelength in the range of 530 nm to 590 nm or separately using a lower wavelength excitation at 405 nm. The PtOEPK luminescence will change based on the quenching of oxygen, meaning that higher oxygen concentrations correlate to a lower observed emission intensity when the dye is excited. The DiD dye, however, is oxygen independent – meaning no change in signal is observed as the oxygen concentration is changed. Fig. 1 shows the luminescence spectra of the dual probe nanosensor, normalized to the DiD luminescence when excited by a 532 nm light emitting diode. In these spectra, we can see a clear response of the oxygen dye as the concentration of dissolved oxygen in the sample is changed. The raw luminescence spectra, Fig. S5,† are similar.For quenching based approaches like this, oxygen concentration in relation to signal intensity can be described by the Stern–Volmer relationship. The Stern–Volmer equation for collisional quenching of luminophores is given by eqn (1).
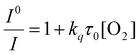 | (1) |
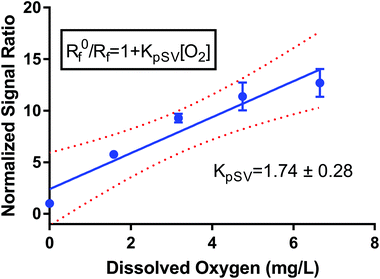
I 0 and I refer to the intensity of the luminescence signal in the absence and presence of oxygen at a given concentration, respectively. The quenching constant (kq) and the decay lifetime (τ0) combined to form the Stern–Volmer constant or KSV. We created a pseudo-Stern–Volmer plot (Fig. 2) using the ratio of the signal of the two dyes (PtOEPK and DiD) instead of the oxygen-quenched dye alone so any variation in dye concentration could be accounted for. To generate a curve, the ratiometric signal in the absence of oxygen is divided by the ratiometric signal at the individual oxygen concentrations to obtain a linear correlation with respect to the dissolved oxygen concentration. The pseudo-Stern Volmer constant (KpSV) is obtained from the slope of the linear regression of this plot demonstrating the relationship between the increasing concentration of the quenching molecule and the decrease in signal. While the pseudo-Stern–Volmer correlation can vary based on the instrument setup, a single instrument setup can be used to determine the consistency in the performance of the sensors over several samples or batches. For our air and nitrogen bubbling setup (with n = 3), we found the KpSV to be 1.74 ± 0.29 when plotted from 0 to 6.65 mg L−1. The result of this value is that the difference in signal between atmospheric oxygen levels and anoxic conditions is roughly 10-fold. The R2 value of the fit was 0.93 indicating some level of non-linearity. The degree of linearity is usually determined by the extent of dynamic quenching vs. static quenching and whether the quenching step is limited by diffusion of oxygen to the dye,39 however, when compared to the Stern Volmer of just the oxygen dye (see Fig. S6†) which shows a high degree of linearity, it indicates the reference dye could be the source of the minor non-linearity. Sensitivity of the sensor was also tested specifically for lower concentrations of oxygen at 0, 0.40, 0.79, 1.19 and 1.58 mg L−1 consecutively (see Fig. S7†). The sensors responded linearly in this range suggesting it is well suited for measuring low oxygen concentrations.
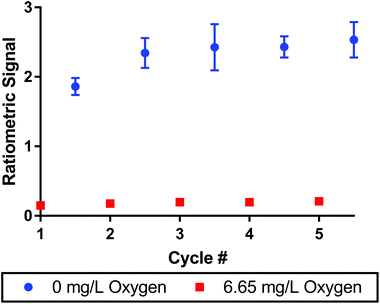
To monitor oxygen concentrations in biological systems, the sensors must be reversible to measure both increases and decreases in oxygen concentrations. We tested reversibility by alternating the composition of the bubbling solution between pure nitrogen and air streams. The signal was measured over the course of 5 cycles between the two conditions as seen in Fig. 3. After a slight increase in the ratiometric signal from cycle 1 to cycle 2, the signal was consistent across the remaining 4 cycles. The maximum variance was 8% between cycle 2 and 5 for the deoxygenated sample. Error bars indicate that no data points had a standard deviation greater than 11% for the deoxygenated sample (cycle 3) and 14% for the oxygenated sample (cycle 5) over n = 3 replicate sensor batches. The oxygen dye alone showed very similar results demonstrating the dye's excellent reversibility (see Fig. S8†).
Temperature, pH and concentration dependence were also tested at oxygenated and deoxygenated levels. As expected from similar dyes, the signal decreased significantly as the temperature of the sensors was increased from 25 °C to 40 °C. Phosphorescence has been shown to be sensitive to temperature in the past due to thermal quenching which decreases the luminescence lifetime of dye.40 However, both dyes were impacted similarly by temperature resulting in a consistent ratiometric signal and showing that oxygen could be measured across biologically relevant temperatures (see Fig. S9†). We tested the pH response of the nanosensors from 5.8 to 8.2 (see Fig. S10†). The sensors showed the highest luminescence at pH 7.6 and 8.2 for the PtOEPK dye and pH 7.0 for the DiD dye though both showed easily measurable signal across the tested range. We determined the concentration dependence of the nanosensors (see Fig. S11†) by diluting a batch of nanosensors down to 5% of the original nanosensor concentration through a series of dilutions. While, as expected, luminescence is not a linear function of concentration, the ratiometric signal between the oxygen dye and the reference dye stayed constant in both oxygenated and deoxygenated samples showing the value of ratiometric measurements to correct for nanosensor concentration. Response time was determined by measuring the ratiometric signal of the sensor as the oxygen in solution was consumed using glucose and GOx (see Fig. S12†). The data showed that response was limited by the speed of oxygen depletion (at low enzyme concentrations) or the delay time of instrument (∼15 s). The actual response time of the sensors is likely much faster as similar fluorescent nanosensors have millisecond scale response times.41 The diameter of the particles was 161.4 ± 1.7 nm, polydispersity was 0.15 ± 0.02 and zeta potential was −25.9 ± 2.3.
We tested the nanosensor function in biological systems by measuring oxygen consumption of Saccharomyces cerevisiae (the yeast commonly used in brewing). Molecular oxygen is a critical component to cellular respiration, which produces many of the precursor molecules essential to the fermentation processes in brewing yeast that lead to alcohol production. Various lipids and growth factors require the high energy yield of oxygen metabolism for production, and are essential components to the yeast in their anaerobic metabolic state.42 We used nanosensors mixed with yeast samples in a microwell plate to assess yeast metabolism. Over the course of ∼60 hours the ratiometric signals of the nanosensors were monitored to track the aerobic respiration process (oxygen consumption) of yeast cells when exposed to fresh aerated wort. Two different strains of yeast were tested: the Kolsch I and Kveik: Oslo strains (Propagate Lab, Golden, CO). Fig. 4A shows a set of ratiometric curves for 3 different concentrations of the Kolsch yeast diluted from stock solutions containing ∼200 billion cells (according to manufacturer) in approximately 200 mL of growth media. At the initial phase of the growth profile, the low ratiometric signal is an indication of the high oxygen concentration, which we would presume to be close to atmospheric levels due to low consumption by the yeast. Samples with lower amounts of initial yeast, saw a greater lag time before a rapid increase in signal was observed at the initial stages of growth. Conversely, the higher initial yeast concentrations saw an increase in the ratiometric signal within the first few hours. Thus, our sensor can distinguish between different oxygen concentrations resulting from different consumption rates based on the number of cells metabolizing oxygen. Eventually each sample reached an equilibrium where the ratiometric curve flattens. This does not indicate where the oxygen level has reached zero, but merely where oxygen consumption and oxygen diffusion from the atmosphere reach a steady state. Larger initial amounts of yeast also correlated to higher ratiometric signals at which the equilibrated state is reached. In Fig. S14,† we see how the importance of the ratiometric signal comes into play as well. The individual dye channels show unexpected behaviour, especially as cell concentration is changed, but when the ratio is taken, the artifacts are removed, and the oxygen concentration profile seems to show a clearer trend. Fig. 4B shows a faster change in oxygen concentrations from the Kveik strain at the same dilution as the Kolsch, though this is likely due to a higher initial cell count from package listed concentrations (Kolsch: 1.82 ± 0.08 billion cells per mL; Kveik 2.79 ± 0.09 billion cells per mL). Similar trends are observed at higher and lower cell dilutions as well (see Fig. S15†). The Kveik showed a similar trend in oxygen metabolism with respect to cell concentration as the Kolsch – with increasing cell density causing a more rapid oxygen concentration change and lower steady state oxygen concentration (see Fig. S16†). We also tested the metabolism of live cells after the addition of an antimicrobial agent. Fig. 4C shows the ratiometric signal of cells spiked with potassium metabisulfite (PMB) at t = 42 h as compared to those with no PMB added. In the Kolsch strain there is clearly a detectable difference between the 2 conditions. The cells that were given PMB in the middle of the run saw a very rapid decrease in signal back to atmospheric conditions as compared to the no PMB samples that continued to hold consistent signal at the equilibrium oxygen concentration. An additional condition was run in which the PMB was added at the start of the run for each yeast dilution (see Fig. S17†). The PMB did not initially slow down the consumption of oxygen, but only at 15 h did the antimicrobial take effect and the oxygen concentration came back to atmospheric levels. The oxygen response in the Kveik: Oslo strain did not respond as dramatically to the addition of PMB (Fig. S18†). In samples where PMB was added at t = 0 h oxygen consumption was slowed for a period of time before recovering to near equilibrium concentrations. Cells exposed to PMB after 42 hours did see an increase in oxygen levels which would occur with diminished metabolism, but they did so much more slowly than the Kolsch strain.
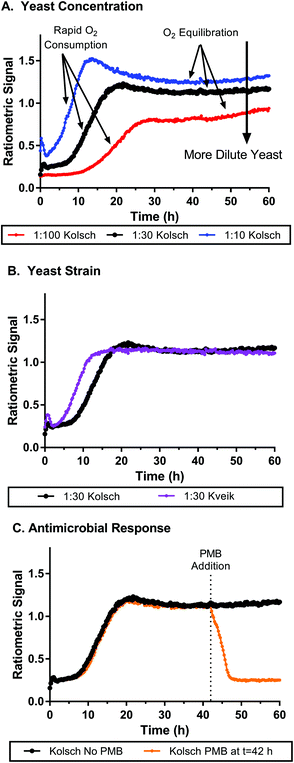 | ||
Fig. 4 Oxygen nanosensors in yeast respond to changes in oxygen metabolism as yeast are grown over time. A. Yeast are diluted at 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100, 1 100, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 30 and 1 30 and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 ratios from stock yeast concentrations showing faster oxygen consumption and lower equilibrium oxygen concentrations for more concentrated samples of yeast. B. Kolsch and Kveik strains at 1 10 ratios from stock yeast concentrations showing faster oxygen consumption and lower equilibrium oxygen concentrations for more concentrated samples of yeast. B. Kolsch and Kveik strains at 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 30 yeast dilution shows similar oxygen consumption behavior though the Kveik strain metabolized oxygen faster likely due to higher initial cell counts in the yeast stock. C. Potassium metabisulfite (PMB), when added to samples (1 30 yeast dilution shows similar oxygen consumption behavior though the Kveik strain metabolized oxygen faster likely due to higher initial cell counts in the yeast stock. C. Potassium metabisulfite (PMB), when added to samples (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 30 dilution, Kolsch strain) at 42 h from inoculation causes an immediate decrease in oxygen metabolism in the samples and a return to initial atmospheric conditions within 6 hours consistent with ceasing of metabolism. The 1 30 dilution, Kolsch strain) at 42 h from inoculation causes an immediate decrease in oxygen metabolism in the samples and a return to initial atmospheric conditions within 6 hours consistent with ceasing of metabolism. The 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 30 Kolsch curve (black) is the same data set in each graph and was used as a standard to compare various conditions. All yeast concentrations were within typical range used for fermentation and brewing. PMB was chosen because it is a common agent used in fermentation processes to inhibit microbial growth. Data shown is average measurement of 4 replicate wells taken every 15 minutes with no irradiation in between measurements. Error bars have been removed for clarity but can be seen in respective graphs in Fig. S13.† 30 Kolsch curve (black) is the same data set in each graph and was used as a standard to compare various conditions. All yeast concentrations were within typical range used for fermentation and brewing. PMB was chosen because it is a common agent used in fermentation processes to inhibit microbial growth. Data shown is average measurement of 4 replicate wells taken every 15 minutes with no irradiation in between measurements. Error bars have been removed for clarity but can be seen in respective graphs in Fig. S13.† | ||
The controls for this experiment showed the expected behaviour when the yeast or the nanosensor were removed from the well (see Fig. S19†). In the absence of yeast, no oxygen is metabolized and the ratiometric signal remains constant. When the nanosensors were removed, background fluorescence from the yeast and wort was minimal. To test if the nanosensors were toxic towards yeast we performed a metabolic assay to assess the impact of the sensors (Fig. S20†). The assay showed that sensors had minimal effect on cell growth and viability as compared to samples grown in its absence, similar to previous reports on similar nanosensors.43 Luminescence spectra for yeast (see Fig. S21†) showed that the longer excitation and emission wavelengths of the nanosensors was beneficial as compared to a shorter wavelength dye combination (such as PtTFPP and DiA)18 where greater background fluorescence was seen at high concentrations of yeast and wort.
Discussion
In this work, we introduce an approach for measuring oxygen dynamics with a ratiometric near-IR sensor tuned for biological applications. These nanoparticles present a sensing method that demonstrates highly desirable traits such as sensitivity, tunability and ease of synthesis.Our calibration data shows that the oxygen-responsive dye is sensitive to changes in oxygen that occur between atmospheric conditions (21% gas, 6.65 mg L−1 in solution) and anoxic conditions (0%, 0 mg L−1). This allows us to identify changes in dissolved oxygen levels with respect to spatial and temporal gradients, and the NIR wavelengths allow use in samples with high background fluorescence in visible wavelengths. Reversibility is also an important aspect of these sensors – the luminescent properties of the nanosensor do not change after an oxygen quenching event. Additionally, PtOEPK and DiD are part of dye families which are known for good photostability. Papkovsky et al. showed that PtOEPK was stable for at least 18 h under continuous illumination.44 While DiD specifically has not been tested, carbocyanines have been shown to demonstrate good photostability as well.45,46 This reversibility and stability are what enables the sensors to be a powerful tool for monitoring both increases and decreases in oxygen concentration as a given sample changes over time as one would see in metabolic monitoring. Also, while the sensors have not been tested under extreme pH or temperature conditions the sensors have shown to work well in the most common physiological and biological conditions. In other biological samples, it is possible that chemicals besides oxygen can affect the luminescent property of the sensor, although the ratiometric nature of the readout does help to mitigate some potential artifacts (e.g. sensor concentration, degradation, and distribution) as both dyes are likely to be impacted similarly.
The application of these sensors in monitoring oxygen in yeast samples shows that the sensors function in a biological setting. In this particular case, we look at how the metabolic activity of yeast can be monitored by tracking the oxygen levels in solution throughout the lifetime of the microorganism. The sensors showed suitable sensitivity in real time to distinguish the relative amount of oxygen being consumed based on the number of yeast cells present as well as the activity level of the strain of yeast cell. Therefore, there is the potential to study subtle differences in the metabolic activity of randomized or specifically tuned yeast samples, or even other types of bacterial or tissue samples, in a high throughput manner. The oxygen sensor method was also able to pick up on distinct response characteristic in the presence of an antimicrobial agent. While most antimicrobial susceptibility tests simply determine whether the microorganism is or isn't susceptible to the agent, our sensors can pinpoint when metabolic activity is activated or slowed based on the changes in oxygen concentration. If the rate of atmospheric oxygen diffusion is known or controlled (to e.g. ∼0 mg cm−2 s−1) then the rate of oxygen consumption could be measured based on the slope of the ratiometric signal at given time points. Even without controlling the oxygen diffusion into the solution, it is likely that the oxygen concentrations were near zero based on the ratiometric signal. As we increase the yeast concentration in the samples, the higher yeast concentrations can more efficiently draw down the oxygen concentration to a lower steady state given the same oxygen transport rate into the solution for all yeast concentrations. This may also explain the “overshoot” in the temporal data at the highest yeast concentrations, where the actively metabolizing yeast are able to bring oxygen below the steady state value before transitioning to fermentative metabolism. This effect is more pronounced at higher yeast density as expected from their increased total oxygen consumption.
Potassium metabisulfite affects yeast viability as sulfur dioxide is formed upon dissolution into aqueous media. The sulfur dioxide and other by-products kill the yeast causing metabolic activities to halt. The PMB experiment highlights the need for a reversible sensor, as without it the sensors would not be able to detect the increase in oxygen concentrations after the cessation of metabolism. Interestingly, the PMB added at different time points (0 h and 42 h) had two very different effects on metabolism. A lag period was observed in samples with PMB added at 0 h where they initially metabolized oxygen but would later die. There are two potential mechanisms which could account for that. The first is that sulfur dioxide converts into sulfurous acid under low pH conditions further increasing the toxicity of the agent to the cells. Yeast are known to decrease the pH in their surrounding environment over time and thus the yeast could be dying as a result of increasing sulfurous acid concentrations over time.47 The second possibility is that sulfur dioxide is bound by glucose and other carbonyl containing sugars, therefore reducing its efficacy. As yeast metabolize the glucose in the surrounding medium, more sulfur dioxide is present to poison the cells.47
The different response to PMB from the two yeast strains used may result from the different lineage of these yeasts, with Kveik yeasts being genetically distinct from traditional brewing yeasts48 and showing improved stress tolerance.49 Resistance to sulfur dioxide in yeast is something that can vary greatly from strain to strain due mostly to genetic factors. Some variants have adapted to survive sulfur dioxide through mechanisms such as (1) entering a dormant, but “protected” state, (2) removal of sulfur dioxide through efflux pumps (3) reduction of sulfur dioxide through sulfur-based metabolic pathways and (4) production of biomolecules which bind and inactivate sulfur dioxide.50
The ratiometric nature of the measurement also gives us several other advantages in biological systems. Not only does it account for well-to-well or spatial variations in nanosensor concentration, but it can also be used to account for optical artifacts which may alter the luminescent measurements when just one dye is used. In yeast, the total amount of light absorbed or scattered in the well increases as the cell density increases. Autofluorescence of microbial species is another property which could change optical aspects of the assay. The advantage of these sensors is that the excitation source will be affected equally by these artifacts given that they use the same excitation wavelength. While there could be optical differences in the emission signals, absorption concerns are less common at near-IR wavelengths. Another aspect of these sensors that we observe is their ability to work effectively in a 200 μL to 2 mL volume range (and presumably both smaller and larger) useful for experiments that require low volumes or high throughput automated screening assays.
In applications such as what we have demonstrated in this paper, ratiometric nanosensors have several advantages over previously developed sensors. In other methods such as electrochemical and optode-based methods there remain several factors which make oxygen-sensing difficult such as disruption of the system, consumption of the analyte and limited spatial information. Microelectrodes generally suffer from all three as the needle must be in direct proximity of the microbial system and will consume the analyte due to the reduction of oxygen needed for the sensing mechanism.9 Our nanosensors overcome these issues as we show that they are non-toxic and reversible without consuming the analyte being measured thus portraying a more accurate depiction of the system under investigation. Both electrochemical and optode-based methods suffer from limited spatial resolution due to them only being able to sense in one (microelectrodes) or two (optodes) dimensions.9,12 In applications, where spatial information is needed, nanosensors are a more suitable method of measurement.
The stability of the reference dye is an aspect of the sensors that needs to be further characterized to fully validate the accuracy of our ratiometric readout. The advantage of the ratiometric approach is that it allows us to account for factors which affect both dyes equally within the sensor. If the DiD dye is less stable to certain factors than the PtOEPK dye, then the ratiometric approach does not work as well under those conditions. Due to the nature of the oxygen-quenching mechanism, platinum porphyrin dyes are known to have poor sensitivity at oxygen levels above atmospheric conditions meaning they would only be suitable for monitoring organisms that consume oxygen and likely not ones that produce oxygen as a part of their metabolic function or only minimally impact oxygen concentrations.
There are a variety of ways that future work could help to overcome these issues. The dye combination can be modified to improve consistency of the ratiometric approach across a wider variety of settings. Ideally, a reference dye that is more stable and further shifted into the NIR emission range should help to overcome factors such as dye susceptibility and sample autofluorescence. Exploring oxygen-sensitive dyes that emit further into the NIR-I (700–1000 nm) or even into the NIR-II (1000–3000 nm) range would open up the possibility of measuring oxygen in tissue samples with high levels of light absorption. The sensitivity and nanometer-scale of our sensors allows scaling down volumes and scaling up throughput to 384 and 1536 well plates that can screen a larger number of samples at once in an automated setting. A similar approach could also be applied to other analytes critical to cellular processes.
Conclusions
In this study we used a dual luminophore ratiometric nanosensor containing a near-IR oxygen-sensitive dye to observe aerobic oxygen consumption in samples of brewing yeast. The sensor demonstrated good sensitivity, linearity, and reversibility in the anoxic to atmospheric oxygen range, making it viable for measuring oxygen changes in temporal experiments.In live samples, the sensitivity of the sensors allows for trends in oxygen metabolism to be tracked for up to 60 hours in yeast samples. We distinguished oxygen consumption differences between samples with varying cell density, cell strain and exposure to antimicrobial agents. The ratiometric aspect of the sensors allows us to overcome optical artifacts such as light absorption and scattering characteristics that change over time due to cell proliferation.
Overall, these sensors are a favorable platform for the testing and screening of biological samples to measure oxygen consumption in high throughput assays. These same principles could potentially be applied to bacterial, tissue, cell, and other yeast cultures in the future.
Author contributions
SCS: conceptualization, investigation, methodology, validation, visualization, writing – original draft, writing – review & editing. KJC: conceptualization, funding acquisition, project administration, supervision, visualization, writing – review & editing.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors would like to thank Tyler Sodia and Adrian Mendonsa for proofreading this manuscript. Bikram Adhikari helped with cell counting protocols and cytotoxicity with equipment provided by the Krebs Lab at Colorado School of Mines. Michael Stadick and Jon Peters at Colorado School of Mines fabricated the gas bubbling system. This research was supported by the US Department of Energy (DOE) office of Science, Office of Biological and Environmental Research Bioimaging Science Program under subcontract B643823 (to KJC) and the LLNL 3DQ Microscope Project, SCW1713.References
- D. B. Papkovsky and A. V. Zhdanov, Free Radicals Biol. Med., 2016, 101, 202–210 CrossRef CAS PubMed.
- J. Kieninger, A. Weltin, H. Flamm and G. A. Urban, Lab Chip, 2018, 18, 1274–1291 RSC.
- S. Jouanneau, L. Recoules, M. J. Durand, A. Boukabache, V. Picot, Y. Primault, A. Lakel, M. Sengelin, B. Barillon and G. Thouand, Water Res., 2014, 49, 62–82 CrossRef CAS PubMed.
- S. H. Lin, T. M. Lin and H. G. Leu, J. Environ. Eng., 1998, 124, 265–271 CrossRef CAS.
- M. Moßhammer, K. E. Brodersen, M. Kühl and K. Koren, Microchim. Acta, 2019, 186, 126 CrossRef.
- J. R. Stetter and J. Li, Chem. Rev., 2008, 108, 352–366 CrossRef CAS.
- S. R. Burge, K. D. Hristovski, R. G. Burge, D. A. Hoffman, D. Saboe, P. Chao, E. Taylor and S. S. Koenigsberg, Sci. Total Environ., 2020, 742, 140528 CrossRef CAS.
- J. Xia and S. Sonkusale, Analyst, 2021, 146, 2983–2990 RSC.
- O. S. Wolfbeis, BioEssays, 2015, 37, 921–928 CrossRef CAS PubMed.
- P. Hartmann, M. J. P. Leiner and M. E. Lippitsch, Sens. Actuators, B, 1995, 29, 251–257 CrossRef CAS.
- S. M. Borisov, in RSC Detection Science, The Royal Society of Chemistry, 2018, vol. 2018-Janua, pp. 1–18 Search PubMed.
- M. Moßhammer, M. Strobl, M. Kühl, I. Klimant, S. M. Borisov and K. Koren, ACS Sens., 2016, 1, 681–687 CrossRef.
- B. D. MacCraith, C. M. McDonagh, G. O'Keeffe, E. T. Keyes, J. G. Vos, B. O'Kelly and J. F. McGilp, Analyst, 1993, 118, 385–388 RSC.
- O. Braissant, M. Astasov-Frauenhoffer, T. Waltimo and G. Bonkat, Front. Microbiol., 2020, 11, 1–25 CrossRef PubMed.
- Y. Amao, Microchim. Acta, 2003, 143, 1–12 CrossRef CAS.
- M. Quaranta, S. M. Borisov and I. Klimant, Bioanal. Rev., 2012, 4, 115–157 CrossRef.
- B. Jana, S. Bhattacharyya and A. Patra, Bull. Mater. Sci., 2018, 41, 122 CrossRef.
- M. P. Jewell, A. A. Galyean, J. K. Harris, E. T. Zemanick and K. J. Cash, Appl. Environ. Microbiol., 2019, 85, 1–12 CrossRef.
- Y. E. K. Lee, E. E. Ulbrich, G. Kim, H. Hah, C. Strollo, W. Fan, R. Gurjar, S. Koo and R. Kopelman, Anal. Chem., 2010, 82, 8446–8455 CrossRef PubMed.
- K. S. Gkika, A. Kargaard, C. S. Burke, C. Dolan, A. Heise and T. E. Keyes, RSC Chem. Biol., 2021, 2, 1520–1533 RSC.
- X. Zheng, H. Tang, C. Xie, J. Zhang, W. Wu and X. Jiang, Angew. Chem., Int. Ed., 2015, 54, 8094–8099 CrossRef CAS PubMed.
- X. Wang, J. A. Stolwijk, T. Lang, M. Sperber, R. J. Meier, J. Wegener and O. S. Wolfbeis, J. Am. Chem. Soc., 2012, 134, 17011–17014 CrossRef CAS.
- L. Ding, W. Zhang, Y. Zhang, Z. Lin and X. Wang, Anal. Chem., 2019, 91, 15625–15633 CrossRef CAS.
- A. Byrne, J. Jacobs, C. S. Burke, A. Martin, A. Heise and T. E. Keyes, Analyst, 2017, 142, 3400–3406 RSC.
- C. Zhou, W. Zhao, F. You, Z. Geng and H. Peng, ACS Sens., 2019, 4, 984–991 CrossRef CAS PubMed.
- S. C. Saccomano, M. P. Jewell and K. J. Cash, Sens. Actuators Rep., 2021, 3, 100043 CrossRef.
- J. Ehgartner, M. Strobl, J. M. Bolivar, D. Rabl, M. Rothbauer, P. Ertl, S. M. Borisov and T. Mayr, Anal. Chem., 2016, 88, 9796–9804 CrossRef CAS PubMed.
- K. J. Cash and H. A. Clark, Anal. Chem., 2013, 85, 6312–6318 CrossRef CAS.
- J. Frangioni, Curr. Opin. Chem. Biol., 2003, 7, 626–634 CrossRef CAS PubMed.
- J.-B. Li, H.-W. Liu, T. Fu, R. Wang, X.-B. Zhang and W. Tan, Trends Chem., 2019, 1, 224–234 CrossRef CAS PubMed.
- A. Jo, J. Sung, S. Lee, H. Nam, H. W. Lee, J. Park, H. M. Kim, E. Kim and S. B. Park, Bioconjugate Chem., 2018, 29, 3394–3401 CrossRef CAS.
- P. W. Barone, S. Baik, D. A. Heller and M. S. Strano, Nat. Mater., 2004, 4, 86–92 CrossRef.
- K. J. Cash, C. Li, J. Xia, L. V. Wang and H. A. Clark, ACS Nano, 2015, 9, 1692–1698 CrossRef CAS.
- T. Hirayama, G. C. Van de Bittner, L. W. Gray, S. Lutsenko and C. J. Chang, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 2228–2233 CrossRef CAS.
- D. B. Papkovsky, G. V. Ponomarev, W. Trettnak and P. O'Leary, Anal. Chem., 1995, 67, 4112–4117 CrossRef CAS.
- X. D. Wang and O. S. Wolfbeis, Chem. Soc. Rev., 2014, 43, 3666–3761 RSC.
- D. B. Papkovsky, G. V. Ponomarev and O. S. Wolfbeis, Spectrochim. Acta, Part A, 1996, 52, 1629–1638 CrossRef.
- T. Doussineau, A. Schulz, A. Lapresta-Fernandez, A. Moro, S. Körsten, S. Trupp and G. J. Mohr, Chem. – Eur. J., 2010, 16, 10290–10299 CrossRef CAS PubMed.
- J. R. Lakowicz and G. Weber, Biochemistry, 1973, 12, 4161–4170 CrossRef CAS PubMed.
- S. Liu, X. Fang, B. Lu and D. Yan, Nat. Commun., 2020, 11, 4649 CrossRef CAS PubMed.
- J. M. Dubach, S. Das, A. Rosenzweig and H. A. Clark, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 16145–16150 CrossRef CAS PubMed.
- Q. Li, Z. Bai, A. O'Donnell, L. M. Harvey, P. A. Hoskisson and B. McNeil, Biotechnol. Lett., 2011, 33, 457–467 CrossRef CAS.
- J. M. Dubach, D. I. Harjes and H. A. Clark, Nano Lett., 2007, 7, 1827–1831 CrossRef CAS PubMed.
- D. B. Papkovsky, Sens. Actuators, B, 1995, 29, 213–218 CrossRef CAS.
- H. Liu and D. Wu, Curr. Drug Delivery, 2016, 13, 40–48 CrossRef CAS.
- M. G. Honig and R. I. Hume, Trends Neurosci., 1989, 12, 333–341 CrossRef CAS PubMed.
- A. D. King, J. D. Ponting, D. W. Sanshuck, R. Jackson and K. Mihara, J. Food Prot., 1981, 44, 92–97 CrossRef CAS PubMed.
- R. Preiss, C. Tyrawa, K. Krogerus, L. M. Garshol and G. van der Merwe, Front. Microbiol., 2018, 9, 1–18 CrossRef.
- B. Foster, C. Tyrawa, E. Ozsahin, M. Lubberts, K. Krogerus, R. Preiss and G. van der Merwe, bioRxiv, 2021 Search PubMed , 2021.07.26.453768.
- S. C. Morgan, J. J. Haggerty, B. Johnston, V. Jiranek and D. M. Durall, Fermentation, 2019, 5, 69 CrossRef CAS.
- M. P. Jewell, S. C. Saccomano, A. A. David, J. K. Harris, E. T. Zemanick and K. J. Cash, Analyst, 2020, 145, 3996–4003 RSC.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1an01855h |
| This journal is © The Royal Society of Chemistry 2022 |