Open Access Article
Open Access ArticleA self-quenching fluorescence probe-mediated exponential isothermal amplification system for highly sensitive and specific detection of microRNAs†
Jun
Zhao
 a,
Jiandong
Wu
d,
Xiaosong
Wu
ab,
Guoqing
Deng
a,
Yong
Liu
a,
Francis
Lin
*c and
Ling
Zhu
*a
a,
Jiandong
Wu
d,
Xiaosong
Wu
ab,
Guoqing
Deng
a,
Yong
Liu
a,
Francis
Lin
*c and
Ling
Zhu
*a
aCenter of Engineering Technology Research for Biomedical Optical Instrument, Hefei Institutes of Physical Science, Chinese Academy of Sciences, Hefei 230031, P. R. China. E-mail: zhul@aiofm.ac.cn
bUniversity of Science and Technology of China, Hefei 230026, P. R. China
cUniversity of Manitoba, Winnipeg R3T2N2, Canada. E-mail: Francis.Lin@umanitoba.ca
dInstitute of Biomedical and Health Engineering, Shenzhen Institutes of Advanced Technology, Chinese Academy of Sciences, Shenzhen 518055, P. R. China
First published on 5th November 2021
Abstract
We designed an efficient self-quenching fluorescence probe and constructed this probe-mediated exponential isothermal amplification system for miRNA detection. Owing to the significant improvement in the detective signal-to-background ratio, a wide dynamic range of 9 orders of magnitude and a limit of detection as low as 0.08 aM can be easily achieved in a single step. Furthermore, benefiting from the additional advantages of high specificity and biocompatibility, the proposed method has been demonstrated to be capable of accurately quantifying miRNA biomarkers in serum, which will provide promising perspectives for clinical diagnosis.
MicroRNAs (miRNAs) are a class of endogenous, noncoding, single-stranded RNAs that have been considered to be important regulators of activity in human life. Since circulating miR-21 was firstly reported as a potential new marker for diffuse large B-cell lymphoma,1 studies on circulating miRNAs have attracted extensive attention from researchers. A considerable number of studies have shown that circulating miRNAs could stay stable in a variety of bodily fluids such as serum, plasma, and urine.2 Meanwhile, owing to their higher specificity in expression and lower invasiveness in sampling, circulating miRNAs have been recognized as ideal biomarkers for early diagnosis of cancer.3 However, due to the particularities of their sequences themselves, effective detection of miRNAs is a great challenge. Therefore, it is of major significance to achieve highly sensitive and highly specific detection of miRNAs.
Nucleic acid amplification-based miRNA detection has always been considered as one of the rapid, specific and sensitive techniques. Quantitative reverse transcription-polymerase chain reaction (qRT-PCR), thereinto, is recognized as the gold standard in detecting miRNAs. Nevertheless, there are still some insurmountable hurdles in the qRT-PCR: (i) reverse transcription is a mandatory step for miRNA detection when employing PCR,4 and this complex process of pretreatments may cause serious degradation of miRNAs; (ii) rapid heating and cooling, and a precise temperature controlling system are critical for efficient PCR amplification,5 and thus it poses fairly high requirements to equipment performance; (iii) the DNA polymerase used in PCR reactions is poorly tolerated in complex biological samples,6 so its reaction can be inhibited by some cellular contents. These are, without a doubt, the most critical factors that have restricted its applications in clinical research and diagnosis for a long time. Recently, with the rapid development of the isothermal amplification technology, lots of new isothermal amplification methods have been developed and proved to be particularly suitable for miRNA detection. Compared with the traditional PCR, isothermal amplification can not only greatly improve the efficiency of miRNA detection, but also disengage itself from sophisticated equipment.7 Furthermore, it is particularly suitable for the scenarios and applications of point-of-care testing (POCT), which can lay a solid foundation for the rapid development of clinical diagnosis.8,9 Hence, it is gradually being recognized as a novel detection method that is comparable to the PCR.
It is generally known that both sensitivity and specificity are the most important indicators to be considered when it comes to the detection performance of one certain methodology. Previously, the problems of poor sensitivity and specificity were always present in the isothermal amplification technology, and these were largely attributed to two facts: (i) some real-time detection assays could merely be monitored using fluorescent dyes owing to their methodological limitations; (ii) multiple primers are required for the amplification reaction, causing serious self-amplification between primers. It is, therefore, a high priority to address these problems. In recent years, the use of an efficient probe has been considered to be an effective solution for improving the specificity and sensitivity in isothermal amplification.10 Furthermore, multiplexed detection of targets can be easily achieved, benefiting from the probes modified by various fluorescence labeling methods. Nevertheless, due to the particularity of the structure in the traditional probe (TaqMan), the signal of the fluorophore cannot be completely quenched before the reaction, and thus a certain background fluorescence is inevitable in the reaction system. It should be pointed out that the higher the background fluorescence is, the lower the signal-to-background ratio of detection will be.11 In other words, some weak signals will be hardly detected if the signal-to-background ratio is fairly low. In this case, this might substantially affect the detection sensitivity of the system. Therefore, researchers have always been devoted to studies on how to decrease the background signal of the probe, and a variety of fluorophores with lower background fluorescence were developed.12–14 Nevertheless, these solutions remain far from satisfactory due to a significant increase in the cost and complexity of probe synthesis.
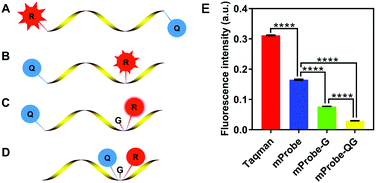
Hence, a new self-quenching fluorescence probe was proposed in this study, and the principle of the designed probe is presented in Fig. 1. Firstly, a modified probe (mProbe) (Fig. 1B) to be used for the isothermal amplification assay was designed, and the spatial distance between the reporter and the quencher was obviously shortened when compared with the TaqMan probe (Fig. 1A). In this way, its background fluorescence could be decreased. Meanwhile, the fluorescence signals of the mProbe could be instantaneously released as soon as the reactions of digestion and replacement occur. Yet, although the difference of background fluorescence between the TaqMan and the mProbe was extremely significant (Fig. 1E, mProbe versus TaqMan, p < 0.0001), the background fluorescence of mProbe was still at a high level. To further reduce the background fluorescence of the mProbe, the guanine (G) base, which has a significant effect on fluorescence quenching,15 was employed for the structural modification of the probe itself for the first time. Consequently, a self-quenching fluorescence probe with an additional quencher of the G base was constructed (mProbe-G) (Fig. 1C), and it was clear that this new structure could contribute to reducing the background fluorescence of mProbe (Fig. 1E, mProbe-G versus mProbe, p < 0.0001). Hence, it is of significant importance to employ G bases as new quenchers in the probe due to their advantages in reducing the cost and avoiding complicated synthesis. Furthermore, an enhanced self-quenching fluorescence probe (mProbe-QG) (Fig. 1D) was designed in which both the quencher and the G base were placed adjacent to the reporter. This unique design could bring the background fluorescence of the probe to almost zero (Fig. 1E, mProbe-QG versus mProbe-G, p < 0.0001), which explained its excellent performance in the self-quenching of fluorescence. More importantly, the proposed mProbe-QG has significant advantages in fluorescence quenching, even compared with previously reported probes with lower background fluorescence (Fig. S1, ESI†).16,17
In this study, a self-quenching fluorescence probe-mediated exponential isothermal amplification system (SqPIA) for miRNA detection was developed, as illustrated in Scheme 1. The SqPIA system is mainly composed of two cycles, and exponential growth of targeted products and signals can be easily achieved through multiple rounds of cascade amplification. The reaction is firstly triggered by targeted miRNA, and the extension of the chain can be launched instantly as soon as the RNA–DNA hybrids (miRNA to mProbe-QG) formed. With the formation of a double strand of RNA–DNA, enzymatic digestion begins to occur, then the DNA part of the RNA–DNA hybrids is nicked, thereby creating a gap. Meanwhile, the sequence at the 5′ end of the incision continues to extend and displace the sequence which labeled a reporter at the 3′ end of the incision. Thus, the reporter will be free from the quenching effects of the quencher and the G base, and its signal could be instantaneously released. Meanwhile, an equal number of single-stranded DNAs (ssDNA, named M) can also be generated. In particular, the sequence of M is reversed complementary to the targeted miRNA sequence. It is worth mentioning that the reactions of extension, nicking, and displacement can be repeated in cycle I and continuously generate lots of single-stranded DNAs (M) and signals. To achieve exponential amplification of products and exponential growth of signals, a hairpin probe (HPP) (Fig. S2, ESI†) with the same self-quenching feature was also designed and primarily involved in cycle II. Once M hybridizes with the 3′ end of HPP, the extension reaction will begin and open the stem of HPP, and thus the reporter can separate from the quencher and the signals will be released. Besides, the reactions of nicking and displacement can be performed simultaneously, generating a new kind of ssDNA, named M*. In particular, the M* sequence is exactly the same as the targeted miRNA, but with all the U bases replaced by T bases, so it can serve as a direct target to trigger cycle I again. In this way, multiple rounds of cascade amplification within or between cycle I and cycle II can proceed continuously until complete consumption of materials.
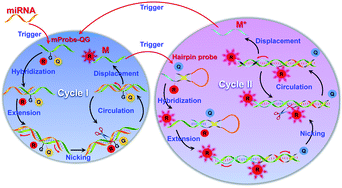 | ||
| Scheme 1 Schematic illustration of a self-quenching probe-mediated exponential isothermal amplification system for miRNA detection. | ||
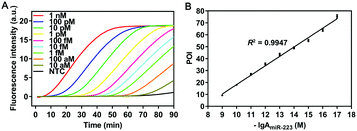
To validate the amplification mechanism of the proposed SqPIA system for miRNA detection, a series of confirmatory trials were conducted. Firstly, an agarose gel electrophoresis assay was performed to validate the specificity of the amplification product generated from the SqPIA system (Fig. S3, ESI†). Besides, the real-time SqPIA assay was performed to investigate the roles of the regular fluorescence probe (mProbe) and the self-quenching fluorescence probes (mProbe-G and mProbe-QG) in improving the signal-to-background ratio (Fig. S4, ESI†). Finally, to determine the sensitivity of the SqPIA system in miRNA detection, the reaction system was optimized in advance (Fig. S5–S8, ESI†). Then, the synthetic miR-223 standard with different concentrations was analyzed under the optimal conditions. As shown in Fig. 2, a good linear correlation was observed between POI values and negative logarithmic values within the range of 10 aM to 1 nM. Furthermore, the correlation equation was POI = 7.8767 (−lg![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AmiR-223) − 60.5410 (R2 = 0.9947), where AmiR-223 is the concentration of miR-223. In addition, the limit of detection (LOD) was calculated to be as low as 0.08 aM (mean ± 3SD, n = 21), particularly the mean value was the average POI value of 21 assays in blank control groups, and the tripled standard deviation was added. Furthermore, to evaluate the overall performance of the SqPIA system, comparisons between our approach and recently proposed ones for miRNA detection were made. As shown in Table S2 (ESI†), the detection efficiency, detection range, LOD, and other performance metrics observed in our approach were better than those of others.18–27 Without a doubt, this ultrasensitive detection threshold can be ascribed to the following reasons: (i) the adoption of self-quenching fluorescence probes greatly improves the detective signal-to-background ratio. Thus, the targets at lower concentrations can be easily detected; (ii) only two kinds of probes are employed in the system, the occurrence of non-specific amplification is avoided, and the exponential growth of products and signals is also guaranteed. By contrast, this is the problem that cannot be addressed by loop-mediated isothermal amplification or other technologies.28 Meanwhile, the complementary region is completely absent between the employed probes, thus minimizing the risk of non-specific amplification. Indeed, these considerations did not attract attention from other researchers;28,29 (iv) the effects of the secondary structure of the sequence on the amplification efficiency are taken into account for the first time, since the secondary structure of the sequence is greatly affected by temperature, as shown in Fig. S9 (ESI†). Without a doubt, the self-hybridization of the sequence can greatly influence the binding efficiency and thus affect the detection sensitivity. In summary, the aforementioned technical solutions can provide particularly favorable conditions for making highly sensitive detection of circulating miRNAs practically applicable.
AmiR-223) − 60.5410 (R2 = 0.9947), where AmiR-223 is the concentration of miR-223. In addition, the limit of detection (LOD) was calculated to be as low as 0.08 aM (mean ± 3SD, n = 21), particularly the mean value was the average POI value of 21 assays in blank control groups, and the tripled standard deviation was added. Furthermore, to evaluate the overall performance of the SqPIA system, comparisons between our approach and recently proposed ones for miRNA detection were made. As shown in Table S2 (ESI†), the detection efficiency, detection range, LOD, and other performance metrics observed in our approach were better than those of others.18–27 Without a doubt, this ultrasensitive detection threshold can be ascribed to the following reasons: (i) the adoption of self-quenching fluorescence probes greatly improves the detective signal-to-background ratio. Thus, the targets at lower concentrations can be easily detected; (ii) only two kinds of probes are employed in the system, the occurrence of non-specific amplification is avoided, and the exponential growth of products and signals is also guaranteed. By contrast, this is the problem that cannot be addressed by loop-mediated isothermal amplification or other technologies.28 Meanwhile, the complementary region is completely absent between the employed probes, thus minimizing the risk of non-specific amplification. Indeed, these considerations did not attract attention from other researchers;28,29 (iv) the effects of the secondary structure of the sequence on the amplification efficiency are taken into account for the first time, since the secondary structure of the sequence is greatly affected by temperature, as shown in Fig. S9 (ESI†). Without a doubt, the self-hybridization of the sequence can greatly influence the binding efficiency and thus affect the detection sensitivity. In summary, the aforementioned technical solutions can provide particularly favorable conditions for making highly sensitive detection of circulating miRNAs practically applicable.
To further evaluate the analytical performance of the proposed SqPIA system in multiplexed detection of miRNAs, a combination of targets including miR-21, miR-27a and miR-223 was detected using the same reaction system. As shown in Fig. S10A (ESI†), the real-time curves for the triplex detection of miR-21, miR-27a and miR-223 showed good homogeneity. Furthermore, these could be validated through a more intuitive linear relationship (R2 > 0.9) between the POI values and dilution of miRNAs (Fig. S10B–D, ESI†), indicating that the SqPIA system was capable of achieving multiplexed detection of miRNAs in a single reaction system. Without a doubt, this will be of immense significance for improving the detection accuracy of circulating miRNA biomarkers clinically.
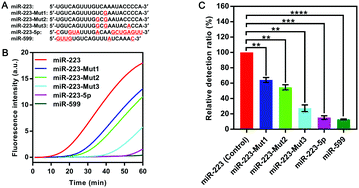
To explore the specificity of the SqPIA method, the same concentration (100 pM) of miR-223 and its homologous sequences and mutant sequences (Fig. 3A) were simultaneously detected using the probes corresponding to miR-223. As shown in Fig. 3B. It was observed that the real-time curve of miR-223 could be effectively distinguished from the other groups under equivalent conditions. To further validate the analytical specificity of the system, the relative detection ratio of the homologous sequences was calculated using the formula: 2−[POI(Mut) − POI(Wet)]/POI(Wet) × 100% (Fig. 3C). Obviously, a significant difference in the value of the relative detection ratio was obtained not only for the two homologous genes, but also for the three mutants, relative to the control group (p < 0.01). More importantly, miR-223-5p (15.22% ± 1.90, p < 0.001) and miR-599 (12.87% ± 0.43, p < 0.0001) as the homologous sequences of miR-223 were barely undetectable in the SqPIA. Hence, these results clearly demonstrate that the proposed SqPIA method has good specificity in miRNA detection.
Finally, in order to determine the applicability in real biological samples and the accuracy in the quantification of the proposed SqPIA method, the expression pattern of the three serum miRNAs was analyzed between 8 HCC patients and 4 healthy subjects. Particularly, prior to the quantitative assay, the primers and probes used in both RT-qPCR and SqPIA were tested and optimized (Fig. S11 and S12, ESI†). Then, the expression of miR-21, miR-27a, and miR-223 in HCC patients and healthy subjects is presented in Fig. S13 (ESI†). It was not difficult to see that the obtained expression patterns of the same sample from two approaches were roughly the same. Therefore, these results have sufficiently proved that the SqPIA method was well tolerated in complex biological samples, and it was capable of accurately quantifying miRNA biomarkers in human serum.
In summary, we developed a self-quenching fluorescence probe-mediated exponential isothermal amplification system for miRNA detection. With a unique probe designed, as well as a cascade amplification system constructed, the SqPIA approach could achieve the goals of an ultrasensitive detection limit of 0.08 aM, an effective distinction of a single-base difference among homologous miRNAs and a triplex detection of miRNAs. Furthermore, compared with previous studies on miRNA assays,30,31 the SqPIA strategy has its significant advantages: (i) the sensitivity and the signal-to-background ratio in miRNA detection were greatly improved; (ii) the usage of self-quenching probes instead of dyes could make multiple target detection in a single sample more feasible; (iii) the use of probes with a special design of structure and strict control of the amount can effectively reduce non-specific amplification. More importantly, the proposed method can get rid of the dependence on sophisticated equipment, when compared with the traditional PCR. In a word, based on the advantages of the SqPIA method, we firmly believe it will make a significant difference in the future if combined with the assay of digital readout, or applied to the POCT system.
This work was financially supported by the Natural Science Foundation of Anhui Province, China (2108085QC96), the Science and Technology Service Network Initiative of Chinese Academy of Sciences (KFJ-STS-ZDTP-080), and the Science and Technology Major Program of Anhui, China (18030801139).
Conflicts of interest
There are no conflicts to declare.Notes and references
- C. H. Lawrie, S. Gal, H. M. Dunlop, B. Pushkaran, A. P. Liggins, K. Pulford, A. H. Banham, F. Pezzella, J. Boultwood, J. S. Wainscoat, C. S. R. Hatton and A. L. Harris, Br. J. Haematol., 2008, 141, 672–675 CrossRef PubMed.
- J. Jeon, E. Olkhov-Mitsel, H. L. Xie, C. Q. Yao, F. Zhao, S. Jahangiri, C. Cuizon, S. Scarcello, R. Jeyapala, J. D. Watson, M. Fraser, J. Ray, K. Commisso, A. Loblaw, N. E. Fleshner, R. G. Bristow, M. Downes, D. Vesprini, S. Liu, B. Bapat and P. C. Boutros, J. Natl. Cancer Inst., 2020, 112, 247–255 CrossRef PubMed.
- X. J. Lin, Y. T. Chong, Z. W. Guo, C. Xie, X. J. Yang, Q. Zhang, S. P. Li, Y. J. Xiong, Y. F. Yuan, J. Min, W. H. Jia, Y. S. Jie, M. S. Chen, M. X. Chen, J. H. Fang, C. X. Zeng, Y. J. Zhang, R. P. Guo, Y. K. Wu, G. L. Lin, L. M. Zheng and S. M. Zhuang, Lancet Oncol., 2015, 16, 804–815 CrossRef CAS PubMed.
- C. F. Chen, D. A. Ridzon, A. J. Broomer, Z. H. Zhou, D. H. Lee, J. T. Nguyen, M. Barbisin, N. L. Xu, V. R. Mahuvakar, M. R. Andersen, K. Q. Lao, K. J. Livak and K. J. Guegler, Nucleic Acids Res., 2005, 33, e179 CrossRef PubMed.
- L. N. Sanford and C. T. Wittwer, Anal. Biochem., 2013, 434, 26–33 CrossRef CAS PubMed.
- A. Kuffel, A. Gray and N. N. Daeid, Leg. Med., 2021, 135, 63–72 Search PubMed.
- P. Fu, M. J. Xu, S. Xing, Y. Zhao and C. Zhao, Analyst, 2021, 146, 3242–3250 RSC.
- Y. R. Cao, J. J. Wu, B. Pang, H. Q. Zhang and X. C. Le, Chem. Commun., 2021, 57, 6871–6874 RSC.
- L. L. Guo, H. Li, R. J. Zhao, Y. D. Tang and B. L. Li, Chem. Commun., 2021, 57, 5714–5717 RSC.
- Y. J. Xu, J. Luo, M. S. Wu, F. F. Hu, Z. S. Lu, H. Jing, R. Chen and H. Zhang, Sens. Actuators, B, 2018, 269, 158–163 CrossRef CAS.
- T. Buckle, S. van der Waals, D. M. van Willigen, G. Aalderink, G. H. KleinJan and F. W. B. van Leeuwen, Theranostics, 2020, 10, 9890–9898 CrossRef CAS PubMed.
- J. Y. Xu, M. Shi, H. K. Huang, K. Hu, W. T. Chen, Y. Huang and S. L. Zhao, Analyst, 2018, 143, 3918–3925 RSC.
- S. Y. Li, K. He, R. Liao, C. Y. Chen, X. M. Chen and C. Q. Cai, Talanta, 2017, 174, 679–683 CrossRef CAS PubMed.
- G. Gines, R. Menezes, K. Nara, A. S. Kirstetter, V. Taly and Y. Rondelez, Sci. Adv., 2020, 6, eaay5952 CrossRef CAS PubMed.
- H. H. Mao, G. H. Luo, Y. X. Zhan, J. Zhang, S. Yao and Y. Yu, Analyst, 2018, 143, 3292–3301 RSC.
- S. W. Tam-Chang, T. D. Carson, L. M. Huang, N. G. Publicover and K. W. Hunter, Anal. Biochem., 2007, 366, 126–130 CrossRef CAS PubMed.
- L. M. Huang, G. H. Aryal, S. W. Tam-Chang, N. G. Publicover and K. W. Hunter, Analyst, 2016, 141, 1376–1382 RSC.
- K. Treerattrakoon, T. Jiemsakul, C. Tansarawiput, P. Pinpradup, T. Iempridee, P. Luksirikul, K. Khoothiam, T. Dharakul and D. Japrung, Anal. Biochem., 2019, 577, 89–97 CrossRef CAS PubMed.
- S. Q. Liu, H. Fang, C. J. Sun, N. N. Wang and J. Li, Analyst, 2018, 143, 5137–5144 RSC.
- X. J. Qu, H. J. Jin, Y. Q. Liu and Q. J. Sun, Anal. Chem., 2018, 90, 3482–3489 CrossRef CAS PubMed.
- W. C. Zhang, F. Hu, Q. Zhang, J. Q. Zhang, Y. Mao, P. Wang and Y. Q. Gu, Talanta, 2018, 179, 685–692 CrossRef CAS PubMed.
- S. Z. Yue, T. T. Zhao, S. Bi and Z. P. Zhang, Biosens. Bioelectron., 2017, 98, 234–239 CrossRef CAS PubMed.
- J. Y. Xu, J. J. Guo, N. Golob-Schwarzl, J. Haybaeck, X. Qiu and N. Hildebrandt, ACS Sens., 2020, 5, 1768–1776 CrossRef CAS PubMed.
- J. J. Li, S. S. Yang, C. Zuo, L. Dai, Y. C. Guo and G. M. Xie, ACS Sens., 2020, 5, 970–977 CrossRef CAS PubMed.
- Z. A. Hu, F. J. Xu, G. W. Sun, S. C. Zhang and X. R. Zhang, Chem. Commun., 2020, 56, 5409–5412 RSC.
- X. W. Wei, F. K. Bian, X. X. Cai, Y. Wang, L. J. Cai, J. Yang, Y. F. Zhu and Y. J. Zhao, Anal. Chem., 2020, 92, 6121–6127 CrossRef CAS PubMed.
- M. Kim, D. M. Kim and D. E. Kim, Analyst, 2020, 145, 6130–6137 RSC.
- W. M. Tian, P. J. Li, W. L. He, C. H. Liu and Z. P. Li, Biosens. Bioelectron., 2019, 128, 17–22 CrossRef CAS PubMed.
- A. A. Abdullah Al-Maskri, J. W. Ye, J. Talap, H. H. Hu, L. L. Sun, L. S. Yu, S. Cai and S. Zeng, Anal. Chim. Acta, 2020, 1126, 1–6 CrossRef PubMed.
- K. J. Gao, P. B. Zhang, H. Wang, H. H. Wang, F. X. Su and Z. P. Li, Chem. Commun., 2021, 57, 5570–5573 RSC.
- L. J. Wang, M. Ren, H. X. Wang, J. G. Qiu, B. H. Jiang and C. Y. Zhang, Anal. Chem., 2020, 92, 8546–8552 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1cc05522d |
| This journal is © The Royal Society of Chemistry 2021 |