Open Access Article
Open Access ArticleIntegration of exonuclease III-powered three-dimensional DNA walker with single-molecule detection for multiple initiator caspases assay†
Meng
Liu‡
a,
Rui
Xu‡
a,
Wenjing
Liu‡
b,
Jian-Ge
Qiu
*b,
Yan
Wang
*a,
Fei
Ma
*c and
Chun-yang
Zhang
 *a
*a
aCollege of Chemistry, Chemical Engineering and Materials Science, Collaborative Innovation Center of Functionalized Probes for Chemical Imaging in Universities of Shandong, Key Laboratory of Molecular and Nano Probes, Ministry of Education, Shandong Provincial Key Laboratory of Clean Production of Fine Chemicals, Shandong Normal University, Jinan 250014, China. E-mail: cyzhang@sdnu.edu.cn; fagong@sdu.edu.cn; Fax: +86-0531-82615258; Tel: +86-0531-86186033
bAcademy of Medical Sciences, The Affiliated Cancer Hospital of Zhengzhou University, Zhengzhou University, Zhengzhou, 450000, China. E-mail: jiangeqiu@zzu.edu.cn
cSchool of Chemistry and Chemical Engineering, Southeast University, Nanjing, 211189, China. E-mail: fei@seu.edu.cn
First published on 8th November 2021
Abstract
Initiator caspases are important components of cellular apoptotic signaling and they can activate effector caspases in extrinsic and intrinsic apoptotic pathways. The simultaneous detection of multiple initiator caspases is essential for apoptosis mechanism studies and disease therapy. Herein, we develop a sensitive nanosensor based on the integration of exonuclease III (Exo III)-powered three-dimensional (3D) DNA walker with single-molecule detection for the simultaneous measurement of initiator caspase-8 and caspase-9. This assay involves two peptide–DNA detection probe-conjugated magnetic beads and two signal probe-conjugated gold nanoparticles (signal probes@AuNPs). The presence of caspase-8 and caspase-9 can induce the cleavage of peptides in two peptide–DNA detection probes, releasing two trigger DNAs from the magnetic beads, respectively. The two trigger DNAs can serve as the walker DNA to walk on the surface of the signal probes@AuNPs powered by Exo III digestion, liberating numerous Cy5 and Texas Red fluorophores which can be quantified by single-molecule detection, with Cy5 indicating caspase-8 and Texas Red indicating caspase-9. Notably, the introduction of the AuNP-based 3D DNA walker greatly reduces the background signal and amplifies the output signals, and the introduction of single-molecule detection further improves the detection sensitivity. This nanosensor is very sensitive with a detection limit of 2.08 × 10−6 U μL−1 for caspase-8 and 1.71 × 10−6 U μL−1 for caspase-9, and it can be used for the simultaneous screening of caspase inhibitors and the measurement of endogenous caspase activity in various cell lines at the single-cell level. Moreover, this nanosensor can be extended to detect various proteases by simply changing the peptide sequences of the detection probes.
Introduction
Apoptosis is programmed cell death which can result in the disappearance of cells without any inflammatory phenomena.1 Caspases are a family of cysteinyl aspartate-directed proteases, and they are the central executioners of apoptosis.2 Once activated by a specific stimulus (e.g., ultraviolet (UV) radiation, γ-irradiation, heat, DNA damage, viral virulence factors),3 caspases execute the part-proteolysis of downstream substrates to trigger a cascade of events that culminates in the desired biological response: disruption of cellular membranes, breaking down of cytoplasmic and nuclear skeletons, extruding of cytosol, degradation of chromosomes, and fragmentation of the nucleus.4 Inappropriate apoptosis and deregulation of caspase activity are implicated in various human diseases, including Alzheimer’s disease,5 ischemic damage,6 autoimmune diseases,7 and cancers.8 The apoptotic-associated caspases are classified into initiators and effectors by their location in the apoptosis cascade signalling pathways. The initiator caspases (e.g., caspase-8, caspase-9, and caspase-10) are responsible for initiating the apoptosis cascade, and the effector caspases (e.g., caspase-3, caspase-6, and caspase-7) can be activated by initiator caspases to destroy cellular proteins and execute apoptosis.9 Extrinsic and intrinsic apoptotic pathways are the only two pathways of apoptosis, and caspase-8 and caspase-9 are critical participants in extrinsic and intrinsic apoptosis, respectively. In the extrinsic apoptotic pathway, caspase-8 is specifically activated by death receptors such as Fas/CD95.10 In the intrinsic apoptotic pathway, caspase-9 is activated by the accumulation of cytochrome C in the cytosol due to dissipation of the mitochondrial membrane potential.11 Therefore, the simultaneous measurement of caspase-8 and caspase-9 are of great importance for the study of the apoptosis mechanism and disease therapy.Conventional methods for caspase assays include enzyme-linked immune sorbent assay (ELISA),12 western blot,13 flow cytometry,14 and mass spectrometry.15 They usually involve multi-step and time-consuming processes,16 and extensive pre-treatment of samples.17 Recently, a variety of new methods have been developed for in vitro and in vivo detection of caspases, including electrochemical,18 colorimetric19 and fluorescence measurements.20,21 Most of these methods rely on either antibodies18 or peptide substrates.19–21 The main limitation of antibody-based assays is the requirement for high quality antibodies.22 Alternatively, peptide substrates are a more attractive choice for caspase assay with the distinct advantages of accessibility, simplicity, cost-effectiveness, and chemical definition.23 In the peptide substrate-based assays, peptide substrates are often labelled with fluorophores and the fluorescence signals are directly measured after caspase cleavage, resulting in limited sensitivity.24 The development of new methods for sensitive caspase assay is highly desirable.
Due to the highly selective and programmable Watson–Crick complementarity, DNA is becoming an increasingly attractive building material for the construction of a series of functional nanodevices (e.g., walker,25–27 gear,28 and tweezer29). In particular, a DNA walker that is able to convert chemical energy to mechanical motion is the most widely studied DNA nanodevice and has been extensively applied for molecular transport,30 DNA computing,31 chemical synthesis,32 and biosensing.33,34 DNA walkers can be classified into one-dimensional (1D), two-dimensional (2D), and three-dimensional (3D) DNA walkers in terms of dimensionality. The 1D DNA walkers move along 1D tracks such as double-stranded DNA (dsDNA) and single walled carbon nanotubes (SWCNTs).35,36 2D DNA walkers can move along the 2D surfaces of gold electrodes and DNA tile.37,38 3D DNA walkers can move along 3D scaffolds on nanoparticles such as gold nanoparticles (AuNPs),39 magnetic beads (MBs),40 SiO2@CdTe,41 Au@Fe3O4,42 hollow carbon nanospheres (HCS),43 and DNA origami.44 Such 3D DNA walkers are usually powered by nicking endonucleases,45,46 exonuclease III (Exo III),47 or the DNA catalytic hairpin assembly reaction,48 and possess large specific surface area and high DNA loading capability,49 and they can greatly enhance the local concentration of track DNA for improved walking efficiency and higher processivity,50 facilitating high signal gain in a given period of time.51
A rolling circle amplification (RCA)-based method has been developed for caspase assay,52 but it is only suitable for single target detection and is prone to a high background due to nonspecific amplification. In addition, it requires laborious procedures for the preparation of the circular template, and its accuracy is challenged by the inner filter effect and the collisional quenching of ensemble fluorescence measurements. Herein, we develop a simple and sensitive caspase nanosensor with the capability of multiplexed assay based on the integration of an Exo III-powered 3D DNA walker with single-molecule detection. The Exo III mediates highly efficient signal amplification through catalyzing the cyclic digestion of one strand of dsDNA from the 3′ end,53 which can be performed under isothermal conditions with the elimination of complex thermal cycling and nonspecific artificial amplification involving in polymerase chain reaction (PCR).54 In comparison with other isothermal amplification methods such as strand displacement amplification (SDA),55 exponential amplification reaction (EXPAR),56 rolling circle amplification,57 and loop mediated isothermal amplification (LAMP),58 the Exo III-mediated amplification takes advantage of a much simpler reaction scheme without the requirement of a specific recognition sequence for nicking endonuclease, complicated procedures for preparation of the circular template, and multiple primers for completing the amplification reaction. Moreover, in comparison with the ensemble measurement, single-molecule detection has the distinct advantages of high sensitivity, simplicity, and low sample consumption.59–65 This assay involves two peptide–DNA detection probe-conjugated magnetic beads and two signal probe-conjugated gold nanoparticles (signal probes@AuNPs). The integration of Exo III-mediated amplification with the AuNP-based DNA walker facilitates facile and efficient amplification of the caspase signal to acquire high sensitivity with reduced background, and the introduction of single-molecule detection further improves the detection sensitivity. The presence of caspase-8 and caspase-9 can induce the cleavage of peptides in two peptide–DNA detection probes, releasing two trigger DNAs from the magnetic beads. The two trigger DNAs can serve as the walker DNA to walk on the surface of signal probes@AuNPs powered by Exo III digestion, liberating numerous Cy5 and Texas Red fluorophores which can be quantified by single-molecule detection, with Cy5 indicating caspase-8 and Texas Red indicating caspase-9. This nanosensor is very sensitive with a detection limit of 2.08 × 10−6 U μL−1 for caspase-8 and 1.71 × 10−6 U μL−1 for caspase-9, and it can be used for simultaneous screening of caspase inhibitors and measurement of endogenous caspase activity in various cell lines at the single-cell level. To the best of our knowledge, the integration of Exo III-powered 3D DNA walker with single-molecule detection for the simultaneous measurement of multiple caspases has not been explored so far.
Results and discussion
The schematic illustration of the nanosensor based on the integration of the Exo III-powered 3D DNA walker with single-molecule detection for the simultaneous measurement of initiator caspase-8 and caspase-9 is shown in Scheme 1. This assay involves two peptide–DNA detection probe-conjugated magnetic beads and two signal probe-conjugated gold nanoparticles (signal probes@AuNPs). The biotinylated peptide–DNA detection probes (Fig. S1, ESI†) can be self-assembled on the streptavidin-coated MBs through the specific streptavidin–biotin interaction. The peptide domain of detection probe 1 (Scheme 1, green color) contains a tetrapeptide sequence Ile–Glu–Thr–Asp (IETD) for caspase-8 recognition, and the peptide domain of detection probe 2 (Scheme 1, magenta color) contains a tetrapeptide sequence Leu–Glu–His–Asp (LEHD) for caspase-9 recognition. The DNA domain of detection probe 1 (Scheme 1, yellow color) is complementary to signal probe 1 (Scheme 1, blue color), and the DNA domain of detection probe 2 (Scheme 1, cyan color) is complementary to signal probe 2 (Scheme 1, red color). Signal probes are attached to the surface of AuNPs via a Au–S bond to form the 3D scaffold of the DNA walker which is powered by Exo III. In the presence of caspase-8, it specifically recognizes the sequence IETD and hydrolyzes the peptide bond adjacent to the carboxylic group of the aspartic acid residue (i.e., the peptide bond between aspartic acid and glycine), releasing the cleaved detection probe 1 from the magnetic beads (Fig. S1A, ESI†). In the presence of caspase-9, it specifically recognizes the sequence LEHD and hydrolyzes the peptide bond between aspartic acid and glycine, releasing the cleaved detection probe 2 from the magnetic beads (Fig. S1B, ESI†). The intact detection probes are separated by magnetic separation. The trigger DNA in the cleaved detection probes (Scheme 1, yellow and cyan color) can serve as the walker DNA to initiate the recyclable cleavage of signal probes on the surface of the AuNPs, releasing numerous Cy5 and Texas Red fluorophores from the signal probes@AuNPs. Specifically, the hybridization of trigger DNA 1 of the cleaved detection probe 1 (Scheme 1, yellow color) and signal probe 1 (Scheme 1, blue color) form a dsDNA with 5′ overhangs, which can be recognized by Exo III. The stepwise catalytic removal of mononucleotides from the 3′-terminus of signal probe 1 (Scheme 1, blue color) leads to the liberation of the Cy5 fluorophore from the signal probe 1@AuNPs, while the digestion of trigger DNA 1 (Scheme 1, yellow color) is blocked by the oligo-T tail. As the digestion of signal probe 1 (Scheme 1, blue color) proceeds, the trigger DNA 1 (Scheme 1, yellow color) is partially released and hybridizes with another signal probe 1 (Scheme 1, blue color) nearby. In this way, the trigger DNA 1 (Scheme 1, yellow color) walks along the AuNP surface, and abundant Cy5 fluorophores are liberated from the signal probe 1@AuNPs, resulting in the restoration of Cy5 fluorescence which can be quantified by single-molecule detection, with Cy5 indicating the presence of caspase-8. Similarly, the hybridization of trigger DNA 2 of the cleaved detection probe 2 (Scheme 1, cyan color) with signal probe 2 (Scheme 1, red color) forms a dsDNA with 5′ overhangs, which can be recognized by Exo III. The Exo III-mediated stepwise catalytic removal of mononucleotides from the 3′-terminus of signal probe 2 (Scheme 1, red color) leads to the liberation of abundant Texas Red fluorophores from the signal probe 2@AuNPs, resulting in the restoration of Texas Red fluorescence which can be used for the quantification of caspase-9. Because there are 120 signal probes per AuNP (ESI†), the high density of the scaffold DNA enhances the movement of trigger DNA along the 3D tracks, greatly amplifying the fluorescence signals. In contrast, in the absence of caspase-8 and caspase-9, the detection probes remain intact, and no trigger DNA is released. Consequently, no Exo III-mediated digestion of signal probes occurs, and neither the Cy5 nor Texas Red signal can be detected.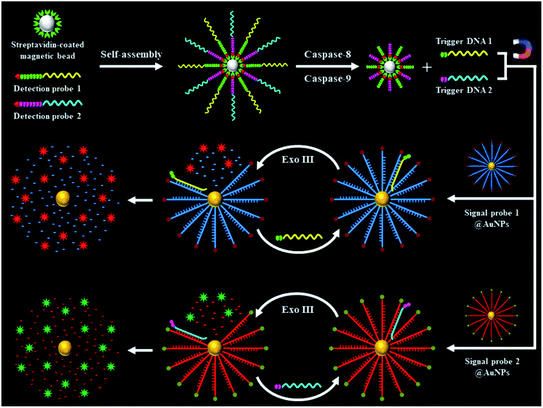 | ||
| Scheme 1 Schematic illustration of the integration of Exo III-powered 3D DNA walker with single-molecule detection for the simultaneous measurement of initiator caspase-8 and caspase-9. | ||
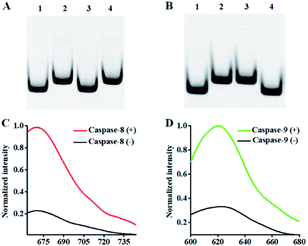
This assay mainly relies on the caspase-mediated cleavage of peptide to initiate the recycling liberation of Cy5/Texas Red fluorescent molecules from the signal probes@AuNPs. We used 12% nondenaturing polyacrylamide gel electrophoresis (PAGE) to verify the cleavage of the detection probes by caspase-8 and caspase-9. For the caspase-8 assay, when caspase-8 is absent, only one distinct band is observed (Fig. 1A, lane 2), which is identical to that of the intact detection probe 1, indicating that the cleavage of detection probe 1 cannot occur in the absence of caspase-8. When caspase-8 is present, a new smaller-sized band is observed (Fig. 1A, lane 3), which corresponds to the cleaved peptide–DNA 1 (Fig. 1A, lane 1), indicating the cleavage of detection probe 1 induced by caspase-8. When only caspase-9 is present, only one identical band to that of the intact detection probe 1 (Fig. 1A, lane 2) is observed (Fig. 1A, lane 4), indicating that the cleavage of detection probe 1 cannot occur in the presence of caspase-9. For the caspase-9 assay, when caspase-9 is absent, only one distinct band is observed (Fig. 1B, lane 2), which is identical to that of intact detection probe 2, indicating that the cleavage of detection probe 2 cannot occur in the absence of caspase-9. When only caspase-8 is present, only one distinct band identical to lane 2 (Fig. 1B) is observed (Fig. 1B, lane 3), indicating that the cleavage of detection probe 2 cannot occur in the presence of caspase-8. When caspase-9 is present, a new smaller-sized band is observed (Fig. 1B, lane 4), which corresponds to the cleaved peptide–DNA 2 (Fig. 1B, lane 1), indicating the cleavage of detection probe 2 induced by caspase-9. To verify the feasibility of the proposed nanosensor, we performed fluorescence measurements (Fig. 1C and D). As shown in Fig. 1C, a distinct Cy5 fluorescence signal with a characteristic emission peak of 670 nm is observed in the presence of 0.025 U μL−1 caspase-8 (Fig. 1C, red line). In contrast, in the control group without caspase-8, a low Cy5 signal (Fig. 1C, black line) is detected. As shown in Fig. 1D, a distinct Texas Red fluorescence signal with a characteristic emission peak of 620 nm is observed in the presence of 0.025 U μL−1 caspase-9 (Fig. 1D, green line). In contrast, in the control group without caspase-9, a low Texas Red signal (Fig. 1D, black line) is detected.
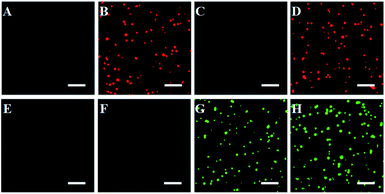
We further employed single-molecule detection technology to detect the fluorescence signals (Fig. 2). In the absence of caspase-8 and caspase-9, neither Cy5 (Fig. 2A) nor the Texas Red fluorescence signal (Fig. 2E) is observed, indicating that the Exo III-mediated recycling liberation of fluorophores cannot occur in the absence of caspase-8 and caspase-9. In contrast, in the presence of 0.025 U μL−1 caspase-8, distinct Cy5 fluorescence signals are observed (Fig. 2B, red color), but no Texas Red fluorescence signal is detected (Fig. 2F), indicating that caspase-8 can induce the recycling liberation of Cy5 fluorophores from the signal probe 1@AuNPs. In the presence of 0.025 U μL−1 caspase-9, distinct Texas Red fluorescence signals are observed (Fig. 2G, green color), but no Cy5 fluorescence signal is detected (Fig. 2C), indicating that caspase-9 can induce the recycling liberation of Texas Red fluorophores from the signal probe 2@AuNPs. In the presence of both caspase-8 and caspase-9, distinct Cy5 (Fig. 2D, red color) and Texas Red fluorescence signals (Fig. 2H, green color) are simultaneously observed. Notably, both the Cy5 and Texas Red fluorescence spots exhibit single-step photobleaching (Fig. S3, ESI†), suggesting that the observed individual fluorescent spot originates from a single dye molecule. These results clearly demonstrate that the proposed nanosensor can be applied for the simultaneous detection of caspase-8 and caspase-9 at the single-molecule level.
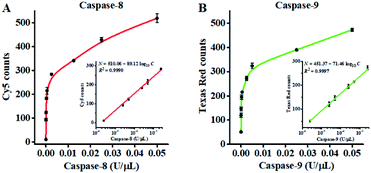
Under the optimized experimental conditions (Fig. S4 and S5, ESI†), we evaluated the sensitivity of the proposed nanosensor by measuring the variance of fluorescent counts with the caspase concentration. As shown in Fig. 3A, the Cy5 counts improve with increasing concentration of caspase-8 from 2.50 × 10−6 to 0.05 U μL−1. In the logarithmic scale, the Cy5 counts show a linear correlation with the concentration of caspase-8 over the range from 2.50 × 10−6 to 2.50 × 10−3 U μL−1. The regression equation is N = 510.06 + 89.12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log10
log10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) C with a correlation coefficient (R2) of 0.9990 (inset of Fig. 3A), where N is the measured Cy5 count and C is the concentration of caspase-8 (U μL−1). By evaluating the average response of the control group plus three times the standard deviation, the limit of detection (LOD) was calculated to be 2.08 × 10−6 U μL−1 (7.51 pM). The sensitivity of the proposed nanosensor is 96-fold higher than the conjugated polymer-based fluorescence assay (0.2 U mL−1),66 and 439-fold higher than the reported fluorescence assay (3.3 nM).67
C with a correlation coefficient (R2) of 0.9990 (inset of Fig. 3A), where N is the measured Cy5 count and C is the concentration of caspase-8 (U μL−1). By evaluating the average response of the control group plus three times the standard deviation, the limit of detection (LOD) was calculated to be 2.08 × 10−6 U μL−1 (7.51 pM). The sensitivity of the proposed nanosensor is 96-fold higher than the conjugated polymer-based fluorescence assay (0.2 U mL−1),66 and 439-fold higher than the reported fluorescence assay (3.3 nM).67
As shown in Fig. 3B, the Texas Red counts improve with increasing concentration of caspase-9 from 2.50 × 10−6 to 0.05 U μL−1. In the logarithmic scale, the Texas Red counts show a linear correlation with the concentration of caspase-9 over a range from 2.50 × 10−6 to 2.50 × 10−3 U μL−1. The regression equation is N = 451.37 + 71.46![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log10
log10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) C with a correlation coefficient (R2) of 0.9997 (inset of Fig. 3B), where N is the measured Texas Red count and C is the concentration of caspase-9 (U μL−1). The LOD was calculated to be 1.71 × 10−6 U μL−1 (92.37 pM). The sensitivity of the proposed nanosensor is 40-fold higher than the up-conversion nanoparticles-based FRET assay (0.068 U mL−1),68 and 650-fold higher than the gold nanoparticle–polydopamine-based electrochemical immunosensor (0.06 μM).18 The improved sensitivity of the proposed nanosensor can be ascribed to (1) the efficient conversion of the caspase activity signal into the DNA signal through the cleavage of the peptide–DNA detection probe, (2) recyclable liberation of fluorescent molecules from the signal probes@AuNPs induced by the Exo III-driven 3D DNA walker, and (3) the high signal-to-noise ratio of single-molecule detection.
C with a correlation coefficient (R2) of 0.9997 (inset of Fig. 3B), where N is the measured Texas Red count and C is the concentration of caspase-9 (U μL−1). The LOD was calculated to be 1.71 × 10−6 U μL−1 (92.37 pM). The sensitivity of the proposed nanosensor is 40-fold higher than the up-conversion nanoparticles-based FRET assay (0.068 U mL−1),68 and 650-fold higher than the gold nanoparticle–polydopamine-based electrochemical immunosensor (0.06 μM).18 The improved sensitivity of the proposed nanosensor can be ascribed to (1) the efficient conversion of the caspase activity signal into the DNA signal through the cleavage of the peptide–DNA detection probe, (2) recyclable liberation of fluorescent molecules from the signal probes@AuNPs induced by the Exo III-driven 3D DNA walker, and (3) the high signal-to-noise ratio of single-molecule detection.
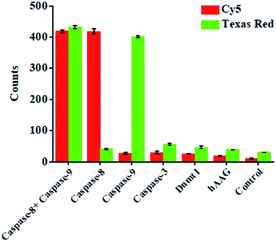
To investigate the selectivity of the proposed nanosensor for caspase assay, we used caspase-3, DNA (cytosine-5)-methyltransferase 1 (Dnmt1) and human alkyladenine DNA glycosylase (hAAG) as the negative controls. Caspase-3 is a core effector caspase that can be activated by caspase-8 in the extrinsic apoptotic pathway and can be activated by caspase-9 in the intrinsic pathway, but it cannot cleave the peptide substrates of caspase-8 and caspase-9.16 Dnmt1 can specifically recognize the hemimethylated sequence 5′-CG-3′, and catalyze the transfer of a methyl group from S-adenosyl-L-methionine (SAM) to the cytosine in genomic DNA.69 hAAG can recognize and excise a diverse group of alkylated purine bases and cleave the N-glycosidic bond between the sugar and the damaged base.70 As shown in Fig. 4, in the presence of caspase-3, Dnmt1 and hAAG, neither Cy5 nor Texas Red fluorescence signal is observed, consistent with the control with only the reaction buffer. In contrast, in the presence of caspase-8, an enhanced Cy5 fluorescence signal is observed, but no Texas Red fluorescence signal is detected. In the presence of caspase-9, an enhanced Texas Red fluorescence signal is detected, but no Cy5 fluorescence signal is detected. Moreover, in the presence of both caspase-8 and caspase-9, both Cy5 and Texas Red fluorescence signals can be simultaneously detected. These results demonstrate that the proposed nanosensor exhibits excellent selectivity toward caspase-8 and caspase-9.
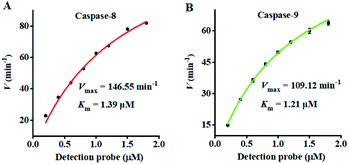
To investigate the feasibility of the proposed nanosensor for kinetic analysis, we measured the initial velocity (V) in response to various concentrations of the detection probes. To evaluate the enzyme kinetic parameters of caspase-8, we measured the V in the presence of 0.025 U μL−1 caspase-8 and different concentrations of detection probe 1 for 5 min at 37 °C. As shown in Fig. 5A, the initial velocity of caspase-8 enhances with the increasing concentration of detection probe 1. Vmax is calculated to be 146.55 min−1 and Km is calculated to be 1.39 μM. The Km value is consistent with that obtained by the rolling circle amplification (RCA)-based fluorescence assay (1 μM).52 To evaluate the enzyme kinetic parameters of caspase-9, we measured the V in the presence of 0.025 U μL−1 caspase-9 and different concentrations of detection probe 2 for 5 min at 37 °C. As shown in Fig. 5B, the initial velocity of caspase-9 enhances with the increasing concentration of detection probe 2. Vmax is calculated to be 109.12 min−1 and Km is calculated to be 1.21 μM. The Km value is consistent with that obtained by the multicolor gold–selenium bonding nanoprobe-based fluorescence assay (8.53 μM).71 These results suggest that the proposed nanosensor can be used to accurately evaluate the kinetic parameters of caspases.
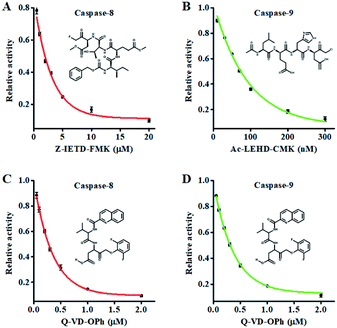
Fluoromethylketone (FMK),72 chloromethylketone (CMK)73 and difluorophenoxymethyl (OPh)74 are competitive caspase inhibitors, and they can irreversibly inactivate caspases by forming a covalent thioether adduct with the cysteine of the active site in caspases. We used the caspase-8 inhibitor Z-IETD-FMK, caspase-9 inhibitor Ac-LEHD-CMK and broad-spectrum caspase inhibitor Q-VD-OPh as the model inhibitors to investigate the feasibility of the proposed nanosensor for the caspase inhibition assay. As shown in Fig. 6A, the relative activity of caspase-8 decreases with the increasing concentration of Z-IETD-FMK from 0 to 20 μM. The concentration of inhibitor required to reduce the activity of caspase-8 by 50% (IC50) is determined to be 1.95 μM, consistent with that obtained by the RCA-based fluorescence assay (0.9656 mM).52 As shown in Fig. 6B, the relative activity of caspase-9 decreases with increasing concentration of Ac-LEHD-CMK from 0 to 300 nM. The IC50 is determined to be 72.80 nM, consistent with the data from the manufacturer (70 nM). As shown in Fig. 6C and D, the relative activities of caspase-8 and caspase-9 decrease with increasing concentration of Q-VD-OPh, respectively. The IC50 is determined to be 0.28 μM for caspase-8 and 0.31 μM for caspase-9. These results suggest that the proposed method can be applied for the screening of caspase inhibitors.
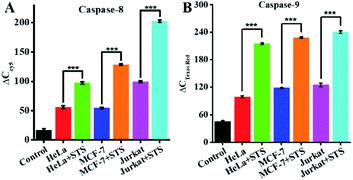
To demonstrate the feasibility of the proposed method for cellular caspase assays, we simultaneously measured endogenous caspase-8 and caspase-9 activity in a human cervical cancer cell line (HeLa cells), a human breast cancer cell line (MCF-7 cells), and a human acute T-lymphocytic leukemia cell line (Jurkat cells). Staurosporine (STS) is a broad-spectrum inhibitor of protein kinases with the capability of inducing in vitro apoptosis.75 To avoid the interference from nonspecific caspases, we employed anti-caspase-8 and anti-caspase-9 antibodies to completely inhibit target caspase-8/9 activity (Fig. S6, ESI†), and calculated the specific caspase-8/9 signal ΔC according to eqn (1).
| ΔC = C − Ca | (1) |
Conclusions
In summary, we develop a sensitive nanosensor based on the integration of exonuclease III-powered 3D DNA walker with single-molecule detection for simultaneous measurement of initiator caspase-8 and caspase-9. The presence of caspase-8 and caspase-9 can induce the cleavage of peptides in two peptide–DNA detection probes, releasing two trigger DNAs from the magnetic beads. The two trigger DNAs can serve as the walker DNA to walk on the surface of signal probes@AuNPs powered by Exo III digestion, liberating numerous Cy5 and Texas Red fluorophores which can be quantified by single-molecule detection, with Cy5 indicating caspase-8 and Texas Red indicating caspase-9. This nanosensor possesses the following distinct advantages: (1) the introduction of the AuNP-based 3D DNA walker greatly reduces the background signal and amplifies the output signals, and the introduction of single-molecule detection further improves the detection sensitivity, endowing this nanosensor with higher sensitivity and less assay time than the reported caspase assays (Table S1, ESI†) and facilitating the accurate detection of low-abundant caspase with low sample consumption for biomedical research and disease diagnosis; (2) the amplification reaction can be conducted at constant temperature without the requirement of complicated thermal cycling, and the signal can be easily quantified by single-molecule counting; (3) the use of peptide–DNA detection probe-conjugated magnetic beads and signal probes@AuNPs greatly simplifies the experimental procedures, facilitating the simultaneous detection of multiple caspases. This nanosensor is very sensitive with a detection limit of 2.08 × 10−6 U μL−1 for caspase-8 and 1.71 × 10−6 U μL−1 for caspase-9, and it can be used for the simultaneous screening of caspase inhibitors and measurement of endogenous caspase activity in various cell lines at the single-cell level. Moreover, this nanosensor can be extended to detect various proteases by simply changing the peptide sequences of the detection probes.Experimental section
Chemicals and materials
All oligonucleotides (Table 1) were HPLC-purified and synthesized by TaKaRa Bio Inc. (Dalian, China). The peptides were obtained from the Chinese Peptide Company (Hangzhou, China), and conjugated to DNA by TaKaRa Bio Inc. (Dalian, China). The caspase-8 (human, recombinant, active), caspase-9 (human, recombinant, active) and caspase-3 (human, recombinant, active) were obtained from Enzo Biochem, Inc. (Farmingdale, NY, USA). According to the datasheet from Enzo Biochem, Inc., the specific activity of caspase-8 is 5000 U mg−1 and its molecular weight is 55![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 391 g mol−1, and thus 1 U μL−1 caspase-8 corresponds to 3.61 μM. Similarly, the specific activity of caspase-9 is 400 U mg−1 and its molecular weight is 46
391 g mol−1, and thus 1 U μL−1 caspase-8 corresponds to 3.61 μM. Similarly, the specific activity of caspase-9 is 400 U mg−1 and its molecular weight is 46![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 281 g mol−1, and thus 1 U μL−1 caspase-9 corresponds to 54.02 μM. Human DNA (cytosine-5)methyltransferase (Dnmt1), human alkyladenine DNA glycosylase (hAAG), exonuclease III (Exo III) and 10× CutSmart buffer were purchased from New England Biolabs (Ipswich, MA, USA). The streptavidin-coated magnetic beads (Dynabeads™ M-280 streptavidin), 1× phosphate buffered saline (1× PBS, pH 7.4) and SYBR gold were bought from Thermo Fisher Scientific (Waltham, MA, USA). The Au nanoparticles (10 nm, 0.01% Au) were bought from Nanocs, Inc. (New York, NY, USA). The caspase-8 inhibitor (Z-IETD-FMK) and caspase-9 inhibitor (Ac-LEHD-CMK) were obtained from Sigma-Aldrich (St. Louis, MO, USA). The broad-spectrum caspase inhibitor Q-VD-OPh was obtained from R&D System (Minneapolis, MN, USA). Staurosporine (STS) was purchased from Millipore Corporation (Billerica, MA, USA). Caspase-8 monoclonal antibody and caspase-9 monoclonal antibody were purchased from Proteintech Group Inc., (Chicago, IL, USA). Human cervical cancer cell line (HeLa cells), human breast cancer cell line (MCF-7 cells), and human acute T-lymphocytic leukemia cell line (Jurkat cells) were obtained from the cell bank of Chinese Academy of Sciences (Shanghai, China). All other reagents were of analytical grade and used as received without further purification. Ultrapure water obtained from a Millipore filtration system (Temecula, CA, USA) was used throughout all experiments.
281 g mol−1, and thus 1 U μL−1 caspase-9 corresponds to 54.02 μM. Human DNA (cytosine-5)methyltransferase (Dnmt1), human alkyladenine DNA glycosylase (hAAG), exonuclease III (Exo III) and 10× CutSmart buffer were purchased from New England Biolabs (Ipswich, MA, USA). The streptavidin-coated magnetic beads (Dynabeads™ M-280 streptavidin), 1× phosphate buffered saline (1× PBS, pH 7.4) and SYBR gold were bought from Thermo Fisher Scientific (Waltham, MA, USA). The Au nanoparticles (10 nm, 0.01% Au) were bought from Nanocs, Inc. (New York, NY, USA). The caspase-8 inhibitor (Z-IETD-FMK) and caspase-9 inhibitor (Ac-LEHD-CMK) were obtained from Sigma-Aldrich (St. Louis, MO, USA). The broad-spectrum caspase inhibitor Q-VD-OPh was obtained from R&D System (Minneapolis, MN, USA). Staurosporine (STS) was purchased from Millipore Corporation (Billerica, MA, USA). Caspase-8 monoclonal antibody and caspase-9 monoclonal antibody were purchased from Proteintech Group Inc., (Chicago, IL, USA). Human cervical cancer cell line (HeLa cells), human breast cancer cell line (MCF-7 cells), and human acute T-lymphocytic leukemia cell line (Jurkat cells) were obtained from the cell bank of Chinese Academy of Sciences (Shanghai, China). All other reagents were of analytical grade and used as received without further purification. Ultrapure water obtained from a Millipore filtration system (Temecula, CA, USA) was used throughout all experiments.
| Note | Sequence (5′–3′) |
|---|---|
| Detection probe 1 | Biotin–(COOH)–Lys–Ser–His–Ser–His–Gly–Asp–Thr–Glu–Ile–Cys–(NH2)–TTT TTC ACT TGA GGC TAA CAC TTT TT |
| Detection probe 2 | Biotin–(COOH)–Lys–Arg–Gly–Arg–Gly–Gly–Asp–His–Glu–Leu–Cys(NH2)–TTT TTC ACA ATC GGA CTA TCG TTT TT |
| Cleaved peptide–DNA 1 | Asp–Thr–Glu–Ile–Cys-TTT TT C ACT TGA GGC TAA CAC TTT TT |
| Cleaved peptide–DNA 2 | Asp–His–Glu–Leu–Cys-TTT TTC ACA ATC GGA CTA TCG TTT TT |
| Signal probe 1 | SH-TTT TTT TTT TGT GTT AGC CTC AAG TG-Cy5 |
| Signal probe 2 | SH-TTT TTT TTT TCG ATA GTC CGA TTG TG-Texas Red |
Preparation of the detection probe-conjugated MBs
The assembly of detection probes onto the MBs was carried out according to the protocol of the manufacturer. 50 μL of the 10 mg mL−1 streptavidin-coated MBs solution was washed three times using 1× PBS. After resuspending with 50 μL of 1× PBS, 10 μL of 10 μM detection probes were added to form the detection probes-MB nanostructure through biotin–streptavidin interaction at room temperature for 30 min. The detection probe-MBs were then washed five times using 1× PBS to remove the uncoupled probes by magnetic separation, followed by resuspending in 50 μL of 1× PBS.Construction of signal probes@AuNPs
AuNPs (10 nm) were functionalized with thiolated signal probes using the freeze-directed methods.76 The method is reagentless without the involvement of extra salts, acids, and surfactants. Specifically, 20 μL of 100 μM signal probes were mixed with 1 mL of AuNPs. The mixture was subsequently placed in a laboratory freezer at −20 °C for 2 h, followed by thawing at room temperature. At last, the signal probe-coated AuNPs were centrifuged and washed three times with ultrapure water to remove excess signal probes, and resuspended in 40 μL of sterile water, and stored at 4 °C. In the signal probes@AuNPs solution, the concentration of DNA was measured using a NanoDrop 2000c Spectrophotometer (Thermo Scientific, Wilmington, Delaware, USA) and the number of signal probes per AuNP is estimated to be 120 ± 1 for both Cy5-labeled signal probe 1 and Texas Red-labeled signal probe 2 (Fig. S2, ESI†).Detection of caspase activities
The caspase activity assay includes two steps: (1) caspase-mediated cleavage of detection probes, and (2) recycling liberation of fluorophores induced by the Exo III-powered 3D DNA walker. The caspase-mediated cleavage of the detection probe was performed in 20 μL of solution containing 3.5 μL of 1× PBS, 1 mM DTT, 8 μL of detection probe 1-conjugated MBs or/and detection probe 2-conjugated MBs, and 0.5 μL of caspase at different concentrations at 37 °C for 1 h. After magnetic separation, the supernatant with cleavage products was collected for the next-step use. The Exo III-powered 3D DNA walker-based recycling liberation of fluorophores was performed in 20 μL of reaction solution containing 1× CutSmart buffer (50 mM potassium acetate, 20 mM tris-acetate, 10 mM magnesium acetate, 100 μg mL−1 BSA, pH 7.9), 0.71 μL of signal probe 1@AuNPs or/and 0.36 μL of signal probe 2@AuNPs, and 0.5 U of Exo III at 37 °C for 1 h.Ensemble fluorescence measurement
The 20 μL of reaction products was diluted to a final volume of 50 μL with ultrapure water for the measurement of fluorescence emission spectra using a 1 cm path length quartz cuvette on a Hitachi F-7000 fluorescence spectrophotometer (Tokyo, Japan). The excitation wavelength was 645 nm for Cy5 and 560 nm for Texas Red, and the emission spectra were recorded in the wavelength range of 660–750 nm for Cy5 and 600–680 nm for Texas Red with a slit width of 5 nm for both excitation and emission.Single-molecule detection and data analysis
The reaction products of caspase-8 were diluted 200-fold in the imaging buffer (1 mg mL−1 glucose oxidase, 0.4% (w/v) D-glucose, 0.04% mg mL−1 catalase, 50 μg mL−1 BSA, 67 mM glycine-potassium hydroxide, 1 mg mL−1 Trolox, 2.5 mM magnesium chloride, pH 9.4), and the reaction products of caspase-9 were diluted 500-fold in the imaging buffer. For total internal reflection fluorescence (TIRF) imaging, 10 μL of the sample was directly pipetted onto the coverslips. The Cy5 and Texas Red fluorescent molecules were excited by the sapphire 640 and 561 nm lasers (Coherent, USA), respectively. The resulting photons were collected by an oil immersion objective (CFI Apochromat TIRF 100×). The Cy5 and Texas Red fluorescence signals were imaged on an Andor ixon Ultra 897 EMCCD camera (Andor, Belfast, UK) with an exposure time of 500 ms. For data analysis, the ImageJ software was used for counting the Cy5 and Texas Red fluorescent molecules from an imaging region of 600 × 600 pixels.Gel electrophoresis
The reaction products were analyzed by 12% nondenaturing polyacrylamide gel electrophoresis (PAGE) in 1× TBE buffer (9 mM Tris–HCl, 9 mM boric acid, 0.2 mM ethylenediaminetetraacetic acid, EDTA, pH 7.9) at a 110 V constant voltage at room temperature for 50 min. The gels were stained by SYBR gold and analyzed by a Bio-Rad ChemiDoc MP Imaging System (Hercules, CA, USA).Kinetic analysis
To evaluate the enzyme kinetic parameters of caspases, we measured the initial velocity in the presence of 0.025 U μL−1 caspase (caspase-8 or caspase-9) and different concentrations of the detection probes at 37 °C for 5 min. The kinetic parameter is fitted to the Michaelis–Menten equation.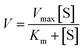 | (2) |
Inhibition assay
To evaluate the effect of inhibitor upon the caspase activity, different concentrations of the inhibitor (Z-IETD-FMK for caspase-8, Ac-LEHD-CMK for caspase-9, and broad-spectrum caspase inhibitor Q-VD-OPh for both caspase-8 and caspase-9) were preincubated with 0.025 U μL−1 caspase (caspase-8 or caspase-9) at room temperature for 15 min, respectively. Then 8 μL of detection probe 1-conjugated MBs or detection probe 2-conjugated MBs were added into the mixture, and the reaction volume was adjusted to 20 μL with 1× PBS, followed by the same detection procedure as described above. The relative activity (RA) of caspase was measured according to eqn (3).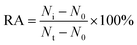 | (3) |
Cell culture and preparation of cell extracts
HeLa cells and MCF-7 cells were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM, Life Technologies, USA) with 10% FBS (Life Technologies, USA) and 1% penicillin–streptomycin (Gibco, USA). Jurkat cell was cultured in 1640 cell medium (Life Technologies, USA) with 10% FBS (Life Technologies, USA) and 1% penicillin–streptomycin (Gibco, USA). The cells were cultured at 37 °C in a humidified atmosphere containing 5% CO2. For real sample analysis, cells in the exponential phase of growth were collected and counted using Countstar BioTech Automated Cell Counter IC1000 (Shanghai, China), washed twice with ice-cold 1× PBS, and centrifuged at 800 rpm for 5 min. To isolate cytoplasmic components from nuclear ones, the cells were treated with a nuclear protein extraction kit (Beyotime Biotechnology, Wuhan, China) and centrifuged at 3400 rpm for 15 min at 4 °C. For STS-induced apoptosis analysis, cells were incubated in 5 mL of cell medium containing 0.4 μM STS for 4 h prior to the cell lysis procedure. The protein concentration was measured using a NanoDrop 2000c Spectrophotometer (Thermo Scientific, Wilmington, Delaware, USA).Author contributions
Meng Liu: conceptualization, methodology, investigation, writing – original draft preparation; Rui Xu: methodology, investigation; Wenjing Liu: conceptualization, writing – review and editing; Jian-Ge Qiu: conceptualization, writing – review and editing; Yan Wang: conceptualization, writing – review and editing; Fei Ma: conceptualization, writing – review and editing; Chun-yang Zhang: conceptualization, funding acquisition, writing – review and editing.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the National Natural Science Foundation of China (Grant No. 21735003, 22174017) and the Award for Team Leader Program of Taishan Scholars of Shandong Province, China.Notes and references
- K. Lauber, S. G. Blumenthal, M. Waibel and S. Wesselborg, Mol. Cell, 2004, 14, 277–287 CrossRef CAS PubMed.
- Y. Fuchs and H. Steller, Nat. Rev. Mol. Cell Biol., 2015, 16, 329–344 CrossRef CAS PubMed.
- S. Ghavami, M. Hashemi, S. R. Ande, B. Yeganeh, W. Xiao, M. Eshraghi, C. J. Bus, K. Kadkhoda, E. Wiechec, A. J. Halayko and M. Los, J. Med. Genet., 2009, 46, 497–510 CrossRef CAS PubMed.
- M. Kurokawa and S. Kornbluth, Cell, 2009, 138, 838–854 CrossRef CAS PubMed.
- E. C. Rui, M. M. S. Maria, M. C. G. Paulo, J. A. R. Carlos, M. S. F. Duarte, M. X. Joana, M. Rui and M. P. R. Cecilia, Curr. Pharm. Des., 2010, 16, 2851–2864 CrossRef PubMed.
- P. J. Barr and L. D. Tomei, Bio/Technology, 1994, 12, 487–493 CrossRef CAS PubMed.
- C. Thompson, Science, 1995, 267, 1456–1462 CrossRef CAS PubMed.
- D. Hanahan and R. A. Weinberg, Cell, 2000, 100, 57–70 CrossRef CAS PubMed.
- C. Pop and G. S. Salvesen, J. Biol. Chem., 2009, 284, 21777–21781 CrossRef CAS PubMed.
- A. Ashkenazi and V. M. Dixit, Science, 1998, 281, 1305–1308 CrossRef CAS PubMed.
- A. G. Porter and R. U. Jänicke, Cell Death Differ., 1999, 6, 99–104 CrossRef CAS PubMed.
- L. H. Russell, E. Mazzio, R. B. Badisa, Z.-P. Zhu, M. Agharahimi, E. T. Oriaku and C. B. Goodman, Anticancer Res., 2012, 32, 1595–1602 CAS.
- X.-M. Sun, M. MacFarlane, J. Zhuang, B. B. Wolf, D. R. Green and G. M. Cohen, J. Biol. Chem., 1999, 274, 5053–5060 CrossRef CAS PubMed.
- S. Yasuhara, Y. Zhu, T. Matsui, N. Tipirneni, Y. Yasuhara, M. Kaneki, A. Rosenzweig and J. A. J. Martyn, J. Histochem. Cytochem., 2003, 51, 873–885 CrossRef CAS PubMed.
- J. Su, T. W. Rajapaksha, M. E. Peter and M. Mrksich, Anal. Chem., 2006, 78, 4945–4951 CrossRef CAS PubMed.
- Y. Yang, Y. Liang and C.-y. Zhang, Anal. Chem., 2017, 89, 4055–4061 CrossRef CAS PubMed.
- G. B. Kim and Y.-P. Kim, Theranostics, 2012, 2, 127–138 CrossRef CAS PubMed.
- Q. Wen, X. Zhang, J. Cai and P.-H. Yang, Analyst, 2014, 139, 2499–2506 RSC.
- V. Gurtu, S. R. Kain and G. Zhang, Anal. Biochem., 1997, 251, 98–102 CrossRef CAS PubMed.
- R. Onuki, A. Nagasaki, H. Kawasaki, T. Baba, T. Q. P. Uyeda and K. Taira, Proc. Natl. Acad. Sci. U. S. A., 2002, 99, 14716–14721 CrossRef CAS PubMed.
- Y. Yuan, R. Zhang, X. Cheng, S. Xu and B. Liu, Chem. Sci., 2016, 7, 4245–4250 RSC.
- C. Köhler, S. Orrenius and B. Zhivotovsky, J. Immunol. Methods, 2002, 265, 97–110 CrossRef.
- H. Qi, M. Li, M. Dong, S. Ruan, Q. Gao and C. Zhang, Anal. Chem., 2014, 86, 1372–1379 CrossRef CAS PubMed.
- A. S. Goryashchenko, M. G. Khrenova and A. P. Savitsky, Methods Appl. Fluoresc., 2018, 6, 022001 CrossRef PubMed.
- P. Yin, H. Yan, X. G. Daniell, A. J. Turberfield and J. H. Reif, Angew. Chem., Int. Ed., 2004, 43, 4906–4911 CrossRef CAS PubMed.
- C. Jung, P. B. Allen and A. D. Ellington, Nat. Nanotechnol., 2016, 11, 157–163 CrossRef CAS PubMed.
- W. B. Sherman and N. C. Seeman, Nano Lett., 2004, 4, 1203–1207 CrossRef CAS.
- H. Brutzer, F. W. Schwarz and R. Seidel, Nano Lett., 2012, 12, 473–478 CrossRef CAS PubMed.
- M. Liu, J. Fu, C. Hejesen, Y. Yang, N. W. Woodbury, K. Gothelf, Y. Liu and H. Yan, Nat. Commun., 2013, 4, 2127 CrossRef PubMed.
- J.-S. Shin and N. A. Pierce, J. Am. Chem. Soc., 2004, 126, 10834–10835 CrossRef CAS PubMed.
- F. Dannenberg, M. Kwiatkowska, C. Thachuk and A. J. Turberfield, Nat. Comput., 2015, 14, 195–211 CrossRef CAS.
- Y. He and D. R. Liu, Nat. Nanotechnol., 2010, 5, 778–782 CrossRef CAS PubMed.
- D. Wang, C. Vietz, T. Schröder, G. Acuna, B. Lalkens and P. Tinnefeld, Nano Lett., 2017, 17, 5368–5374 CrossRef CAS PubMed.
- D. Yao, S. Bhadra, E. Xiong, H. Liang, A. D. Ellington and C. Jung, ACS Nano, 2020, 14, 4007–4013 CrossRef CAS PubMed.
- M. Xiao, X. Wang, L. Li and H. Pei, Anal. Chem., 2019, 91, 11253–11258 CrossRef CAS PubMed.
- Y. Chen, Y. Xiang, R. Yuan and Y. Chai, Nanoscale, 2015, 7, 981–986 RSC.
- S. Cai, M. Chen, M. Liu, W. He, Z. Liu, D. Wu, Y. Xia, H. Yang and J. Chen, Biosens. Bioelectron., 2016, 85, 184–189 CrossRef CAS PubMed.
- Y. Yang, M. A. Goetzfried, K. Hidaka, M. You, W. Tan, H. Sugiyama and M. Endo, Nano Lett., 2015, 15, 6672–6676 CrossRef CAS.
- H. Zhang, X. Xu and W. Jiang, Chem. Sci., 2020, 11, 7415–7423 RSC.
- N. Li, M. Du, S. Tian, X. Ji and Z. He, Anal. Bioanal. Chem., 2019, 411, 4055–4061 CrossRef CAS PubMed.
- X. Peng, Z.-B. Wen, P. Yang, Y.-Q. Chai, W.-B. Liang and R. Yuan, Anal. Chem., 2019, 91, 14920–14926 CrossRef CAS PubMed.
- Z. Xu, L. Liao, Y. Chai, H. Wang and R. Yuan, Anal. Chem., 2017, 89, 8282–8287 CrossRef CAS PubMed.
- X. Li, Q. Qiu, X. Ni, M. N. Hossain, X. Chen, H. Huang and H.-B. Kraatz, Sens. Actuators B Chem., 2020, 319, 128327 CrossRef CAS.
- C. Zhou, X. Duan and N. Liu, Nat. Commun., 2015, 6, 1–6 Search PubMed.
- H. Zhang, M. Lai, A. Zuehlke, H. Peng, X.-F. Li and X. C. Le, Angew. Chem., Int. Ed., 2015, 54, 14326–14330 CrossRef CAS PubMed.
- Y. Li, G. A. Wang, S. D. Mason, X. Yang, Z. Yu, Y. Tang and F. Li, Chem. Sci., 2018, 9, 6434–6439 RSC.
- X. Qu, D. Zhu, G. Yao, S. Su, J. Chao, H. Liu, X. Zuo, L. Wang, J. Shi, L. Wang, W. Huang, H. Pei and C. Fan, Angew. Chem., Int. Ed., 2017, 56, 1855–1858 CrossRef CAS PubMed.
- E. Xiong, D. Zhen, L. Jiang and X. Zhou, Anal. Chem., 2019, 91, 15317–15324 CrossRef CAS PubMed.
- W. Li, L. Wang and W. Jiang, Chem. Commun., 2017, 53, 5527–5530 RSC.
- S. D. Mason, Y. Tang, Y. Li, X. Xie and F. Li, TrAC, Trends Anal. Chem., 2018, 107, 212–221 CrossRef CAS.
- Z. Xu, Y. Chang, Y. Chai, H. Wang and R. Yuan, Anal. Chem., 2019, 91, 4883–4888 CrossRef CAS.
- M. Liu, D. Zhang, X. Zhang, Q. Xu, F. Ma and C.-y. Zhang, Chem. Commun., 2020, 56, 5243–5246 RSC.
- X. Zuo, F. Xia, Y. Xiao and K. W. Plaxco, J. Am. Chem. Soc., 2010, 132, 1816–1818 CrossRef CAS PubMed.
- J. Kim and C. J. Easley, Bioanalysis, 2011, 3, 227–239 CrossRef CAS PubMed.
- G. T. Walker, M. S. Fraiser, J. L. Schram, M. C. Little, J. G. Nadeau and D. P. Malinowski, Nucleic Acids Res., 1992, 20, 1691–1696 CrossRef CAS PubMed.
- F. Ma, Y. Yang and C.-y. Zhang, Anal. Chem., 2014, 86, 6006–6011 CrossRef CAS PubMed.
- M. M. Ali, F. Li, Z. Zhang, K. Zhang, D.-K. Kang, J. A. Ankrum, X. C. Le and W. Zhao, Chem. Soc. Rev., 2014, 43, 3324–3341 RSC.
- T. Notomi, H. Okayama, H. Masubuchi, T. Yonekawa, K. Watanabe, N. Amino and T. Hase, Nucleic Acids Res., 2000, 28, e63 CrossRef CAS PubMed.
- F. Ma, Y. Li, B. Tang and C.-y. Zhang, Acc. Chem. Res., 2016, 49, 1722–1730 CrossRef CAS PubMed.
- C.-c. Li, H.-y. Chen, J. Hu and C.-y. Zhang, Chem. Sci., 2020, 11, 5724–5734 RSC.
- M.-Y. Li, Y.-Q. Wang, Y.-L. Ying and Y.-T. Long, Chem. Sci., 2019, 10, 10400–10404 RSC.
- L.-J. Wang, X. Han, J.-G. Qiu, B. Jiang and C.-Y. Zhang, Chem. Sci., 2020, 11, 9675–9684 RSC.
- W. Ma, H. Ma, J.-F. Chen, Y.-Y. Peng, Z.-Y. Yang, H.-F. Wang, Y.-L. Ying, H. Tian and Y.-T. Long, Chem. Sci., 2017, 8, 1854–1861 RSC.
- Z.-y. Wang, L.-j. Wang, Q. Zhang, B. Tang and C.-y. Zhang, Chem. Sci., 2018, 9, 1330–1338 RSC.
- Z. Ye, H. Liu, F. Wang, X. Wang, L. Wei and L. Xiao, Chem. Sci., 2019, 10, 1351–1359 RSC.
- Y. Xie, R. Zhao, Y. Tan, X. Zhang, F. Liu, Y. Jiang and C. Tan, ACS Appl. Mater. Interfaces, 2012, 4, 405–410 CrossRef CAS PubMed.
- M. S. Rahman, T. Kabashima, H. Yasmin, T. Shibata and M. Kai, Anal. Biochem., 2013, 433, 79–85 CrossRef CAS PubMed.
- L. Liu, H. Zhang, Z. Wang and D. Song, Biosens. Bioelectron., 2019, 141, 111403 CrossRef CAS PubMed.
- Z.-y. Wang, P. Li, L. Cui, Q. Xu and C.-y. Zhang, Anal. Chem., 2020, 92, 13573–13580 CrossRef CAS PubMed.
- C.-c. Li, W.-x. Liu, J. Hu and C.-y. Zhang, Chem. Sci., 2019, 10, 8675–8684 RSC.
- X. Liu, X. Song, D. Luan, B. Hu, K. Xu and B. Tang, Anal. Chem., 2019, 91, 5994–6002 CrossRef CAS PubMed.
- C. J. F. Van Noorden, Acta Histochem., 2001, 103, 241–251 CrossRef CAS PubMed.
- H. Fauvel, P. Marchetti, C. Chopin, P. Formstecher and R. Nevière, Am. J. Physiol. Heart Circ. Physiol., 2001, 280, H1608–H1614 CrossRef CAS.
- K. Kuželová, D. Grebeňová and B. Brodská, J. Cell. Biochem., 2011, 112, 3334–3342 CrossRef.
- G. Feng and N. Kaplowitz, Am. J. Physiol. Gastrointest. Liver Physiol., 2002, 282, G825–G834 CrossRef CAS.
- B. Liu and J. Liu, J. Am. Chem. Soc., 2017, 139, 9471–9474 CrossRef CAS.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1sc05115f |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2021 |