Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Dithiolane quartets: thiol-mediated uptake enables cytosolic delivery in deep tissue†
Rémi
Martinent
,
Salman
Tawffik
,
Javier
López-Andarias
,
Dimitri
Moreau
 ,
Quentin
Laurent
and
Stefan
Matile
,
Quentin
Laurent
and
Stefan
Matile
 *
*
Department of Organic Chemistry, University of Geneva, Geneva, Switzerland. E-mail: stefan.matile@unige.ch; Web: https://www.unige.ch/sciences/chiorg/matile/ Tel: +41 22 379 6523
First published on 6th October 2021
Abstract
The cytosolic delivery of various substrates in 3D multicellular spheroids by thiol-mediated uptake is reported. This is important because most orthodox systems, including polycationic cell-penetrating peptides, fail to deliver efficiently into deep tissue. The grand principles of supramolecular chemistry, that is the pH dependence of dynamic covalent disulfide exchange with known thiols on the transferrin receptor, are proposed to account for transcytosis into deep tissue, while the known but elusive exchange cascades along the same or other partners assure cytosolic delivery in kinetic competition. For quantitative detection in the cytosol, the 2D chloroalkane penetration assay (CAPA) is translated to 3D deep tissue. The targeted delivery of quantum dots, otherwise already troublesome in 2D culture, and the controlled release of mechanophores are realized to exemplify the power of thiol-mediated uptake into spheroids. As transporters, dithiolane quartets on streptavidin templates are introduced as modular motifs. Built from two amino acids only, the varied stereochemistry and peptide sequence are shown to cover maximal functional space with minimal structural change, i.e., constitutional isomers. Reviving a classic in peptide chemistry, this templated assembly of β quartets promises to expand streptavidin biotechnology in new directions, while the discovery of general cytosolic delivery in deep tissue as an intrinsic advantage further enhances the significance and usefulness of thiol-mediated uptake.
Introduction
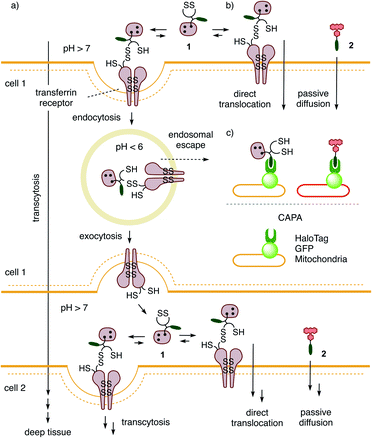
The general delivery of substrates into the cytosol of cells is a persistent challenge in life sciences and calls for input from chemistry. A rich and diverse collection of transporters for delivery in 2D cell culture exists. Their general usefulness is difficult to judge because their activities often depend strongly on conditions (substrates, cells, localization, degradation, etc.), detection methods, and limited synthetic availability. What many existing transporters have in common is a poor ability to penetrate deep tissue.1–13 Besides some remarkable exceptions,14–20 this includes also the popular cell-penetrating peptides (CPPs).7,8 Access to deep tissue is challenging because it requires a balanced combination of transcytosis and release into the cytosol. Transcytosis is best known from the transferrin receptor (TfR), which enters cells by endocytosis and returns to the surface by exocytosis (Fig. 1a).2,21–27 This mechanism allows the TfR to efficiently penetrate tumors and cross the blood–brain barrier to deliver iron to the brain. The attachment of transferrin or ligands of other transcytosis receptors to operational transport systems has been shown to improve delivery into deep tissue.2,21–27Evidence is emerging that the TfR also participates in thiol-mediated uptake28–32 and viral entry,28,33,34 including oligonucleotide phosphorothioates35 and SARS-CoV-2,28,34 respectively (Fig. 1b). It was thus conceivable that thiol-mediated uptake intrinsically assures cytosolic delivery into deep tissue. Thiol-mediated uptake28–39 operates with transporters that are equipped with thiol-reactive motifs, such as poly- or oligochalcogenides, often disulfides, at best cyclic ones (COCs), for dynamic covalent exchange40–49 with cellular thiols. Dynamic covalent disulfide exchange of transporters such as 1 (ref. 50) with thiols C556 and C558 of the TfR and tether the transporters through at least one disulfide bond to the membrane protein.29 Then, direct translocation occurs by thiolate-disulfide exchange cascades along the TfR or other protein partners to deliver COCs like 1 into the cytosol (Fig. 1b). Coinciding TfR endocytosis brings the tethered COCs like 1 into endosomes. The punctate patterns characteristic of endosomes have been identified during COC uptake and co-localization with fluorescently-labeled TfRs.29,51 Unless assisted by enzymes,52 release from endosomes is less favorable because low pH53 inactivates dynamic covalent exchange. It was thus conceivable that transporters like 1 could return to the surface of the first cell, where the higher pH reactivates dynamic covalent release and exchange with TfRs on the next cell to assure continuing transcytosis in kinetic competition with direct translocation. Taken together, these facts imply that thiol-mediated uptake should excel in cytosolic delivery into deep tissue. Here, we report experimental evidence that this is correct and introduce the 3D CAPA method with reporter 2 for detection (Fig. 1c) as well as protein-templated dithiolane quartets as privileged scaffolds for transport (Fig. 2).
Results and discussion
Design and synthesis of transporters
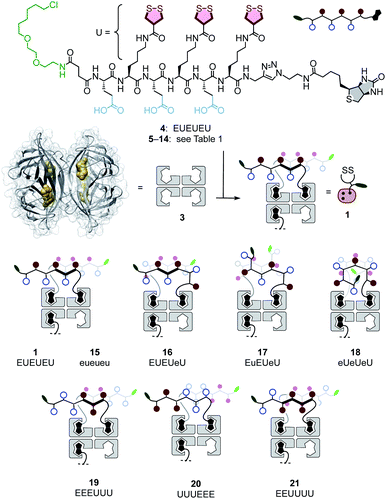
Transporter 1 is a native streptavidin tetramer 3 that has three binding pockets filled with COC oligomers 4 (Fig. 2).50 The trifunctional hexapeptide 4 is equipped with biotin at the C and a chloroalkane at the N terminus for binding to streptavidin and HaloTags, respectively. The sequence contains two amino acids only: glutamates E are placed to hinder uptake by cationic-CPP-like or other mechanisms,54 and to assure solubility. Uptake amino acids U are lysines coupled with COCs. AspA was selected as the COC because its activity was the highest of the series in our introductory studies, and dynamic covalent exchange with TfRs and transit through endosomes together with TfRs have been demonstrated.29,32,50,51 Peptides 5–14 were designed, synthesized, and then complexed with streptavidin 3 to produce a collection of transporters 15–26 that covers wide functional space with minimal structural change (Schemes S1–S10,†Fig. 2 and Table 1). The availability of such a set of isostructural COC transporters with different activities was desirable to elaborate on the intrinsic ability of thiol-mediated uptake to deliver into deep tissue (vide infra).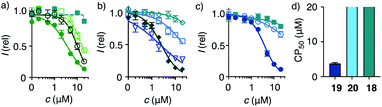 | ||
| Fig. 3 2D HC-CAPA dose–response curves for (a) 1 (filled circles), 15 (empty circles), 16 (empty squares), 17 (filled squares), (b) 21 (filled diamonds), 23 (empty diamonds), 25 (triangles), 26 (squares), (c) 18 (squares), 19 (filled circles), and 20 (empty circles), with (d) CP50 values for comparison with SP50 values (Fig. 4n). | ||
| Tb | Pc | Sequenced | CP50e (μM) | SP50f (μM) | |
|---|---|---|---|---|---|
a From 2D and 3D CAPA in HGM cells.
b Transporters, composed of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 streptavidin 3/peptide, unless stated.
c Peptides. See Fig. 2 for general structures.
d Sequences of hexapeptide units. E: L-glutamate, U: L-ε-AspA–lysine, K: L-lysine. Lower-case letters represent D-isomers.
e Half maximal cell-penetration concentrations, obtained by Hill analysis of DRCs (Fig. 3).
f Half maximal spheroid-penetration concentrations, obtained by Hill analysis of DRCs (Fig. 4n).
g Streptavidin 3/peptide 1 3 streptavidin 3/peptide, unless stated.
c Peptides. See Fig. 2 for general structures.
d Sequences of hexapeptide units. E: L-glutamate, U: L-ε-AspA–lysine, K: L-lysine. Lower-case letters represent D-isomers.
e Half maximal cell-penetration concentrations, obtained by Hill analysis of DRCs (Fig. 3).
f Half maximal spheroid-penetration concentrations, obtained by Hill analysis of DRCs (Fig. 4n).
g Streptavidin 3/peptide 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2.
h Equal equivalents of peptides were used. 2.
h Equal equivalents of peptides were used.
|
|||||
| 1 | 1 | 4 | EUEUEU | 3.6 ± 0.3 | |
| 2 | 15 | 5 | eueueu | 10.0 ± 0.6 | |
| 3 | 16 | 6 | EUEUeU | 16 ± 2 | |
| 4 | 17 | 7 | EuEUeU | >20 | |
| 5 | 18 | 8 | eUeUeU | >20 | 20 ± 8 |
| 6 | 19 | 9 | EEEUUU | 3.8 ± 0.2 | 5.3 ± 0.9 |
| 7 | 20 | 10 | UUUEEE | >20 | 17 ± 6 |
| 8 | 21 | 11 | EEUUUU | 2.5 ± 0.2 | |
| 9 | 22 | 12 | UUUUUU | >20 | |
| 10 | 23 | 13 | EEEEEE | >20 | |
| 11 | 24 | 12 + 13h | >20 | ||
| 12 | 25 | 14 | KKKKKK | 2.8 ± 0.3 | |
| 13 | 26 | 14 + 13h | 22 ± 4 | ||
Thiol-mediated cytosolic delivery in 2D cultured cells
Cytosolic delivery into 2D cell culture was determined first using the chloroalkane penetration assay (CAPA).55–57 This assay has been introduced recently by Kritzer et al. as a reliable tool to quantitatively measure cytosolic delivery.55–57 The CAPA method operates with HGM cells, i.e. HeLa cells that stably express a fusion protein of HaloTag and GFP on mitochondria (Fig. 1c). Chloroalkylated transporters that reach the cytosol will react with these HaloTags, while all unused HaloTags will react with subsequently added chloroalkylated TAMRA 2. The result is decreasing TAMRA fluorescence with increasing uptake of transporters. Automated high-content high-throughput imaging has been introduced to measure TAMRA fluorescence only inside of healthy cells and generate high-precision dose–response curves (DRCs, Fig. 3) while simultaneously reporting on cell viability.50 The uptake efficiency is reported as CP50 value, that is the transporter concentration needed to inhibit HaloTag labeling with 2 by 50%, determined by the Hill analysis of DRCs. In conventional 2D cell culture, complex 1 afforded CP50 = 3.6 ± 0.3 μM (Fig. 2, 3a, filled circles, Table 1).With CP50 = 10.0 ± 0.6 μM, thiol-mediated uptake of the all-D eueueu complex 15 was only slightly worse than that of the all-L isomer 1, suggesting that enantioselectivity and enzymatic peptide degradation contribute little to uptake (Fig. 2, 3a, empty circles, Table 1). One D amino acid in position 5 caused a similar decrease in CP50 = 16 ± 2 μM for EUEUeU 16 (Fig. 2, 3a, empty squares). However, all activity was lost upon two and three inversions of absolute configuration in EuEUeU 17 and eUeUeU 18 (Fig. 2, 3a and c, filled squares, Table 1). This pronounced diastereoselectivity was consistent with homochiral UXU/uxu triads as the consensus sequence. The formation of short antiparallel β sheets templated on the surface of streptavidin emerged as the most plausible active structure, producing a quartet of proximal dithiolanes with the dimensions of 5 × 7 Å (Fig. 2). Such β quartets are central to β sheet chemistry, known, for example, from the early TASP concept of Mutter, Dumy, Ulrich and coworkers,49,58 and from related functional motifs.59,60 The proximity of COCs in dithiolane β quartets was appealing with regard to the exchange cascades expected for thiol-mediated uptake.28
The dithiolane quartet hypothesis was in full agreement with introductory studies focusing on EU peptide length and complex stoichiometry.50 With regard to peptide stereochemistry covered in this study, the high activity of all-D eueueu 15 was consistent with the dithiolane quartet hypothesis and the absence of important enantioselectivity (Fig. 2). In EUEUeU 16, the original β quartet won't form but can be replaced by an alternative β quartet, which could explain a reasonably preserved activity (Fig. 2). More stereochemical inversions in the isomers EuEUeU 17 and eUeUeU 18 formally roll peptides up into S-shaped and β helix-like pseudo-cycles (Fig. 2). These motifs block any possible β quartet formation, which was consistent with their inactivity.
Consistent with the dithiolane quartet hypothesis, EEEUUU 19 showed preserved activity (Fig. 2, 3c, filled circles). The inverse UUUEEE 20 was inactive because the repulsive tethers between the protein template and possible dithiolane quartets are too long (Fig. 2, 3c, empty circles). Beyond constitutional isomers, EEUUUU 21 was the most active of all transporters, perhaps because a second dithiolane quartet is available on the other face of the formal β sheet (Fig. 2, 3b, filled diamonds). However, this interpretation should not be overestimated because the CP50 = 2.5 ± 0.2 μM remained near those of single dithiolane quartets in 19 and 1. Further increase of dithiolane units in UUUUUU 12 was detrimental because complex 22 was insoluble in water (the same for EUUUUU, not finalized). The complementary complex 23 with EEEEEE 13 was soluble but inactive, as expected without U (Fig. 3b, empty diamonds, Table 1). Consistent with the dithiolane quartet hypothesis, solubilization of all-U 12 with all-E 13 in mixed complex 24 did not restore activity although the EU ratio was as in 1 and 15–18, and solubility remained low. For the same reason, all other mixed complexes tested also failed to improve their uniform counterparts (not shown).
With the β quartet hypothesis, two amino acids of different stereochemistry suffice for accessing activities ranging from the best to worst with constitutional isomers, even diastereomers. Such functional extremes in isostructural systems were important to explore delivery into 3D tissue reliably. Also important was the availability of a formally isostructural CPP control. In 2D cell culture, complex 25 with KKKKKK 14 was in the range of the best dithiolane quartets 1, 19 and 21, but the cooperativity in the DRCs of thiol-mediated uptake was much reduced (Table 1, Fig. 3b, triangles). Neutralization or overcompensation of the positive charges from K614 with E613 was considered for a more adequate comparison with multianionic COC quartets. However, already the mixed complex 26 was poorly active (CP50 > 20 μM, Table 1, Fig. 3b, squares).
Thiol-mediated cytosolic delivery in spheroids
With the required set of transporters in hand, quantitative detection of cytosolic delivery in deep tissue was considered next. Multicellular spheroids or organoids are emerging as attractive models to study penetration across deep tissues and barriers. Uptake into spheroids is mostly monitored by attaching fluorophores to transporters.1–26 However, this convenient method is known as unreliable from studies with 2D cell culture because information of intracellular localization, including surface binding and endosomal capture, is missing, and intensities can change for reasons unrelated to transport. The introduction of CAPA has transformed the uptake field because it allows for unambiguous, reliable and quantitative detection of cytosolic delivery in 2D cell culture.55–57 It was thus of interest to use the same CAPA for uptake studies in 3D cell culture, where co-localization experiments are technically challenging for several reasons, and functional response from HaloTags stably expressed on mitochondria would thus be even more important to determine cytosolic delivery in deep tissue quantitatively and unambiguously.Spheroids of HGM cells with an average size of 350–400 μm were prepared in cell-repellent U-bottom 96-well plates following standard procedures (Fig. 4a).1–26 In cross-sectional confocal spinning disc microscopy (CSDM) images taken 80 μm from the surface, GFP emission was homogeneously distributed (Fig. 4b). Similarly the homogeneous cross-sectional TAMRA emission upon addition of 2 was consistent with unhindered passive diffusion of the small, cationic CAPA probe across the spheroid (Fig. 4c, m, red circles, 1c). Incubation of the HGM spheroids with 5 μM 19 for 6 h before the addition of CAPA probe 2 did not affect GFP emission (Fig. 4e) but produced a gradually decreasing TAMRA emission from the center to the periphery (Fig. 4f). Line-scan analysis of the TAMRA intensity along the cross-section in the CSDM image gave an almost linear decrease (Fig. 4m, green squares). Repetition of the same experiment with 20 μM 19 resulted in almost complete disappearance of all fluorescence (Fig. 4i, m, dark blue circles). Line-scan analysis of cross-sectional CSDM images taken already after 2 h rather than 6 h incubation revealed a penetration gradient similar to the one observed with 5 μM 19 after 6 h (Fig. 4l, m, blue circles). Comparison of the line-scan analyses for 20 μM 19 after 2 h and 6 h incubation illustrated how transporter 19 penetrates the spheroids with time from the periphery to the center as expected for operational transcytosis (Fig. 4m, l vs. i). These results demonstrated that at concentrations c > SP50, the CP50 of spheroids (Table 1), transporter 19 enters the cytosol homogeneously throughout the spheroid within 6 hours. At c ∼ SP50, thiol-mediated delivery decreases linearly with increasing depth because the limited amount of transporter 19 provided is gradually consumed by kinetically competing direct translocation and irreversible reaction with the HaloTags on the mitochondria of the cells on the way into the spheroids.
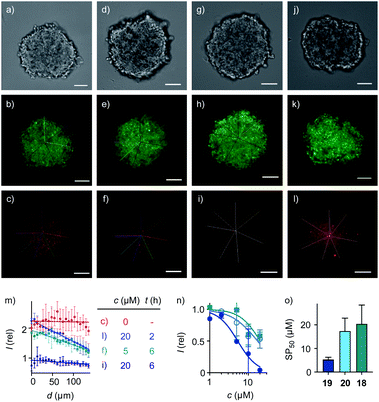 | ||
| Fig. 4 Cross-sectional CSDM images of HGM spheroids incubated with (a–c) 2 for 45 min; (d–f) 5 μM and (g–l) 20 μM 19 for 2 h (j–l) or 6 h (d–i) followed by 2 for 45 min, showing TL (a, d, g, j), GFP (b, e, h, k) and TAMRA channels (c, f, i, l) at 80 μm from the surface (scale bars: 100 μm). (m) Line-scan analysis of the TAMRA intensity corresponding to the cross-sections in c (red), f (green), i (dark blue) and l (blue), with intensities as a function of the distance from the center of the spheroid (error bars: standard deviation, n = 7). (n) 3D CAPA DRCs for 18 (squares), 19 (filled circles), and 20 (empty circles), with (o) SP50 values for comparison with CP50 values (Fig. 3d). | ||
Quantitative DRCs for 3D CAPA integrating the entire spheroid gave for EEEUUU complex 19 an SP50 = 5.3 ± 0.9 μM that was nearly identical with the 2D CP50 = 3.8 ± 0.2 μM (Fig. 4n, o, vs.3c, d, Table 1). The decreasing activities of isomers 20 and 18 were cleanly translated from 2D to 3D, with preserved order. Compared to 2D CP50 > 20 μM, their weak activities even increased slightly to SP50 = 17 ± 6 μM for 20 and SP50 = 20 ± 8 μM for 18, but errors naturally increased as well in the multicellular spheroids (Fig. 4n, o, vs.3c, d). Roughly identical DRCs for differently active transporters in 2D and 3D cell culture suggested that the uptake mechanisms are the same, that is a kinetic competition between thiol-mediated transcytosis and direct translocation (Fig. 1). Importantly, their similarity up to the shape of the DRC implied that the cytosolic delivery into deep tissue is an intrinsic characteristic of thiol-mediated uptake (Fig. 4nvs.3c).
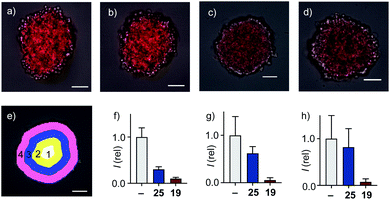
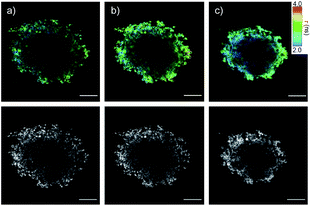
In 2D CAPA, the polycationic CPP complex 25 was as active as the best polyanionic dithiolane quartets 21, 1 and 19 (Fig. 3b). In sharp contrast, CPP complex 25 failed to penetrate HGM spheroids (Fig. 5b). Emission of the CAPA probe 2 was suppressed only at the periphery (Fig. 5b), while dithiolane quartets 1 and 19 suppressed fluorescence throughout the spheroid (Fig. 5c and d). Layer analysis confirmed this impression quantitatively (Fig. 5e), revealing that CPP 25 reaches the cytosol only in the peripheral layer 4 (Fig. 5f), while activity in the middle layers 2–3 was weak and nearly negligible in the central layer 1 (Fig. 5g and h). Different activity in 3D cell culture (Fig. 5b–d) for isostructural transporters with similar activity in 2D (Fig. 3c) evinced that cytosolic delivery into deep tissue is a distinctive advantage of thiol-mediated uptake compared to classics such as CPPs.
Thiol-mediated delivery of quantum dots in spheroids
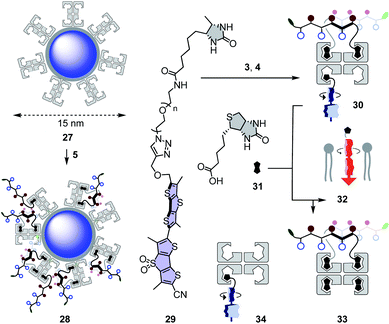
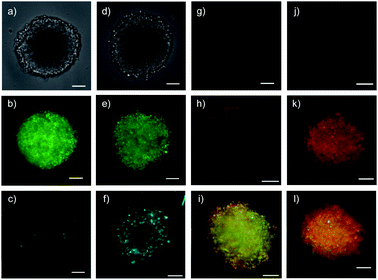
Quantum dots (QDs) are notoriously difficult to deliver to the cytosol already in 2D culture.61,62 Most CPP conjugates end up trapped in endosomes. Thiol-mediated uptake has recently been shown with CPDs (cell-penetrating poly(disulfide)s)61 and diselenolanes61,62 to solve this problem in 2D culture. To probe for the even more ambitious delivery of QDs to the cytosol in multicellular spheroids, streptavidin-coated Qdot™ 605 27 were loaded with 80 equivalents of eueueu peptide 5 (Fig. 6). This all-D peptide was selected among several candidates (Fig. 2) because aggregation of the resulting cell-penetrating system 28 was negligible. Aggregation of coated QDs is quite common, and more aggregation with L peptides than that with D enantiomers was interesting with regard to the diastereomeric active quartets formed with the chiral streptavidin templates.Six hours after the addition of control QDs 27 without COC transporters, the spheroid was intact (Fig. 7a) and the QD channel was essentially dark (Fig. 7c) compared to the internal standard in the GFP channel (Fig. 7b). This result confirmed the inability of protein-coated QDs to penetrate also HGM spheroids. In clear contrast, QDs 28 combined through streptavidin with dithiolane quartets produced brightly fluorescent spheroids (Fig. 7f). The fluorescence was very homogeneous, and the bright spots detected were minor compared to the images reported with other approaches.1–26 This homogeneous fluorescence reaching the center of the spheroids confirmed that spheroid penetration by thiol-mediated uptake is powerful also for most demanding substrates,61 and both the transporter and streptavidin are intact despite passing through endosomes multiple times.
Thiol-mediated targeted delivery and controlled release of mechanophores in spheroids
Functional delivery into deep tissue with thiol-mediated uptake was probed with SupraFlippers 29 (Fig. 6).63 The mechanosensitive flipper probes have been introduced recently to image membrane tension in live cells. SupraFlippers 29 in particular were designed for the controlled release of the mechanophore in the membrane of interest within cells. They are equipped with a desthiobiotin, which binds to streptavidin 3 but not as strongly as biotin. Combined with a chloroalkylated dithiolane quartet in complex 30, SupraFlippers should thus be delivered to the mitochondria in HGM spheroids. Addition of biotin 31 should then place the flipper into the surrounding mitochondrial membrane (32) for mechanosensing, and afford complex 33 as a side product. Because of the ease of expression of HaloTags, this approach allows us to target in principle any membrane of interest within cells.64The addition of control flipper complex 34 did not produce any fluorescence in HGM spheroids (Fig. 7g). Consistent with efficient penetration of deep tissue, homogeneously fluorescent HGM spheroids were obtained from the addition of COC-flipper complex 30 (Fig. 7h). Flipper emission correlated well with GFP emission (green, Fig. 7i).
The addition of biotin 31 further increased the intensity of flipper emission (Fig. 7h vs. k, S11†). The change of color in the merged images of flipper and GFP emission confirmed that the increase in emission upon biotin addition occurred only with flippers and not with GFP (Fig. 7i vs. l). The selective increase of flipper emission was consistent with flipper release from complex 30 by ligand exchange with 31 and the formation of partially planarized, more emissive flippers 32 in the mitochondrial membranes, as previously confirmed by co-localization with GFP in 2D images.63
Preliminary results with fluorescence lifetime imaging microscopy (FLIM) confirmed detectability quite deep into spheroids (Fig. 8a, τav = 2.83 ns), increasing counts and lifetimes upon flipper release (Fig. 8b, τav = 2.95 ns), and decreasing lifetimes upon hyperosmotic stress (Fig. 8c, τav = 2.75 ns). Although average flipper lifetimes, sensitive to experimental conditions, were slightly shorter in 3D than in 2D culture, the decrease in response to tension was similar (3.3 to 3.1 ns in 2D (ref. 65)). This decrease is the key characteristic for operational mechanosensing within cells. Although there is much room for technical improvements for tension imaging by FLIM in spheroids, these results provide experimental support for the targeted delivery and spatio-temporal control of the release of SupraFlippers 29 on the mitochondria in the cytosol of multicellular spheroids by external chemical stimulation. Interestingly, only a few recent studies exist on substrate release in spheroids, triggered by changes in pH,15,16 disulfide reduction14,19 and enzymatic cleavage13 but not external chemical stimulation.
Conclusions
We report that cytosolic delivery in deep tissue is a distinct, intrinsic property of thiol-mediated uptake, compatible with difficult substrates such as quantum dots, targeting and controlled release. Classical systems such as CPPs are confirmed not to meet this challenge convincingly. To secure these results, we also translated the CAPA method from 2D to 3D cell culture, which will be useful for the community. As for transport systems, the streptavidin-templated assembly of β quartets is introduced as a privileged motif, a finding that revives an old classic and expands streptavidin biotechnology in a new direction.Cytosolic delivery into deep tissue is of broad interest in biology and medicine, from tumor treatment to crossing the blood–brain barrier. The here introduced thiol-mediated spheroid penetration is based on the grand principles of supramolecular chemistry. The pH dependence of dynamic covalent disulfide exchange with known surface thiols on the transferrin receptor conceivably accounts for thiol-mediated transcytosis, while dynamic covalent exchange cascades assure kinetically competing direct translocation. The transferrin receptor is known for canonical, transferrin-mediated transcytosis to deliver iron into the brain, but also to mediate thiol-mediated uptake and viral entry into cells. The perspectives emerging from this study are thus broad and substantial. They will further increase the general significance, scope, appreciation and use of thiol-mediated uptake.
Experimental section
See the ESI.†Data availability
Data for this paper are available at Zenodo at https://doi.org/10.5281/zenodo.5515808.Author contributions
R. M. and S. T. synthesized the transporters; R. M., J. L.-A., D. M. and Q. L. developed spheroid imaging; S. M. directed the study, and all co-authors contributed to experiment design, data analysis and interpretation, and manuscript writing.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We thank K. Straková for contributions to synthesis, the NMR, MS and Bioimaging platforms for services, J. A. Kritzer (Tufts University), T. R. Ward and J. Klehr (University of Basel) for providing materials, and the University of Geneva, the National Centre for Competence in Research (NCCR) Chemical Biology, the NCCR Molecular Systems Engineering and the Swiss NSF for financial support.Notes and references
- K. Kostarelos, D. Emfietzoglou, A. Papakostas, W.-H. Yang, Å. Ballangrud and G. Sgouros, Int. J. Cancer, 2004, 112, 713–721 CrossRef CAS PubMed.
- L. Wei, X.-Y. Guo, T. Yang, M.-Z. Yu, D.-W. Chen and J.-C. Wang, Int. J. Pharm., 2016, 510, 394–405 CrossRef CAS PubMed.
- D. Cao, S. Tian, H. Huang, J. Chen and S. Pan, Mol. Pharm., 2015, 12, 240–252 CrossRef CAS PubMed.
- H. Lu, R. H. Utama, U. Kitiyotsawat, K. Babiuch, Y. Jiang and M. H. Stenzel, Biomater. Sci., 2015, 3, 1085–1095 RSC.
- H. R. Mellor, L. A. Davies, H. Caspar, C. R. Pringle, S. C. Hyde, D. R. Gill and R. Callaghan, J. Gene Med., 2006, 8, 1160–1170 CrossRef CAS PubMed.
- H. Wang, Y. Wang, Y. Wang, J. Hu, T. Li, H. Liu, Q. Zhang and Y. Cheng, Angew. Chem., Int. Ed., 2015, 54, 11647–11651 CrossRef CAS PubMed.
- D. van den Brand, C. Veelken, L. Massuger and R. Brock, Biochim. Biophys. Acta, Biomembr., 2018, 1860, 1342–1349 CrossRef CAS PubMed.
- T. Zong, L. Mei, H. Gao, W. Cai, P. Zhu, K. Shi, J. Chen, Y. Wang, F. Gao and Q. He, Mol. Pharm., 2014, 11, 2346–2357 CrossRef CAS PubMed.
- T. Liu, I. Kempson, M. de Jonge, D. L. Howard and B. Thierry, Nanoscale, 2014, 6, 9774–9782 RSC.
- G. Jarockyte, D. Dapkute, V. Karabanovas, J. V. Daugmaudis, F. Ivanauskas and R. Rotomskis, Biochim. Biophys. Acta, 2018, 1862, 914–923 CrossRef CAS PubMed.
- M. Ulusoy, A. Lavrentieva, J.-G. Walter, F. Sambale, M. Green, F. Stahl and T. Scheper, Toxicol. Res., 2016, 5, 126–135 CrossRef PubMed.
- H. Ma, Q. Jiang, S. Han, Y. Wu, J. C. Tomshine, D. Wang, Y. Gan, G. Zou and X.-J. Liang, Mol. Imaging, 2012, 11, 487–498 CrossRef CAS PubMed.
- M. Oishi, A. Tamura, T. Nakamura and Y. Nagasaki, Adv. Funct. Mater., 2009, 19, 827–834 CrossRef CAS.
- V. M. Gaspar, P. Baril, E. C. Costa, D. de Melo-Diogo, F. Foucher, J. A. Queiroz, F. Sousa, C. Pichon and I. J. Correia, J. Control. Release, 2015, 213, 175–191 CrossRef CAS PubMed.
- S. Ohta, S. Hiramoto, Y. Amano, M. Sato, Y. Suzuki, M. Shinohara, S. Emoto, H. Yamaguchi, H. Ishigami, Y. Sakai, J. Kitayama and T. Ito, Bioconjugate Chem., 2016, 27, 504–508 CrossRef CAS PubMed.
- J. S. Basuki, H. T. T. Duong, A. Macmillan, R. B. Erlich, L. Esser, M. C. Akerfeldt, R. M. Whan, M. Kavallaris, C. Boyer and T. P. Davis, ACS Nano, 2013, 7, 10175–10189 CrossRef CAS PubMed.
- C. Ju, R. Mo, J. Xue, L. Zhang, Z. Zhao, L. Xue, Q. Ping and C. Zhang, Angew. Chem., Int. Ed., 2014, 53, 6253–6258 CrossRef CAS PubMed.
- C.-N. Zhu, L.-Y. Zang, D.-Y. Zheng, H.-M. Cao and X.-J. Liu, Chem. Commun., 2020, 56, 2000–2003 RSC.
- A. W. Du, H. Lu and M. Stenzel, Mol. Pharm., 2016, 13, 3648–3656 CrossRef CAS PubMed.
- S. J. Zamolo, T. Darbre and J.-L. Reymond, Chem. Commun., 2020, 56, 11981–11984 RSC.
- A. Kohata, P. K. Hashim, K. Okuro and T. Aida, J. Am. Chem. Soc., 2019, 141, 2862–2866 CrossRef CAS PubMed.
- C.-F. Cho, J. M. Wolfe, C. M. Fadzen, D. Calligaris, K. Hornburg, E. A. Chiocca, N. Y. R. Agar, B. L. Pentelute and S. E. Lawler, Nat. Commun., 2017, 8, 15623 CrossRef CAS PubMed.
- Y. J. Yu, Y. Zhang, M. Kenrick, K. Hoyte, W. Luk, Y. Lu, J. Atwal, J. M. Elliott, S. Prabhu, R. J. Watts and M. S. Dennis, Sci. Transl. Med., 2011, 3, 84ra44 Search PubMed.
- Z. Baranyai, B. Biri-Kovács, M. Krátký, B. Szeder, M. L. Debreczeni, J. Budai, B. Kovács, L. Horváth, E. Pári, Z. Németh, L. Cervenak, F. Zsila, E. Méhes, É. Kiss, J. Vinšová and S. Bősze, J. Med. Chem., 2021, 64, 2982–3005 CrossRef CAS PubMed.
- R. Beard, D. C. A. Gaboriau, A. D. Gee and E. W. Tate, Chem. Sci., 2019, 10, 10772–10778 RSC.
- C. L. Waite and C. M. Roth, Bioconjugate Chem., 2009, 20, 1908–1916 CrossRef CAS PubMed.
- P. Kumari, S. V. K. Rompicharla, O. S. Muddineti, B. Ghosh and S. Biswas, Int. J. Biol. Macromol., 2018, 116, 1196–1213 CrossRef CAS PubMed.
- Q. Laurent, R. Martinent, B. Lim, A.-T. Pham, T. Kato, J. López-Andarias, N. Sakai and S. Matile, JACS Au, 2021, 1, 710–728 CrossRef CAS PubMed.
- D. Abegg, G. Gasparini, D. G. Hoch, A. Shuster, E. Bartolami, S. Matile and A. Adibekian, J. Am. Chem. Soc., 2017, 139, 231–238 CrossRef CAS PubMed.
- J. Guo, T. Wan, B. Li, Q. Pan, H. Xin, Y. Qiu and Y. Ping, ACS Cent. Sci., 2021, 7, 990–1000 CrossRef CAS PubMed.
- Z. Shu, I. Tanaka, A. Ota, D. Fushihara, N. Abe, S. Kawaguchi, K. Nakamoto, F. Tomoike, S. Tada, Y. Ito, Y. Kimura and H. Abe, Angew. Chem., Int. Ed., 2019, 58, 6611–6615 CrossRef CAS PubMed.
- A. Tirla and P. Rivera-Fuentes, Biochemistry, 2019, 58, 1184–1187 CrossRef CAS PubMed.
- Y. Cheng, A.-T. Pham, T. Kato, B. Lim, D. Moreau, J. López-Andarias, L. Zong, N. Sakai and S. Matile, Chem. Sci., 2021, 12, 626–631 RSC.
- X. Tang, M. Yang, Z. Duan, Z. Liao, L. Liu, R. Cheng, M. Fang, G. Wang, H. Liu, J. Xu, P. M. Kamau, Z. Zhang, L. Yang, X. Zhao, X. Peng and R. Lai, bioRxiv, 2020 DOI:10.1101/2020.10.23.350348.
- Q. Laurent, R. Martinent, D. Moreau, N. Winssinger, N. Sakai and S. Matile, Angew. Chem., Int. Ed., 2021, 60, 19102–19106 CrossRef CAS PubMed.
- J. Lu, H. Wang, Z. Tian, Y. Hou and H. Lu, J. Am. Chem. Soc., 2020, 142, 1217–1221 CrossRef CAS PubMed.
- J. Zhou, L. Sun, L. Wang, Y. Liu, J. Li, J. Li, J. Li and H. Yang, Angew. Chem., Int. Ed., 2019, 58, 5236–5240 CrossRef CAS PubMed.
- S. Du, S. S. Liew, L. Li and S. Q. Yao, J. Am. Chem. Soc., 2018, 140, 15986–15996 CrossRef CAS PubMed.
- X. Meng, T. Li, Y. Zhao and C. Wu, ACS Chem. Biol., 2018, 13, 3078–3086 CrossRef CAS PubMed.
- S. Ulrich, Acc. Chem. Res., 2019, 52, 510–519 CrossRef CAS PubMed.
- M. Juanes, I. Lostalé-Seijo, J. R. Granja and J. Montenegro, Chem.–Eur. J., 2018, 24, 10689–10698 CrossRef CAS PubMed.
- C. Gehin, J. Montenegro, E.-K. Bang, A. Cajaraville, S. Takayama, H. Hirose, S. Futaki, S. Matile and H. Riezman, J. Am. Chem. Soc., 2013, 135, 9295–9298 CrossRef CAS PubMed.
- J. M. Priegue, D. N. Crisan, J. Martínez-Costas, J. R. Granja, F. Fernandez-Trillo and J. Montenegro, Angew. Chem., Int. Ed., 2016, 55, 7492–7495 CrossRef CAS PubMed.
- J. F. Reuther, S. D. Dahlhauser and E. V. Anslyn, Angew. Chem., Int. Ed., 2019, 58, 74–85 CrossRef CAS PubMed.
- B. Liu, C. G. Pappas, E. Zangrando, N. Demitri, P. J. Chmielewski and S. Otto, J. Am. Chem. Soc., 2019, 141, 1685–1689 CrossRef CAS PubMed.
- S. Otto, Acc. Chem. Res., 2012, 45, 2200–2210 CrossRef CAS PubMed.
- J. G. Felber, L. Zeisel, L. Poczka, K. Scholzen, S. Busker, M. S. Maier, U. Theisen, C. Brandstädter, K. Becker, E. S. J. Arnér, J. Thorn-Seshold and O. Thorn-Seshold, J. Am. Chem. Soc., 2021, 143, 8791–8803 CrossRef CAS PubMed.
- C. Bravin and C. A. Hunter, Chem. Sci., 2020, 11, 9122–9125 RSC.
- E. Bartolami, Y. Bessin, V. Gervais, P. Dumy and S. Ulrich, Angew. Chem., Int. Ed., 2015, 54, 10183–10187 CrossRef CAS PubMed.
- R. Martinent, D. Du, J. López-Andarias, N. Sakai and S. Matile, ChemBioChem, 2021, 22, 253–259 CrossRef CAS PubMed.
- N. Chuard, A. I. Poblador-Bahamonde, L. Zong, E. Bartolami, J. Hildebrandt, W. Weigand, N. Sakai and S. Matile, Chem. Sci., 2018, 9, 1860–1866 RSC.
- G. Saito, J. A. Swanson and K.-D. Lee, Adv. Drug Delivery Rev., 2003, 55, 199–215 CrossRef CAS PubMed.
- F. Piazzolla, V. Mercier, L. Assies, N. Sakai, A. Roux and S. Matile, Angew. Chem., Int. Ed., 2021, 60, 12258–12263 CrossRef CAS PubMed.
- B. R. McNaughton, J. J. Cronican, D. B. Thompson and D. R. Liu, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 6111–6116 CrossRef CAS PubMed.
- L. Peraro, K. L. Deprey, M. K. Moser, Z. Zou, H. L. Ball, B. Levine and J. A. Kritzer, J. Am. Chem. Soc., 2018, 140, 11360–11369 CrossRef CAS PubMed.
- K. Deprey and J. A. Kritzer, Meth. Enzymol., 2020, 641, 277–309 CrossRef CAS PubMed.
- J. López-Andarias, J. Saarbach, D. Moreau, Y. Cheng, E. Derivery, Q. Laurent, M. González-Gaitán, N. Winssinger, N. Sakai and S. Matile, J. Am. Chem. Soc., 2020, 142, 4784–4792 CrossRef PubMed.
- M. Mutter, P. Dumy, P. Garrouste, C. Lehmann, M. Mathieu, C. Peggion, S. Peluso, A. Razaname and G. Tuchscherer, Angew. Chem., Int. Ed., 1996, 35, 1482–1485 CrossRef CAS.
- S. M. Butterfield and M. L. Waters, J. Am. Chem. Soc., 2003, 125, 9580–9581 CrossRef CAS PubMed.
- S. Hagihara, H. Tanaka and S. Matile, J. Am. Chem. Soc., 2008, 130, 5656–5657 CrossRef CAS PubMed.
- E. Derivery, E. Bartolami, S. Matile and M. Gonzalez-Gaitan, J. Am. Chem. Soc., 2017, 139, 10172–10175 CrossRef CAS PubMed.
- E. Bartolami, D. Basagiannis, L. Zong, R. Martinent, Y. Okamoto, Q. Laurent, T. R. Ward, M. Gonzalez-Gaitan, N. Sakai and S. Matile, Chem.–Eur. J., 2019, 25, 4047–4051 CrossRef CAS PubMed.
- J. López-Andarias, K. Straková, R. Martinent, N. Jiménez-Rojo, H. Riezman, N. Sakai and S. Matile, JACS Au, 2021, 1, 221–232 CrossRef PubMed.
- K. Straková, J. López-Andarias, N. Jiménez-Rojo, J. E. Chambers, S. J. Marciniak, H. Riezman, N. Sakai and S. Matile, ACS Cent. Sci., 2020, 6, 1376–1385 CrossRef PubMed.
- A. Goujon, A. Colom, K. Straková, V. Mercier, D. Mahecic, S. Manley, N. Sakai, A. Roux and S. Matile, J. Am. Chem. Soc., 2019, 141, 3380–3384 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Detailed procedures and results for all reported experiments. See DOI: 10.1039/d1sc04828g |
| This journal is © The Royal Society of Chemistry 2021 |