Open Access Article
Open Access ArticleNleB/SseK-catalyzed arginine-glycosylation and enteropathogen virulence are finely tuned by a single variable position contiguous to the catalytic machinery†
Ana
García-García
a,
Thomas
Hicks
b,
Samir
El Qaidi
c,
Congrui
Zhu
c,
Philip R.
Hardwidge
c,
Jesús
Angulo
 *bde and
Ramon
Hurtado-Guerrero
*bde and
Ramon
Hurtado-Guerrero
 *afg
*afg
aInstitute of Biocomputation and Physics of Complex Systems (BIFI), University of Zaragoza, Mariano Esquillor s/n, Campus Rio Ebro, Edificio I+D, Zaragoza, Spain. E-mail: rhurtado@bifi.es
bSchool of Pharmacy, University of East Anglia, Norwich Research Park, Norwich, NR4 7TJ, UK
cCollege of Veterinary Medicine, Kansas State University, Manhattan, KS 66506, USA
dDepartamento de Química Orgánica, Universidad de Sevilla, Sevilla, 41012, Spain. E-mail: jangulo@us.es
eInstituto de Investigaciones Químicas (CSIC-US), Sevilla, 41092, Spain
fCopenhagen Center for Glycomics, Department of Cellular and Molecular Medicine, School of Dentistry, University of Copenhagen, Copenhagen, Denmark
gFundación ARAID, Zaragoza, Spain
First published on 19th August 2021
Abstract
NleB/SseK effectors are arginine-GlcNAc-transferases expressed by enteric bacterial pathogens that modify host cell proteins to disrupt signaling pathways. While the conserved Citrobacter rodentium NleB and E. coli NleB1 proteins display a broad selectivity towards host proteins, Salmonella enterica SseK1, SseK2, and SseK3 have a narrowed protein substrate selectivity. Here, by combining computational and biophysical experiments, we demonstrate that the broad protein substrate selectivity of NleB relies on Tyr284NleB/NleB1, a second-shell residue contiguous to the catalytic machinery. Tyr284NleB/NleB1 is important in coupling protein substrate binding to catalysis. This is exemplified by S286YSseK1 and N302YSseK2 mutants, which become active towards FADD and DR3 death domains, respectively, and whose kinetic properties match those of enterohemorrhagic E. coli NleB1. The integration of these mutants into S. enterica increases S. enterica survival in macrophages, suggesting that better enzymatic kinetic parameters lead to enhanced virulence. Our findings provide insights into how these enzymes finely tune arginine-glycosylation and, in turn, bacterial virulence. In addition, our data show how promiscuous glycosyltransferases preferentially glycosylate specific protein substrates.
Introduction
Glycosyltransferases (GTs) are ubiquitous enzymes that transfer a sugar moiety from either sugar nucleotides or lipid phosphate sugars to small molecules, lipids, carbohydrates, small RNAs, and proteins.1–3 Glycosylation of proteins is generally a multistep process in most eukaryotic organisms,3 and likely the most abundant posttranslational modification (PTM), at least in humans.3 Glycans are generally attached to proteins in four different ways – N-linked to asparagine (Asn), O-linked to the hydroxyl groups of serine (Ser) threonine (Thr), tyrosine (Tyr), or C-linked to tryptophan (Trp) and glypiation.3 Each type of protein glycosylation is initiated by one or more unique protein GTs that in turn define the different glycosylation pathways. While most of these pathways occur post-translationally, initiation of N-glycosylation and likely POMT-directed O-mannosylation occur co-translationally.3,4A rare PTM described a few years ago, arginine-glycosylation, is catalyzed by Gram-negative bacterial GTs.5 This unusual PTM occurs on the arginine guanidinium group, a very poor nucleophile. In Pseudomonas and Neisseria species, arginine-glycosylation is catalyzed by EarP, while in enteropathogens, it is catalyzed by the type III secretion system effectors arginine GTs NleB and SseK.6–9 EarP is a rhamnosyl-transferase that uniquely glycosylates the bacterial translation elongation factor P (EF-P) to activate its function and drive bacterial pathogenicity.6 The NleB/SseK GTs transfer GlcNAc to arginines of several mammalian proteins and to at least five bacterial proteins.7–12 The NleB/SseK GTs are not classified in the CAZy database (GTnc).13
While C. rodentium only encodes one NleB, most E. coli strains encode two NleB proteins named NleB1 and NleB2. For several years, the role of NleB2 was unclear10 until a recent publication reported that NleB2 is an arginine GT that preferably transfers glucose to RIPK1, inhibiting host protein function similarly to other NleB/SseK GTs.14 The change in sugar donor preference was attributed to Ser252NleB2, which corresponds to a Gly residue in all homologous sequences.14CrNleB is highly conserved in the attaching/effacing pathogens enterohemorrhagic Escherichia coli (EHEC) and enteropathogenic E. coli (EPEC) NleB1s.15 Particularly, the identity between these enzymes is ∼89% between CrNleB and NleB1s, and 98% between NleB1s (Fig. 1a). Salmonella enterica strains encode up to three functional NleB orthologs named SseK1, SseK2, and SseK3. When the CrNleB/NleB1sEPEC/EHEC are compared to SseK1/2/3, the sequence identities drop significantly, ranging from 51 to 57% (Fig. 1a). At the structural level, these enzymes show a high degree of similarity and are built by two conserved major domains and a C-terminal lid, which is also required for the catalytic activity of the enzyme. The GT-A fold-adopting catalytic domain is the largest domain and includes the essential DxD and HEN (His–Glu–Asn) motifs. The helix–loop–helix (HLH) domain comprises two helices, α3 and α4, connected by a loop16–19 (Fig. 1a and b).
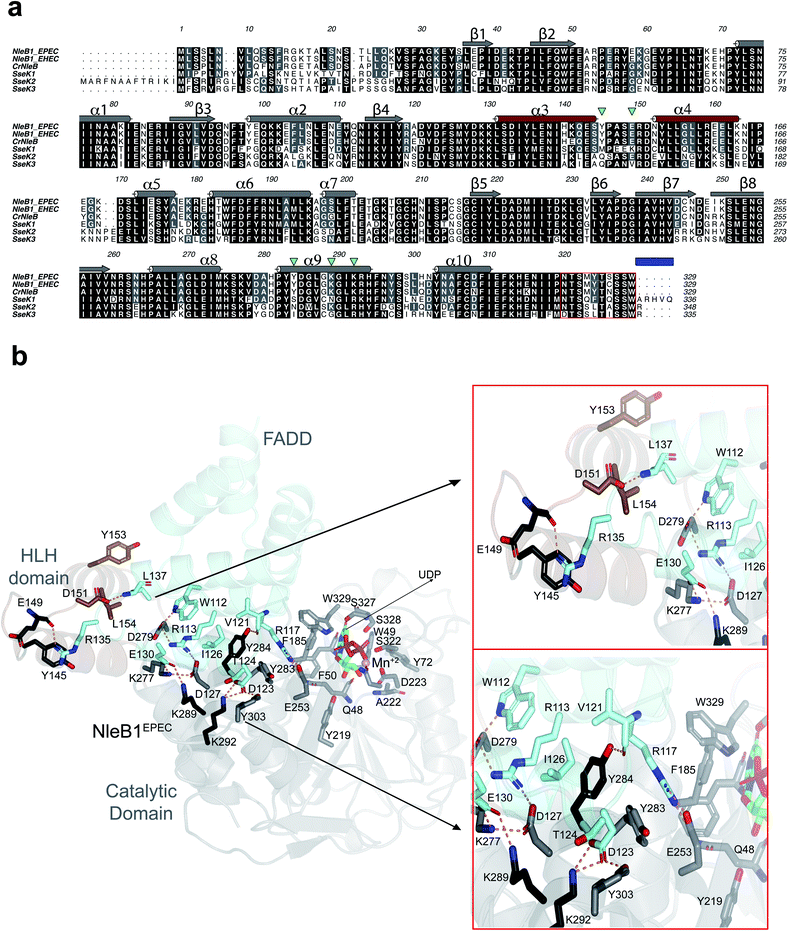 | ||
| Fig. 1 Analysis of the interacting residues in the NleB1EPEC–FADDDD interface. (a) Multiple sequence alignment of CrNleB, NleB1EPEC, NleB1EHEC, SseK1wt, SseK2wt and SseK3wt. Residues are color-coded by their degree of sequence conservation where black, grey and white colors denote identity, high similarity and dissimilarity, respectively. Shown above the NleB1EPEC sequence, in gray (catalytic domain) and brown (HLH domain), are the secondary structure elements (α-helices and β-strands) based on the NleB1EPEC–UDP–Mn2+–FADDDD structure (PDB entry 6ACI19). The residues forming part of the C-terminal lid are indicated within a red box while a blue rectangle determines the five C-terminal residues. The five inverted green triangles indicate the residues in NleB GTs that are non-conserved or partly conserved with the SseK GTs and are engaged in FADDDD interaction. These residues were targeted for site-directed mutagenesis in SseK1wt and SseK2wt. (b) Cartoon representation of the NleB1EPEC–UDP–Mn2+–FADDDD. The catalytic and HLH domains of NleB1EPEC are shown in gray and brown, respectively. The FADDDD is shown in cyan. Residues are shown as sticks with carbon atoms with the corresponding colors indicated above. UDP and Mn2+ are shown as green carbon atoms and as a pink sphere, respectively; hydrogen bond interactions are shown as dotted orange lines but only for residues interacting at the interface of the complex. The interactions with UDP and Mn2+ have been extensively discussed before16,17 and will not be further discussed. | ||
NleB GlcNAc-transferase activity is essential to bacterial virulence.7 Multiple host protein substrates for the CrNleB and NleB1EPEC/EHEC have been described and include the death domains (DD) of tumor necrosis factor receptor 1 (TNFR1), the TNFR1-associated death domain protein (TRADD), the receptor-interacting serine/threonine-protein kinase 1 (RIPK1), the TNF-receptor superfamily member 25 (DR3 or TNFRSF25), and FAS-associated death domain protein (FADD).9,19,20 In addition, these enzymes glycosylate other proteins non-containing DDs. CrNleB and NleB1EHEC, but not NleB1EPEC, glycosylate GAPDH,10 and NleB1EPEC glycosylates HIF-1α.21 Overall, NleB1 disrupts TNFR-associated factor (TRAF) signaling, leading to inhibition of the pro-inflammatory NF-κB pathway.7,9,10 SseK1, SseK2, and SseK3 have a narrower protein substrate selectivity; SseK1 glycosylates TRADD9 and GAPDH,10 but not FADD;10 SseK2 glycosylates FADD10 but not TRADD17 or GAPDH;10 SseK3 glycosylates TNRF1,18 TRAIL,18 and the small GTPase Rab1,22 but not GAPDH10 or FADD.10 This illustrates that although the SseK and NleB GTs are highly similar at the sequence and structural level, they display dissimilarities in their protein substrate selectivity. Furthermore, the SseK GTs are inactive towards some NleB-specific protein substrates.10,17
To address the molecular basis of NleB/SseK protein substrate selectivity and to determine why some SseK GTs do not glycosylate particular protein substrates, we report herein a multidisciplinary approach on wild type (wt) NleB1EHEC, SseK1, and SseK2, combined with the characterization of different SseK1 and SseK2 mutants, which reveals that a single second-shell residue near to the catalytic machinery, finely tunes substrate selectivity and catalysis. We also show that optimal kinetic parameters are accomplished by the mutants S286YSseK1 and N302YSseK2, which contain a Tyr residue that replaces SseK1wt Ser286 and SseK2wt Asn302. We recover full activity with S286YSseK1 on FADDDD and N302YSseK2 on DR3DD. Finally, we demonstrate that the integration of S286YSseK1 and N302YSseK2 mutants in a Salmonella enterica strain devoid of all SseK enzymes increases Salmonella survival in macrophages, suggesting that better enzymatic kinetic parameters lead to enhanced virulence.
Results and discussion
Bioinformatics and structural analysis of NleB/SseK GTs
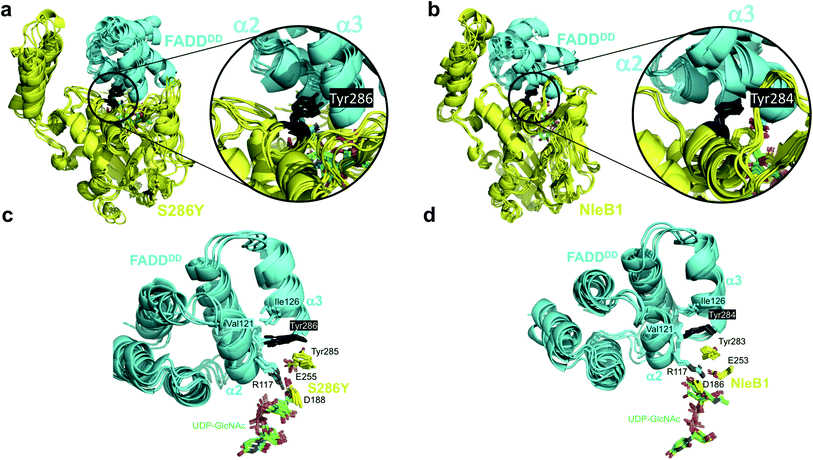
To determine the molecular basis of why SseK1wt does not glycosylate FADDDD, we performed a multiple alignment analysis of CrNleB, NleB1EPEC/EHEC, and SseK GTs, and also carefully inspected the residues of NleB1EPEC engaged in recognition of FADDDD (Fig. 1a and b; PDB entry 6ACI19). Fourteen NleB1EPEC residues established diverse types of interactions with FADDDD residues, with salt bridge interactions being the most prevalent, followed by hydrogen bond/hydrophobic interactions (Fig. 1b and Table S1†). Five of these 14 residues were in the HLH domain (Tyr145, Glu149, Asp151, Tyr153 and Leu154), while the others (Glu253, Lys277, Asp279, Tyr283, Tyr284, Asp285, Lys289, Lys292, and Tyr303) were located in the catalytic domain (Fig. 1b and Table S1†). Interestingly, not a single residue of the C-terminal lid interacted with FADDDD residues. In addition, but with the exception of Glu149, which interacted through its backbone with the Arg135FADD side chain, the other residues interacted with FADD residues through their side chains. A smaller number of FADDDD residues were engaged in recognition of NleB1EPEC. All these nine residues were located along α2, α3, α4, and the loops connecting them (Fig. S1 and Table S1†). Again, and as found for NleB1EPEC residues, most of the FADDDD residues involved in recognition of NleB1EPEC utilized their side chains. Four of these nine residues (Trp112FADD, Arg117FADD, Val121FADD, and Ile126FADD) were completely conserved between different DDs and likely comprise the minimal epitope for NleB/SseK GTs recognition, as suggested before.17,19 Note that Arg117FADD is the acceptor site and is engaged in a salt bridge with the proposed catalytic base Glu253 (ref. 19) (Fig. 1b).A thorough comparison of the grade of conservation of the NleB1EPEC fourteen interacting residues with aligned residues from other orthologs indicated that only five residues might be responsible for the null activity of SseK1wt on FADDDD (Table S1†). These residues were mostly non-conserved or partially conserved with SseK1wt residues. Tyr145 and Glu149 were quite close in the structure and located in a loop connecting α3 with α4 of the HLH domain (Fig. 1b). The other three residues, Tyr284, Lys289, and Lys292, were located in the catalytic domain and exclusively in α9 (Fig. 1a). While both Lys residues were proximal in the structure and established salt bridges with Asp123FADD and Glu130FADD, Tyr284 was more isolated from them and interacted through its side chain to Val121FADD backbone and Ile126FADD side chain (Fig. 1 and Table S1†). Tyr284 was also engaged in a CH–π interaction with the proximal Tyr283 side chain, which likely controls the interaction between the acceptor Arg117FADD and the catalytic base Glu253 (Fig. 1b). Hence, Tyr284 is a second-shell residue with respect to the catalytic machinery in which Glu253 is one of the key players.
The S286YSseK1 mutation switches on activity against FADDDD
Based on the above analysis, we selected five residues from NleB GTs to perform site-directed mutagenesis on the corresponding aligned residues in SseK1wt and SseK2wt. To simplify the number of mutations in SseK1wt and to minimize the potential combinations among them, we established three groups of residues: the two pairs Tyr145–Glu149NleB1, and Lys289–Lys292NleB1, based on their proximity at structural level, and Tyr284NleB1 as one unique and independent residue from the others.To initiate this study, we determined the kinetic parameters of NleB1EHEC on UDP-GlcNAc and FADDDD (see Fig. 2a, left panel, ESI, Fig. S2, and Table S2†). NleB1EHEC displayed a clear substrate inhibition profile under the presence of variable concentrations of FADDDD (Fig. 2a; Ki = 793 ± 160 μM) that was not present in the other SseK2wt and mutant proteins. This behavior might be attributed to a different non-productive FADDDD structural arrangement under high concentrations of this substrate. On the contrary, substrate inhibition under variable concentrations of UDP-GlcNAc was barely present (Fig. S2†). The Kms for UDP-GlcNAc and FADDDD were 125 ± 33 and 13 ± 2.5 μM, respectively, and the kcat was ∼100 min−1 (Fig. 2b, left and middle panels, and Table S2†), a value in agreement with other previous reported kcat values for a similar protein GT such as EarP23 (kcat of 35 min−1), and an unrelated one such as PoFUT2 (ref. 24) (kcat of 144 min−1), both of which are glycosyltransferases that glycosylate other types of folded domains. Furthermore, these kcat values also match reported kcat values for unstructured peptides in the presence of other protein GTs (a range between 46–400 min−1, and 18–300 min−1 for different peptides using GalNAc-T3 (ref. 25) and N-glycosyltransferase,26,27 respectively). As expected from previous8,9 and our own studies,10 SseK1wt was inactive on FADDDD and SseK2wt was slow on FADDDD (Fig. 2a, middle and right panel, and Table S2†). Particularly, the Km, kcat, and catalytic efficiency of SseK2wt were 95-, 6.7-, and 630-fold worse than those of NleB1EHEC (Fig. 2b). Note that the kinetic parameters for SseK2wt are estimated due to its poor binding to FADDDD.
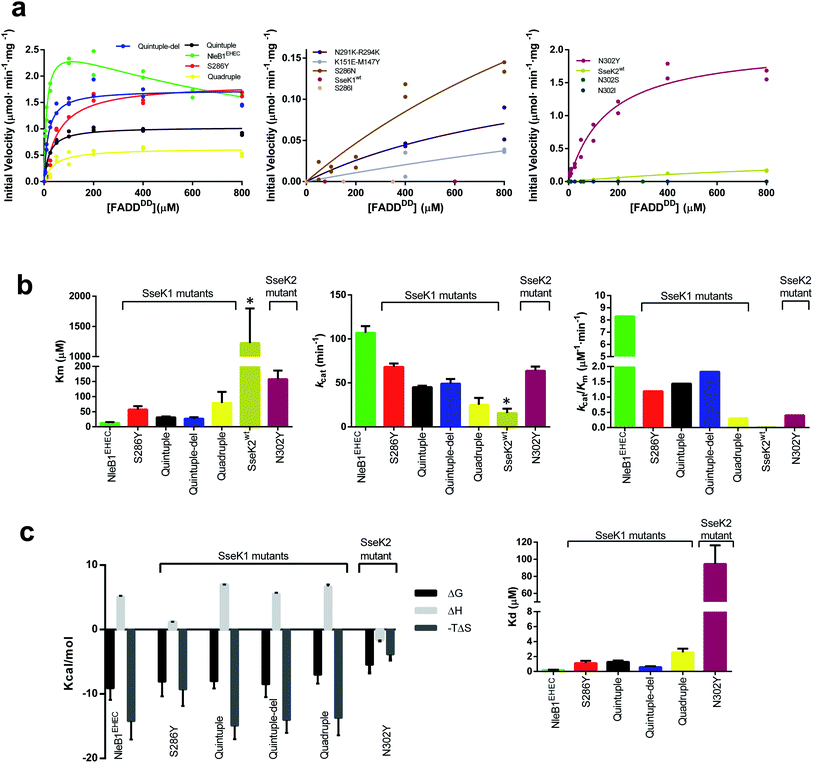 | ||
| Fig. 2 Enzyme kinetics and ITC experiments of NleB1EHEC/SseK1wt/SseK2wt and mutants on FADDDD. (a) Glycosylation kinetics of NleB1EHEC/SseK1wt/SseK2wt and mutants against FADDDD. (b) Plots comparing the Km, kcat and catalytic efficiency (kcat/Km) of the different NleB1EHEC/SseK1wt/SseK2wt and mutants. Additional kinetic data are given in Table S2.† Asterisks indicate that the kinetic parameters for SseK2wt are estimated due to its poorer binding to FADDDD. (c) (left) Thermodynamic dissection of the interaction of the different enzyme forms with FADDDD. The binding Gibbs energy (ΔG), enthalpy (ΔH), and entropy (−TΔS) are in kcal mol−1. Any negative value represents a favorable contribution to the binding, whereas a positive value represents an unfavorable contribution (right) graph depicting the Kds of the different enzymes. | ||
Seeking to switch on the activity of SseK1wt towards FADDDD, we characterized the double mutants M147Y–K151ESseK1 and N291K–R294KSseK1, and the single mutant S286YSseK1. In all of these mutants, we replaced the SseK1wt residues by the corresponding positions in NleB1EPEC/EHEC. While the initial velocity of the double mutants was very slow (∼46-fold worse than the NleB1EHEC initial velocity at 800 μM FADDDD; Fig. 2a, middle panel), strikingly, the kcat for the single mutant S286YSseK1 matched that of NleB1EHEC (only 1.38-fold worse; Fig. 2b, middle panel), implying that with a single mutation we reached the optimal kcat found for NleB1EHEC. On the contrary, Km and the catalytic efficiency for S286YSseK1 were slightly worse (4.4- and 6.7-fold worse than the ones reported for NleB1EHEC; Fig. 2b, left and right panels, and Table S2†). To obtain an SseK1 mutant with similar kinetic parameters as those of NleB1EHEC, we combined the double mutants to generate a quadruple mutant (M147Y-K151E-N291K-R294KSseK1). We further added the S286YSseK1 mutation to the latter mutant generating a quintuple mutant (M147Y-K151E-S286Y-N291K-R294KSseK1). Additionally, we made another mutant combining the quintuple mutant with a deletion of the C-terminal last 5 residues only found in SseK1wt (Fig. 1a) named as quintuple-del mutant. The kcat, Km and catalytic efficiency for the quadruple mutant were 4.3-/2.7-, 6.1-/1.4-, and 27-/4-fold worse than the ones for NleB1EHEC and S286YSseK1, respectively (Fig. 2a, b, and Table S2†). This demonstrates that the kinetic parameters of the quadruple mutant are closer to those of S286YSseK1 than those of NleB1EHEC. Hence, either the combination of multiple changes (quadruple mutant) or just a single mutation (S286YSseK1 mutant) leading to higher affinity of SseK1 towards FADDDD are two different approaches to achieve kcat values close to that of NleB1EHEC. The kinetic parameters of the quintuple and quintuple-del, mainly kcat and kcat/Km, were highly similar to each other and very close to those of S286YSseK1 (Fig. 2b, middle and right panels). However, they differed slightly more in their Kms, with the quintuple/quintuple-del mutants Kms being ∼2-fold lower than that of S286Y (Fig. 2b, left panel). The reduction in Km for the quintuple/quintuple-del mutants enhanced their catalytic efficiencies, approximating their values to that of NleB1EHEC. Overall, the removal of the C-terminal 5 amino acids in SseK1wt and the addition of 5 mutations led to an SseK1 form with the closest kinetic parameters to those of NleB1EHEC, mainly due to the lowest Km towards FADDDD of these SseK1 mutants.
Having established that a single mutation (S286YSseK1) was sufficient to achieve optimal kinetic parameters and render the best kcat of all the characterized mutants, we mutated Ser286SseK1 and Asn302SseK2 to Asn/Ile, and Ser/Ile/Tyr, respectively, rendering the mutants S286NSseK1, S286ISseK1, N302SSseK2, N302ISseK2 and N302YSseK2. These single mutants are derived from the alignment of Ser286SseK1 with Asn302SseK2, Ile289SseK3 and Tyr284CrNleB/NleB1 (Fig. 1a). As expected from the previous results10 of SseK3wt on FADDDD, the S286ISseK1 and N302ISseK2 were completely inactive, and N302SSseK2 was also completely inactive on FADDDD, implying that both Ser or Ile residues are likely deleterious for catalysis (Fig. 2a, middle and right panel). In addition, S286NSseK1 showed poor glycosylation of FADDDD (∼15-fold worse initial velocity than the one reported for NleB1EHEC at 800 μM FADDDD) as found for SseK2wt (Fig. 2a, middle panel). Although N302YSseK2 achieved a highly similar kcat to those of NleB1EHEC and S286YSseK1, its Km towards FADDDD was worse (3- and 12-fold higher than those of S286YSseK1 and NleB1EHEC, respectively). This caused a drop in catalytic efficiency compared to that of NleB1EHEC that was more drastic than that for S286Y (3- and 20-fold worse to those of S286YSseK1 and NleB1EHEC, respectively; Fig. 2a, b, and Table S2†).
Overall, our data indicate that a single mutation, either from Ser286SseK1 or Asn302SseK2 to Tyr, is sufficient to switch on and improve SseK1 and SseK2 glycosylation on FADDDD, respectively. This mutation allows reaching kinetic parameters very close to those of NleB1EHEC.
Binding of SseK1 to FADDDD is dramatically increased by the S286Y mutation
To determine the thermodynamic parameters of the mutants to FADDDD, we performed isothermal titration calorimetry (ITC) experiments. First, we determined the Kd of UDP for binding to NleB1EHEC in the presence of MnCl2 (Kd = 14.92 ± 1.11 μM) (Table S3 and Fig. S3†). Then, we evaluated whether these enzymes require UDP binding prior to binding FADDDD. While in the absence of UDP, NleB1EHEC or S286YSseK1 did not show any binding to FADDDD, this turned out to be the opposite in the presence of excess UDP (Fig. 2c, Table S3 and Fig. S3†). This provides compelling evidence that the NleB/SseK GTs likely follow an ordered bi–bi kinetic mechanism. In this mechanism, these enzymes are likely in an inactive state in the apo form (open C-terminal lid) that only shifts to the active state (closed C-terminal lid) in the presence of UDP-GlcNAc/MnCl2. This mechanism also implies an induced-fit mechanism by UDP-GlcNAc, in which this sugar nucleotide induces the closure of the C-terminal lid leading to the active state (Fig. S4†). The induced-fit mechanism has been also proposed for unrelated glycosyltransferases such as GalNAc-T2, B4GALT1 and the lactose synthase.28–30Once we determined that FADDDD binding to these enzymes requires prior UDP binding, we measured the thermodynamic parameters for all mutants versus FADDDD under an excess of UDP. We could only get titration for NleB1EHEC and the S286YSseK1, N302YSseK2, quadruple, quintuple, and quintuple-del mutants (Fig. S3†).
Detailed analysis of the thermodynamic parameters of the interaction showed that the binding of FADDDD to NleB1EHEC and the mutants was largely entropy-driven (−TΔS), while the binding of UDP was favored by a gain in enthalpy (ΔH), with a reduced entropic component (Fig. 2c and Table S3†), implying distinct interaction behaviors between these molecules. The unique thermodynamic profile exhibited by FADDDD might be due to the release of a vast number of surface water molecules from both FADDDD and NleB1EHEC/SseK1/SseK2 mutants upon binding, promoting a favorable desolvation entropy. On the contrary, the significant reduction in donor substrate mobility upon binding to the enzyme, along with the large number of hydrogen bonds between UDP or UDP-GlcNAc and these enzymes are largely the major factors explaining the reduction in the entropic component and the favorable enthalpy.17,19 Interestingly, the single mutants S286YSseK1 and N302YSseK2 achieve favorable binding Gibbs energy to FADD by reducing the beneficial entropy component of the interaction, what is accompanied by a more favorable enthalpy. Binding to FADD thus globally follows a pattern of enthalpy–entropy compensation where multiple mutants show similar thermodynamic profiles to that of NleB1, with single mutants benefitting from enthalpy, suggesting that the solvation/desolvation process at the interface of interaction with FADD is more similar to NleB1 for the multiple mutants than for the single mutants (Fig. S5†).
The Kds are in the low μM range except for that of N302YSseK2. Although the Kds are much lower than the Kms, there is some correlation between the Kms and the Kds: those enzymes with lower Km values also possess lower Kds (Table S3†). Again, NleB1EHEC displays the highest affinity (Kd = 0.2 ± 0.04 μM), being 3-, ∼6-, 13-, and −472-fold better than those of quintuple-del, quintuple/S286YSseK1, quadruple, and N302YSseK2 mutants, respectively (Fig. 2c, right panel, and Table S3†).
Overall, our data show that the improvement in binding of SseK1 mutants and N302YSseK2 to FADDDD is essential to promote catalysis. Strikingly, this can be achieved by a single-mutation, S286YSseK1 or N302YSseK2, or by a combination of multiple mutations in different regions of the enzyme. Nevertheless, these single mutations are enough to account for the best kcat of all mutants, implying that the Tyr residue in that position might also play a catalytic role.
Molecular dynamics simulations determine that the mutation S286YSseK1 couples binding to catalysis
To understand the structural role of the single mutation S286YSseK1 in enhancing binding and enabling glycosylation of the FADDDD, we carried out Gaussian accelerated Molecular Dynamics simulations (GaMD) on 3D molecular models of the complexes of NleB1EHEC, SseK1wt and mutants with UDP-GlcNAc-Mn2+ and FADDDD (see ESI and Fig. S6†).In the case of S286YSseK1, Tyr286 maintains similar favorable contacts as those in the complex of NleB1EPEC with FADDDD (Fig. 3), where its aromatic side chain is inserted into the groove formed by helices α2 and α3 and the loop connecting them, making close contacts with the backbone of Val121FADD and the side chain of Ile126FADD. However, for SseK1wt and S286ISseK1 the side chain at the point of mutation is either too small (Ser286) or too bulky (Ile286) to be properly allocated in the groove between helices α2 and α3, respectively (Fig. S7†). In the case of S286NSseK1, the side chain is equally not well allocated in the FADD groove, although a persistent hydrogen bond with Asp123FADD at helix α2 was observed (Fig. S7†).
These GaMD simulations results correlate very well with the kinetics measurements for SseK1wt and mutants, supporting a key role of the interaction of the side chain at the point of mutation with Ile126FADD from the FADDDD α2–α3 groove, most likely in the form of a favorable enthalpy contribution (Fig. S8†).
The GaMD simulations also allowed us to identify an important correlation between the side chain present at the point of mutation and catalysis, by analyzing the internal dynamics of the acceptor site (Arg117FADD side chain). In NleB1EPEC, the salt-bridge interaction between the proposed catalytic base (Glu253NleB1) and the guanidinium of Arg117FADD is maintained and holds the Arg117 in an orientation appropriate for the nucleophilic attack over the beta face. That interaction is not very conserved in the simulations of the other enzymes, and the role of Glu253NleB1 (Glu255SseK1) is replaced by the carboxylate of another residue, Asp188SseK1, which holds the Arg117 side chain in a rather rigid proper orientation all along the simulation time. This only occurs for the mutants that show glycosylating activity, S286YSseK1 and S286NSseK1, whereas that interaction is absent in the cases of S286ISseK1 and SseK1wt (Fig. S8†) where the Arg117 is more dynamic. This is reflected in the root-mean-square-fluctuations (RMSF) values of Arg117 in the complexes with NleB1EPEC, S286YSseK1, and S286NSseK1, which showed the lowest values (below 1 Å; Table S4†).
GaMD simulations also show that Phe187SseK1, Asp188SseK1, and Arg191SseK1 form a stable network of interactions with Arg117FADD (acceptor) and the sugar nucleotide (donor, Fig. S9†). Asp188SseK1 and Arg191SseK1 constitute the Asp/Arg dyad present in other bacterial GTs effectors.19 These results explain the need for a GT-bound sugar nucleotide to have efficient FADDDD binding (ordered bi–bi mechanism). The identified network of interactions leads to favorable contacts of the carboxylate side chain of Asp188SseK1 with the acceptor Arg117FADD, and of this with the sugar nucleotide. These interactions are conserved in NleB1-FADDDD complex (Fig. S9†).
The GaMD simulations support that a single mutation on Ser286SseK1 to Tyr leads to a favorable coupling between increased affinity and stability of Arg117FADD orientation, appropriated towards the nucleophilic attack of the anomeric carbon to render inversion of the configuration. This is achieved by a favorable interaction of the Tyr286SseK1 residue with Ile126FADD, leading to a stable salt-bridge of the Arg117FADD guanidinium polar head with the carboxylate of Asp188SseK1. The fact that Glu255SseK1 is far away from Arg117FADD and that Asp188SseK1 takes the role of the leading carboxylate in keeping the guanidinium on a proper orientation over the beta face of the GlcNAc residue of the donor substrate, strongly suggests that Asp188SseK1 might function as the catalytic base in S286YSseK1 and S286NSseK1 mutants. In fact, the D186A mutation in NleB1EPEC (the aligned residue in NleB1) has been also reported to be detrimental for NleB1 activity.19
The N302YSseK2 mutant is active against DR3DD
Having established the importance of a Tyr residue in binding/catalysis at position 284 of CrNleB/NleB1EPEC/EPEC, 286 of SseK1 and 302 of SseK2, we reasoned that the null activity of the SseK2wt on a NleB substrate could likely be rescued by replacing Asn302 by Tyr. To achieve that, we first evaluated the activity of NleB1EHEC, SseK1wt and SseK2wt on DR3DD, finding out that while NleB1EHEC, SseK1wt were active on DR3DD, SseK2wt was not (Fig. 4a). In these experiments, the Km, kcat and the catalytic efficiency for NleB1EHEC were 2.5-, 2.2- and 5.4-fold better than those of SseK1wt (Fig. 4b and Table S5†).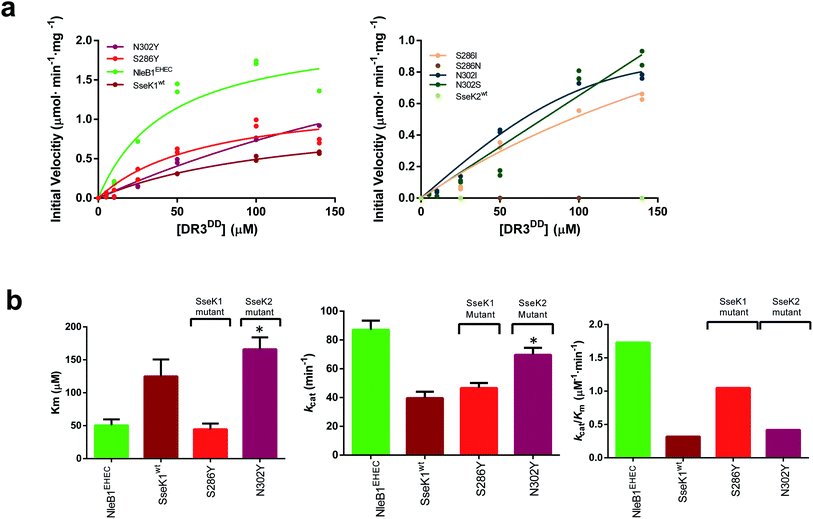 | ||
| Fig. 4 Enzyme kinetics of NleB1EHEC/SseK1wt/SseK2wt and mutants on DR3DD. (a) Glycosylation kinetics of NleB1EHEC/SseK1wt/SseK2wt and mutants against DR3DD. (b) Plots comparing the Km, kcat and catalytic efficiency (kcat/Km) of the different NleB1EHEC/SseK1wt/SseK2wt and mutants. Additional kinetic data are shown in Table S5.† Asterisks indicate that the kinetic parameters for N302YSseK2 are estimated due to its poorer binding to DR3DD. | ||
We then performed enzyme kinetics assays on mutants S286I/N/YSseK1 and N302I/S/YSseK2. As found for SseK2wt on DR3DD, S286NSseK1 was also inactive on DR3DD (Fig. 4a, right panel). The other mutants displayed different degrees of initial velocities. Although the initial velocity for the mutants S286ISseK1 and N302I/SSSeK2 was approximately half that of NleB1EHEC at 140 μM DR3DD, these mutants did not reach saturated kinetics, preventing us from determining their kinetic parameters. However, we could obtain kinetic parameters for S286YSseK1 and N302YSseK2 (Fig. 4a, b, Table S5†). Again, the mutation to Tyr in both enzymes provided DR3DD saturation curves. The Kms for NleB1EHEC and S286YSseK1 were similar while kcat and catalytic efficiency were ∼1.75-fold better for NleB1EHEC than S286YSseK1. On the contrary, kcat values were similar for NleB1EHEC and N302YSseK2, differing more in Km and catalytic efficiency (3.3- and 4.1-fold better constants for NleB1EHEC than those for N302YSseK2; Table S5†). Overall, our data with DR3DD are slightly more complex than the ones for FADDDD, and in particular suggest that a Tyr residue in positions 284CrNleB/NleB1, 286SseK1 and 302SseK2 is more beneficial for enzyme kinetics than a Ser, and the latter over Ile, being an Asn residue in those positions deleterious for activity.
The S286YSseK1 and N302YSseK2 mutants promote Salmonella enterica survival in macrophages
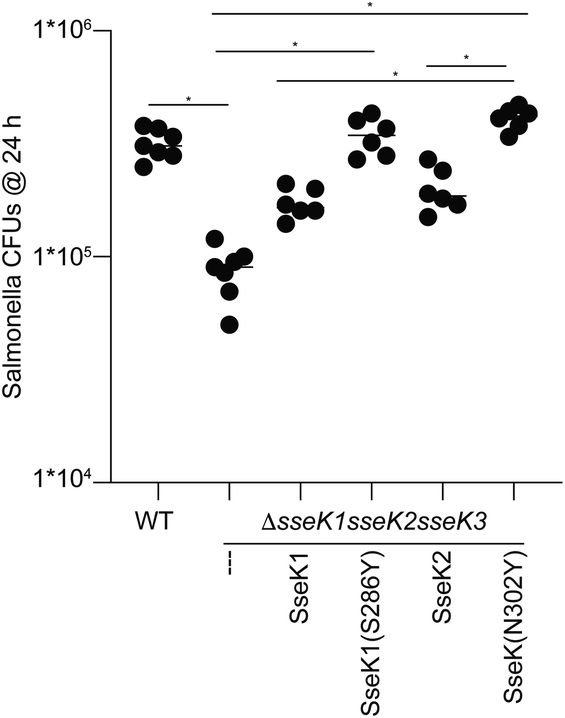
Finally, we rationalized that if Tyr286SseK1 and Tyr302SseK2 strikingly improve the kinetic and the thermodynamic parameters, these mutants might also increase the survival of Salmonella enterica strains in infected macrophages. To test this hypothesis, we introduced in a Salmonella strain lacking all SseK GTs the constructs encoding for the expression of SseK1wt, SseK2wt, S286YSseK1 and N302YSseK2. The survival of these mutated strains was evaluated in macrophages showing that S286YSseK1 and N302YSseK2 improved the proliferation of this bacterium compared to the strains lacking the expression of all SseK GTs or expressing specifically either SseK1wt or SseK2wt. In addition, the level of proliferation reached by strains expressing S286YSseK1 and N302YSseK2 was highly similar to the wild type bacteria that expresses all three GTs (Fig. 5). This result again demonstrates that SseK GTs containing a Tyr residue in 286SseK1, 302SseK2, and potentially 289SseK3 are more robust enzymes that lead to bacterial proliferation in macrophages and in turn, virulence.Conclusions
Why Salmonella enterica strains encode three functional NleB orthologs with a narrowed substrate selectivity is puzzling. Salmonella enterica differs from EHEC/EPEC E. coli and C. rodentium both in its life cycle and also in the different location of its GTs in host cells. While EHEC/EPEC E. coli or C. rodentium are extracellular pathogens, Salmonella enterica strains are intracellular pathogens. Furthermore, CrNleB/NleB1EPEC/EHEC/SseK1 are cytoplasmic proteins, while SseK2/SseK3 are Golgi-associated GTs.15 This provides another way of regulating the types of protein substrates being glycosylated, which for SseK3 is unique as recently reported.22 Hence, the SseK narrowed substrate selectivity together with the location-dependent glycosylation of SseK1/2/3wt might endow Salmonella enterica strains with novel ways of hijacking host signaling pathways. However, the selection pressures, if any, driving the differing substrate selectivities among these enzymes, have yet to be investigated.Here, we have addressed the molecular basis of this narrowed substrate selectivity, which relies on a unique second-shell residue, variable between the SseK GTs and located in the interface of the NleB/SseK-protein substrate complex. The mutation of this second-shell residue, either Ser286SseK1 or Asn302SseK2, to TyrNleB/NleB1, leads to mutants with optimal kinetic and thermodynamic parameters. The mutants S286YSseK1 and N302YSseK2 become active on particular protein substrates such as FADDDD and DR3DD, respectively, leading potentially to mutants with a broader substrate selectivity as that of CrNleB/NleB1EPEC/EHEC. This is also supported by the increase in Salmonella abundance in macrophages by strains expressing these mutants. These mutants promote binding and catalysis, likely because the binding of this second-shell residue surrounding residues is coupled to the stability of the interaction between the acceptor Arg with the catalytic base residue. Therefore, the identity of the second-shell residue finely tunes protein substrate selectivity and, in turn, glycosylation, and might explain whether GT substrate selectivity is narrow or broad.
Promiscuous GTs act on multiple protein substrates and are found in all animal kingdoms.4 Several GTs mechanisms have been discovered by combining X-ray crystallography experiments with other biophysical and biochemical techniques. Initiating GTs such as POFUT1/POGLUT1 (also called Rumi) and POFUT2, require folded EGF and TSR repeats, respectively. These GTs share in common that the EGF and TSR repeats are tethered by direct hydrogen bonds, and require EGF and TSR repeats containing minimal consensus sequences that encompass mostly variable residues located in between two cysteine residues. However, POFUT2, and to a lesser extent POFUT1, apply additional strategies to recognize many different protein substrates, leveraging the water molecules present in the interface of the complexes to recognize different repeats. In addition, POGLUT1 and POFUT2 also interact with the repeats by hydrophobic interactions.31–33 Other initiating GTs, such as OGT and the large family of GalNAc-T isoenzymes (20 in humans) recognize mostly unfolded extended and compact structures of the peptide acceptor substrates, respectively, by establishing hydrogen bonds. Thus, for example, while for OGT a proline (Pro) residue in −2 (promoting the extended conformation) is required for optimal glycosylation,34 a Pro-X-Pro motif (promoting the compact conformation) contiguous to the preceding Ser/Thr residue usually favors glycosylation.35 In addition, GalNAc-Ts contain a flexible linker located in between the catalytic and the lectin domains, and a flexible loop that provides them with different behaviors due to their dissimilar amino acid sequences. While the flexible linker is behind the dynamics of these isoenzymes and the location of the GalNAc-binding site in the lectin domain,36,37 the flexible loop controls the catalytic cycle and is also behind the recognition of protein substrates.38,39 Together, both the flexible linker and loop determine whether several GalNAc-T isoenzymes are highly specific for particular protein substrates.38,40 The SseK GTs have evolved different features at molecular level to selectively recognize particular protein substrates. These features rely on variable residues located in the HLH domain and mostly in a particular variable second-shell residue with respect to the conserved Tyr284NleB/NleB1 residue.
In conclusion, and to our knowledge, this is a unique example of restoring the activity of enzymes that are inactive on particular protein substrates by a single site-directed mutagenesis. Overall, our finding provides the molecular basis of the differences between NleB/SseK GTs substrate selectivity and offers clues on the molecular pathogenesis of enteropathogens.
Data availability
Data are available from the corresponding authors upon reasonable request.Author contribution
R. H.-G., J. A. and T. H. performed the bioinformatic analysis of the enzymes and R. H.-G. selected the positions for site-directed mutagenesis experiments. A. G.-G. purified the enzymes and characterized them by enzyme kinetics and ITC experiments. T. H. and J. A. performed the molecular dynamics simulations. S. E. Q. and C. Z. performed the macrophage experiments and made the Salmonella mutants. R. H.-G. wrote the article with the contribution of J. A. and P. R. H. All authors read and approved the final manuscript.Conflicts of interest
The authors declare no competing interests.Acknowledgements
We thank ARAID, the Spanish Ministry of Science, Innovation and Universities (BFU2016-75633-P and PID2019-105451GB-I00 to R. H-G., and PID2019-109395GB-I00 to J. A.), and Gobierno de Aragón (E34_R17 and LMP58_18 to R. H-G.) with FEDER (2014–2020) funds for “Building Europe from Aragón” for financial support. A. G.-G. thanks Gobierno de Aragón for a predoctoral fellowship. J. A. also acknowledges funding support from the Biotechnology and Biological Sciences Research Council (BBSRC; BB/P010660/1). T. H. acknowledges a BBSRC DTP studentship (BB/M011216/1). Support from the Universidad de Sevilla (Acciones Especiales del VI Plan Propio de Investigación y Transferencia) is also acknowledged. The project described was supported by grant numbers AI127973 and AI153202 from the National Institute of Allergy and Infectious Diseases (NIAID) and by the National Institute of General Medical Sciences (NIGMS) of the National Institutes of Health under award number P20GM130448 to P. R. H. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health.References
- R. A. Flynn, K. Pedram, S. A. Malaker, P. J. Batista, B. A. H. Smith, A. G. Johnson, B. M. George, K. Majzoub, P. W. Villalta, J. E. Carette and C. R. Bertozzi, Cell, 2021, 184, 3109–3124 e3122 CrossRef CAS.
- A. Biswas and M. Thattai, Biochem. Soc. Trans., 2020, 48, 891–900 CrossRef CAS.
- K. T. Schjoldager, Y. Narimatsu, H. J. Joshi and H. Clausen, Nat. Rev. Mol. Cell Biol., 2020, 21, 729–749 CrossRef CAS PubMed.
- H. J. Joshi, Y. Narimatsu, K. T. Schjoldager, H. L. P. Tytgat, M. Aebi, H. Clausen and A. Halim, Cell, 2018, 172, 632–632 e632 CrossRef CAS PubMed.
- X. Pan, J. Luo and S. Li, Front. Cell. Infect. Microbiol., 2020, 10, 185 CrossRef CAS PubMed.
- J. Lassak, E. C. Keilhauer, M. Furst, K. Wuichet, J. Godeke, A. L. Starosta, J. M. Chen, L. Sogaard-Andersen, J. Rohr, D. N. Wilson, S. Haussler, M. Mann and K. Jung, Nat. Chem. Biol., 2015, 11, 266–270 CrossRef CAS PubMed.
- X. Gao, X. Wang, T. H. Pham, L. A. Feuerbacher, M. L. Lubos, M. Huang, R. Olsen, A. Mushegian, C. Slawson and P. R. Hardwidge, Cell Host Microbe, 2013, 13, 87–99 CrossRef CAS.
- J. S. Pearson, C. Giogha, S. Y. Ong, C. L. Kennedy, M. Kelly, K. S. Robinson, T. W. Lung, A. Mansell, P. Riedmaier, C. V. Oates, A. Zaid, S. Muhlen, V. F. Crepin, O. Marches, C. S. Ang, N. A. Williamson, L. A. O'Reilly, A. Bankovacki, U. Nachbur, G. Infusini, A. I. Webb, J. Silke, A. Strasser, G. Frankel and E. L. Hartland, Nature, 2013, 501, 247–251 CrossRef CAS PubMed.
- S. Li, L. Zhang, Q. Yao, L. Li, N. Dong, J. Rong, W. Gao, X. Ding, L. Sun, X. Chen, S. Chen and F. Shao, Nature, 2013, 501, 242–246 CrossRef CAS.
- S. El Qaidi, K. Chen, A. Halim, L. Siukstaite, C. Rueter, R. Hurtado-Guerrero, H. Clausen and P. R. Hardwidge, J. Biol. Chem., 2017, 292, 11423–11430 CrossRef CAS PubMed.
- S. El Qaidi, N. E. Scott, M. P. Hays, B. V. Geisbrecht, S. Watkins and P. R. Hardwidge, Sci. Rep., 2020, 10, 1073 CrossRef CAS.
- S. El Qaidi, N. E. Scott and P. R. Hardwidge, Sci. Rep., 2021, 11, 3834 CrossRef CAS PubMed.
- V. Lombard, H. Golaconda Ramulu, E. Drula, P. M. Coutinho and B. Henrissat, Nucleic Acids Res., 2014, 42, D490–D495 CrossRef CAS PubMed.
- C. Giogha, N. E. Scott, T. Wong Fok Lung, G. L. Pollock, M. Harper, E. D. Goddard-Borger, J. S. Pearson and E. L. Hartland, PLoS Pathog., 2021, 17, e1009658 CrossRef CAS.
- J. L. Araujo-Garrido, J. Bernal-Bayard and F. Ramos-Morales, Microorganisms, 2020, 8, 1–23 CrossRef.
- D. Esposito, R. A. Gunster, L. Martino, K. El Omari, A. Wagner, T. L. M. Thurston and K. Rittinger, J. Biol. Chem., 2018, 293, 5064–5078 CrossRef CAS.
- J. B. Park, Y. H. Kim, Y. Yoo, J. Kim, S. H. Jun, J. W. Cho, S. El Qaidi, S. Walpole, S. Monaco, A. A. Garcia-Garcia, M. Wu, M. P. Hays, R. Hurtado-Guerrero, J. Angulo, P. R. Hardwidge, J. S. Shin and H. S. Cho, Nat. Commun., 2018, 9, 4283 CrossRef.
- J. P. M. Newson, N. E. Scott, I. Yeuk Wah Chung, T. Wong Fok Lung, C. Giogha, J. Gan, N. Wang, R. A. Strugnell, N. F. Brown, M. Cygler, J. S. Pearson and E. L. Hartland, Mol. Cell. Proteomics, 2019, 18, 1138–1156 CrossRef CAS PubMed.
- J. Ding, X. Pan, L. Du, Q. Yao, J. Xue, H. Yao, D. C. Wang, S. Li and F. Shao, Mol. Cell, 2019, 74, 922–935 e926 CrossRef CAS PubMed.
- X. Gao, T. H. Pham, L. A. Feuerbacher, K. Chen, M. P. Hays, G. Singh, C. Rueter, R. Hurtado-Guerrero and P. R. Hardwidge, J. Biol. Chem., 2016, 291, 18232–18238 CrossRef CAS.
- C. Xu, X. Liu, H. Zha, S. Fan, D. Zhang, S. Li and W. Xiao, PLoS Pathog., 2018, 14, e1007259 CrossRef PubMed.
- J. Gan, N. E. Scott, J. P. M. Newson, R. R. Wibawa, T. Wong Fok Lung, G. L. Pollock, G. Z. Ng, I. van Driel, J. S. Pearson, E. L. Hartland and C. Giogha, Front. Cell. Infect. Microbiol., 2020, 10, 419 CrossRef CAS.
- R. Krafczyk, J. Macosek, P. K. A. Jagtap, D. Gast, S. Wunder, P. Mitra, A. K. Jha, J. Rohr, A. Hoffmann-Roder, K. Jung, J. Hennig and J. Lassak, mBio, 2017, 8, e01412-17 CrossRef PubMed.
- C. I. Chen, J. J. Keusch, D. Klein, D. Hess, J. Hofsteenge and H. Gut, EMBO J., 2012, 31, 3183–3197 CrossRef CAS PubMed.
- M. de Las Rivas, E. J. Paul Daniel, Y. Narimatsu, I. Companon, K. Kato, P. Hermosilla, A. Thureau, L. Ceballos-Laita, H. Coelho, P. Bernado, F. Marcelo, L. Hansen, R. Maeda, A. Lostao, F. Corzana, H. Clausen, T. A. Gerken and R. Hurtado-Guerrero, Nat. Chem. Biol., 2020, 16, 351–360 CrossRef CAS.
- A. Naegeli, G. Michaud, M. Schubert, C. W. Lin, C. Lizak, T. Darbre, J. L. Reymond and M. Aebi, J. Biol. Chem., 2014, 289, 24521–24532 CrossRef CAS.
- W. Kightlinger, L. Lin, M. Rosztoczy, W. Li, M. P. DeLisa, M. Mrksich and M. C. Jewett, Nat. Chem. Biol., 2018, 14, 627–635 CrossRef CAS PubMed.
- E. Lira-Navarrete, J. Iglesias-Fernandez, W. F. Zandberg, I. Companon, Y. Kong, F. Corzana, B. M. Pinto, H. Clausen, J. M. Peregrina, D. J. Vocadlo, C. Rovira and R. Hurtado-Guerrero, Angew. Chem., Int. Ed. Engl., 2014, 53, 8206–8210 CrossRef CAS.
- B. Ramakrishnan, E. Boeggeman and P. K. Qasba, Biochem. Biophys. Res. Commun., 2002, 291, 1113–1118 CrossRef CAS.
- M. de Las Rivas, H. Coelho, A. Diniz, E. Lira-Navarrete, I. Companon, J. Jimenez-Barbero, K. T. Schjoldager, E. P. Bennett, S. Y. Vakhrushev, H. Clausen, F. Corzana, F. Marcelo and R. Hurtado-Guerrero, Chemistry, 2018, 24, 8382–8392 CrossRef CAS.
- Z. Li, K. Han, J. E. Pak, M. Satkunarajah, D. Zhou and J. M. Rini, Nat. Chem. Biol., 2017, 13, 757–763 CrossRef CAS PubMed.
- H. Yu, H. Takeuchi, M. Takeuchi, Q. Liu, J. Kantharia, R. S. Haltiwanger and H. Li, Nat. Chem. Biol., 2016, 12, 735–740 CrossRef CAS.
- J. Valero-González, C. Leonhard-Melief, E. Lira-Navarrete, G. Jiménez-Osés, C. Hernández-Ruiz, M. C. Pallarés, I. Yruela, D. Vasudevan, A. Lostao, F. Corzana, H. Takeuchi, R. S. Haltiwanger and R. Hurtado-Guerrero, Nat. Chem. Biol., 2016, 12, 240–246 CrossRef.
- S. Pathak, J. Alonso, M. Schimpl, K. Rafie, D. E. Blair, V. S. Borodkin, A. W. Schüttelkopf, O. Albarbarawi and D. M. F. van Aalten, Nat. Struct. Mol. Biol., 2015, 22, 744–750 CrossRef CAS PubMed.
- M. de las Rivas, E. Lira-Navarrete, T. A. Gerken and R. Hurtado-Guerrero, Curr. Opin. Struct. Biol., 2019, 56, 87–96 CrossRef CAS.
- E. Lira-Navarrete, M. de las Rivas, I. Compañón, M. C. Pallarés, Y. Kong, J. Iglesias-Fernández, G. J. L. Bernardes, J. M. Peregrina, C. Rovira, P. Bernadó, P. Bruscolini, H. Clausen, A. Lostao, F. Corzana and R. Hurtado-Guerrero, Nat. Commun., 2015, 6, 6937 CrossRef CAS PubMed.
- M. de las Rivas, E. Lira-Navarrete, E. J. P. Daniel, I. Compañón, H. Coelho, A. Diniz, J. Jiménez-Barbero, J. M. Peregrina, H. Clausen, F. Corzana, F. Marcelo, G. Jiménez-Osés, T. A. Gerken and R. Hurtado-Guerrero, Nat. Commun., 2017, 8, 1959 CrossRef.
- M. de las Rivas, E. J. Paul Daniel, H. Coelho, E. Lira-Navarrete, L. Raich, I. Compañón, A. Diniz, L. Lagartera, J. Jiménez-Barbero, H. Clausen, C. Rovira, F. Marcelo, F. Corzana, T. A. Gerken and R. Hurtado-Guerrero, ACS Cent. Sci., 2018, 4, 1274–1290 CrossRef CAS PubMed.
- M. de las Rivas, H. Coelho, A. Diniz, E. Lira-Navarrete, I. Compañón, J. Jiménez-Barbero, K. T. Schjoldager, E. P. Bennett, S. Y. Vakhrushev, H. Clausen, F. Corzana, F. Marcelo and R. Hurtado-Guerrero, Chem. - Eur. J., 2018, 24, 8382–8392 CrossRef CAS PubMed.
- M. de las Rivas, E. J. Paul Daniel, Y. Narimatsu, I. Compañón, K. Kato, P. Hermosilla, A. Thureau, L. Ceballos-Laita, H. Coelho, P. Bernadó, F. Marcelo, L. Hansen, R. Maeda, A. Lostao, F. Corzana, H. Clausen, T. A. Gerken and R. Hurtado-Guerrero, Nat. Chem. Biol., 2020, 16, 351–360 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available: Supporting methodology, 9 supporting figures and 5 supporting tables. See DOI: 10.1039/d1sc04065k |
| This journal is © The Royal Society of Chemistry 2021 |