Open Access Article
Open Access ArticleAn endogenous stimulus detonated nanocluster-bomb for contrast-enhanced cancer imaging and combination therapy†
Huanhuan
Sun
,
Wenjie
Ma
,
Shuangdi
Duan
,
Jin
Huang
,
Ruichen
Jia
,
Hong
Cheng
,
Biao
Chen
,
Xiaoxiao
He
* and
Kemin
Wang
 *
*
State Key Laboratory of Chemo/Biosensing and Chemometrics, College of Biology, College of Chemistry and Chemical Engineering, Hunan University, Key Laboratory for Bio-Nanotechnology and Molecule Engineering of Hunan Province, Changsha 410082, China. E-mail: xiaoxiaohe@hnu.edu.cn; kmwang@hnu.edu.cn
First published on 17th August 2021
Abstract
Exploitation of stimuli-responsive nanoplatforms is of great value for precise and efficient cancer theranostics. Herein, an in situ activable “nanocluster-bomb” detonated by endogenous overexpressing legumain is fabricated for contrast-enhanced tumor imaging and controlled gene/drug release. By utilizing the functional peptides as bioligands, TAMRA-encircled gold nanoclusters (AuNCs) endowed with targeting, positively charged and legumain-specific domains are prepared as quenched building blocks due to the AuNCs' nanosurface energy transfer (NSET) effect on TAMRA. Importantly, the AuNCs can shelter therapeutic cargos of DNAzyme and Dox (Dzs-Dox) to aggregate larger nanoparticles as a “nanocluster-bomb” (AuNCs/Dzs-Dox), which could be selectively internalized into cancer cells by integrin-mediated endocytosis and in turn locally hydrolyzed in the lysosome with the aid of legumain. A “bomb-like” behavior including “spark-like” appearance (fluorescence on) derived from the diminished NSET effect of AuNCs and cargo release (disaggregation) of Dzs-Dox is subsequently monitored. The results showed that the AuNC-based disaggregation manner of the “nanobomb” triggered by legumain significantly improved the imaging contrast due to the activable mechanism and the enhanced cellular uptake of AuNCs. Meanwhile, the in vitro cytotoxicity tests revealed that the detonation strategy based on AuNCs/Dzs-Dox readily achieved efficient gene/chemo combination therapy. Moreover, the super efficacy of combinational therapy was further demonstrated by treating a xenografted MDA-MB-231 tumor model in vivo. We envision that our multipronged design of theranostic “nanocluster-bomb” with endogenous stimuli-responsiveness provides a novel strategy and great promise in the application of high contrast imaging and on-demand drug delivery for precise cancer theranostics.
Introduction
“Stimulus-triggered cargo release” is a well-established strategy by now and has been broadly utilized as the foundation for precise and effective cancer theranostics.1–4 Based on the internal or external stimulus, various types of nanomaterials have been developed as smart nanocarriers endowed with different cargo release mechanisms5–8 including stimulus mediated direct release,9 expansion,10 gatekeeping,11 cavitation11 and disassembly/disaggregation.13–15 Among them, destructive behavior like cavitation and disassembly/disaggregation has attracted much attention due to the high payload capacity, concentrated payload release and size-tunable behavior of the nanomaterials,16 endowing them with prolonged circulation lifetime,16 enhanced localized cytotoxicity18 and renal clearance efficiency.19 The typical nanomaterials working in this manner are the polymersomes and liposomes, whose complex synthetic procedure or tedious modification is inevitable to their theranostic applications for the purpose of improving their stability and target therapy.12,18,20,21 Additionally, the imaging units in these nanosystems are always the encapsulated contrast agents or fluorescein carried in an “always on” manner.17,22 Hence, for precise diagnosis and efficient cancer therapy, it is of pioneering significance to develop an accessible and imageable nanomaterial for constructing a theranostic nanoplatform with a stimuli-triggered destruction mechanism.Gold nanoclusters (AuNCs), known for their ultrasmall size and biocompatible properties, may offer an elegant solution to the need for simultaneously achieving stimuli-responsive contrast-enhanced imaging as a reporter, and maximizing the therapeutic efficacy and minimizing the side effect and toxicity as a nanovector.17,23,24 As for a reporter, in the past few years, gold nanoclusters were mostly utilized for biosensing25 and bioimaging,26 relying on their fluorescence emission properties.27,28 Additionally, as a high-z-element based material, the AuNCs displayed strong absorption of radiation energy, and have been used as radiosensitizers in X-ray imaging and cancer radiotherapy.29,30 Besides that, some other inherent superiorities of AuNCs have rarely been employed in the bioimaging process. Actually, the ultrasmall AuNCs exhibit significant energy-absorption capability to quench a fluorophore on its surface (<20 nm) through the interaction of the dipole-metal plane coupling, which was reported by the Strouse group and named nanosurface energy transfer (NSET) of AuNCs.31,32 Based on this finding, AuNCs were synthesized to serve as an acceptor rather than a donor to design a nanosensor for stimuli-responsive contrast-enhanced imaging (the ratio of signal/noise was up to 20 in vitro) of caspase 3 activity in live cells.24 The results displayed new potential of AuNCs which served as an activable reporter. More importantly, as for a nanocarrier, a simple synthetic route and multiple bio-ligands are available for AuNC synthesis,33,34 such as SH-containing peptides,35,36 proteins37 and polymers.38 They provide AuNCs with not only biocompatible shells and controllable surface charge to bind composite charged cargos to cross bio-barriers, but also multiple functional regions such as a tumor target domain and site-specific substrate to respond at a desired site by rational designing and regulating the proportion of ligands. It implies that AuNCs can also be engineered as multifunctional and biocompatible building blocks of nanocarriers synchronously when needed. For the combination of diagnosis and therapy based on AuNCs, although conceptually feasible, practically applying the NEST effect and ligand properties of AuNCs to achieve activable imaging, followed by controllable concentrated payload release via stimuli-detonated explosion has remained challenging until now.
To fulfill the stimuli-triggered on-demand cargo release in a detonation manner, the choice of stimulus is also a key factor. For the purpose of endowing the formulations with safe and intelligent performance, an endogenous stimulus such as protease overexpressed and activated in some particular way could be considered in the design of the theranostic nanoplatform on account of the exploitable relationship between the peptide ligand and substrate of protease. Legumain, a cysteine protease overexpressed in most human tumors, and particularly distributed in the lysosome,39 is identified as a tumor marker, and has been largely utilized in the design of legumain-sensitive nanosensors or nanotheranostics for tumors.40–45 The short peptides of alanine–alanine–asparagine (AAN), as a specific substrate of legumain, had a perfect attendance presented in different forms in these nanosystems. It inspired us that a fluorophore tagged AAN moiety could be incorporated with the peptide ligand to fabricate a turn-on legumain sensor with the aid of the NSET effect of AuNCs. The specific expression pattern of legumain lends itself not only to be an ideal stimulus for cancer cell imaging but also as an initiator for on-demand drug release.
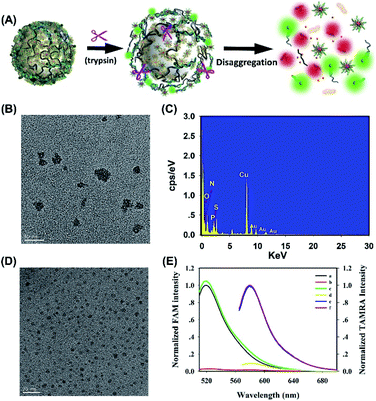
Herein, we present the delicate design of stimuli-responsive nanotheranostics based on the multifunctional peptide-protected AuNCs, which acted as the activated nanosensor and cationic nanocarrier for legumain-triggered specific imaging and combinational therapy of cancer cells (Scheme 1). In this nanosystem, we synthesized multifunctional peptide-coated AuNCs by a one-step biomineralization method, where the peptides provided templates and reducing agent domain (CCY in PL1, PL2 and P3), targeted domain (RGD in PL1, PL2 and P3), positively charged domain (R9 in PL1), legumain-specific substrate domain (AAN in PL1) and TAMRA labeled legumain-specific substrate domain (AAN-TAMRA in PL2), respectively. The fluorescence of TAMRA was absolutely quenched by the formed AuNCs through the NSET effect. The classic anticancer drug of Dox was inserted into the GC base pairs of functional nucleic acid DNAzyme-S (Dzs). Taking advantage of the positive charge on the AuNC surface, the negative therapeutic Dzs-Dox could be absorbed by AuNCs via electrostatic attraction to form large nanoparticles (AuNCs/Dzs-Dox). With the aid of the RGD peptide, AuNCs/Dzs-Dox nanoparticles preferentially target integrin αvβ3 (a biomarker of various cancer cells) and enter into the cancer cells by receptor-mediated endocytosis. The fluorescence recovery and disaggregation of AuNCs/Dzs-Dox nanoparticles were successively triggered by the specific hydrolyzation of overexpressed legumain in the lysosome, achieving legumain-responsive imaging, specific cleavage of EGR-1 (regulates cancer cell proliferation) and Dox release for combinational therapy of cancer cells definitively. The compatibility and the use of AuNCs/Dzs for activated fluorescence imaging, combination therapy of cancer cells in vitro and a tumor model in vivo were systematically tested.
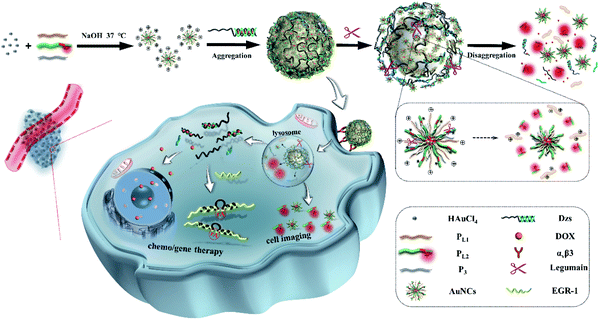 | ||
| Scheme 1 Schematic illustration of nanocluster-bomb system (L-AuNCs/Dzs) synthesis and the detonation strategy for contrast enhanced cancer imaging and combination therapy. | ||
Results and discussion
Synthesis, characterization and optimization of trypsin-responsive AuNCs (T-AuNCs)
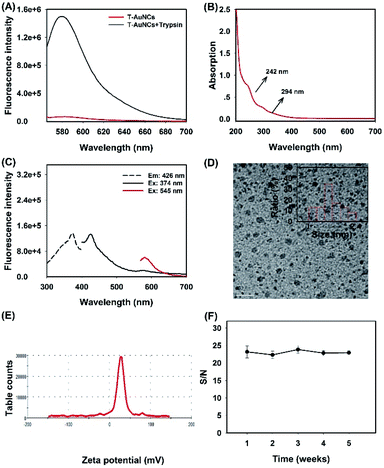
In order to facilitate the investigation of feasibilities of this work, trypsin was used as the model enzyme at the beginning of the work (for pretest). The peptide-functionalized AuNCs were synthesized by one-step reduction of Au3+ according to a previous study.31 In our synthesis system, three kinds of peptides, positive peptide PT1 (CCYRRRRRRRRRGD), signal peptide PT2 (CCYGGGGRGRK (TRMRA) RGD) and helper peptide P3 (CCYGGGGRGRKRGD) with multifunctional domains were utilized to form the trypsin-responsive AuNCs (T-AuNCs). In detail, all three of the peptides were sharing the same functional domains: (1) ligand domain CCY, in which tyrosine (Y) provides phenolic groups as the reducing agent for the reduction of Au3+ under alkaline conditions and cysteine (C) provides –SH as the anchoring site for Au atom clustering via the Au–S bond; (2) targeted domain RGD which has high-affinity to integrin αvβ3 which is overexpressed in breast, lung and pancreatic cancer cells.46 Besides, PT1 contains an oligoarginine segment (R8: eight arginine residues) which bears a positive charge to serve as a cationic carrier for nucleic acid delivery; PT2 contains a trypsin specific substrate labeled with a fluorophore tag, and is utilized as a signal unit for trypsin activity detection; P3 acts as a helper peptide to synthesize AuNCs so as to enhance the fluorescence signal-to-noise (S/N) ratio of enzyme activity assay.To obtain the T-AuNCs with a higher S/N ratio of trypsin cleavage, the fluorescence emission spectra of T-AuNCs synthesized in different molar ratios of PT1, P3 (non-fluorescence peptides were collectively named P0) and PT2 were investigated separately, and the fluorescence S/N ratios of their products in response to trypsin cleavage were calculated (Table S2†). We also tested the fluorescence stability of T-AuNCs in a month, and the ratio of P0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) PT2 at 30
PT2 at 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 60
1 and 60![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 showed better quality and a higher S/N ratio (data not shown). Then T-AuNCs synthesized with different concentrations of NaOH in the peptide ratio of P0
1 showed better quality and a higher S/N ratio (data not shown). Then T-AuNCs synthesized with different concentrations of NaOH in the peptide ratio of P0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) PT2 at 30
PT2 at 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 60
1 and 60![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 were obtained and the corresponding S/N ratio was evaluated (Table S3†). The results demonstrated that a crucial control of pH of the reaction system was necessary for the synthesis of AuNCs due to the isoelectric point around 10–12 of the peptides, as well as demand for proper alkaline conditions for reduction. Therefore, we finally decided to use PT1, P3, and PT2 at a molar ratio of 30
1 were obtained and the corresponding S/N ratio was evaluated (Table S3†). The results demonstrated that a crucial control of pH of the reaction system was necessary for the synthesis of AuNCs due to the isoelectric point around 10–12 of the peptides, as well as demand for proper alkaline conditions for reduction. Therefore, we finally decided to use PT1, P3, and PT2 at a molar ratio of 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 30
30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and the concentration of NaOH at 0.5 M to synthesize T-AuNCs. The fluorescence spectra of the optimal T-AuNCs (PT2-TAMRA) and their trypsin-responsive performance (cleaved PT2-TAMRA) are illustrated in Fig. 1A, where the S/N ratio was calculated to be about 20. We then tested the optical properties of the as-prepared T-AuNCs. It was demonstrated that there were broad absorption bands from 200 nm to 700 nm with two shoulder peaks at 242 nm and 294 nm derived from the phenoxide structure of tyrosine (Fig. 1B), which was consistent with those previously reported.47 As shown in Fig. 1C and Fig. A, it was demonstrated that T-AuNCs exhibited almost no fluorescence but only strong quenching ability to TAMRA due to the NSET effect. For the morphology characterization, a typical TEM image showed that the obtained T-AuNCs were well-dispersed with an average size around 1.76 nm (Fig. 1D). As expected, the T-AuNCs had a positive surface charge of 26.6 mV owing to the oligoarginine segments on the peptides (Fig. 1E), which endowed themselves with the potential of acting as a cationic carrier to deliver negative therapeutic cargos (drug loaded nucleic acids). The stability test of T-AuNCs revealed that the S/N ratio remains almost unchanged upon keeping at 4 °C for a month (Fig. 1F). All of these results suggested that the peptide-coated AuNCs were successfully obtained with expected properties such as stimuli-responsiveness, high NSET efficiency and positive charge, showing the potential of AuNCs to act as a high contrast reporter and a nanocarrier.
1 and the concentration of NaOH at 0.5 M to synthesize T-AuNCs. The fluorescence spectra of the optimal T-AuNCs (PT2-TAMRA) and their trypsin-responsive performance (cleaved PT2-TAMRA) are illustrated in Fig. 1A, where the S/N ratio was calculated to be about 20. We then tested the optical properties of the as-prepared T-AuNCs. It was demonstrated that there were broad absorption bands from 200 nm to 700 nm with two shoulder peaks at 242 nm and 294 nm derived from the phenoxide structure of tyrosine (Fig. 1B), which was consistent with those previously reported.47 As shown in Fig. 1C and Fig. A, it was demonstrated that T-AuNCs exhibited almost no fluorescence but only strong quenching ability to TAMRA due to the NSET effect. For the morphology characterization, a typical TEM image showed that the obtained T-AuNCs were well-dispersed with an average size around 1.76 nm (Fig. 1D). As expected, the T-AuNCs had a positive surface charge of 26.6 mV owing to the oligoarginine segments on the peptides (Fig. 1E), which endowed themselves with the potential of acting as a cationic carrier to deliver negative therapeutic cargos (drug loaded nucleic acids). The stability test of T-AuNCs revealed that the S/N ratio remains almost unchanged upon keeping at 4 °C for a month (Fig. 1F). All of these results suggested that the peptide-coated AuNCs were successfully obtained with expected properties such as stimuli-responsiveness, high NSET efficiency and positive charge, showing the potential of AuNCs to act as a high contrast reporter and a nanocarrier.
Specific cleavage ability and drug loading capacity of Dzs
To construct a theranostic nanoplatform, a segment of EGR-1 specific DNAzyme and a CG-rich region were ligated to serve as a part of the negative therapeutic unit. The former region was a typical 10–23 DNAzyme for gene therapy via specific cleavage of mRNA in the presence of Mg2+48 while another S4 complementary to the latter region was introduced to form partially double-stranded DNA with Dz4 for Dox loading, finally to be engineered as the whole negative therapeutic unit. 12% native polyacrylamide gel electrophoresis (PAGE) was used for determining the feasibility of DNAzyme specific cleavage of EGR-1 and strand assembly. As shown in Fig. S1A,† Mg2+ could trigger DNAzyme to completely cleave S1 but had no effect on S2 (no rArU site) within 2 h, while neither the single base mutant DNAzyme nor the typical 10–23 DNAzyme without the cofactor showed catalytic activity. Then we found that the assembly of three major nucleic acid strands (Dz, S5 (prolonged S4), and S2) and the specific cleavage of S2 of the assembly with the aid of Mg2+ were also feasible (Fig. S1B†). These results demonstrated that the Dzs endowed it with the specific cleavage ability of target mRNA. We then explored the drug loading capacity of Dzs. As illustrated in Fig. S2A,† the fluorescence intensity of Dox was gradually decreased with the increased concentration of Dzs, which saturated at a concentration of 370 nM (Dzs/Dox = 0.37), indicating the successful drug loading of Dzs.49Aggregation and disaggregation of T-AuNCs and Dzs
We further explored the potential of the T-AuNCs as a nanovector for gene delivery. The as obtained T-AuNCs were embedded with abundant positive charges because of the oligoarginine segments of the peptides at a physiological pH value (pH < PI). In general, cationic T-AuNCs can effectively absorb negative therapeutic cargos to form larger nanoparticles via electrostatic attraction. Hence, the TEM images were recorded and shown in Fig. 2B, which revealed that a large amount of nanoparticles was formed with a diameter of 17.8 ± 5.0 nm. We further confirmed the binding of Dzs to T-AuNCs by comparing the differences in the element composition with EDS spectra in Fig. 2C and S3.† Owing to the involvement of Dzs, the aggregates exhibit substantial surface P element, which absolutely comes from the phosphate of nucleotides. Gel electrophoresis was also performed, as illustrated in Fig. S4,† and there was an obvious trend where the free Dzs band gradually got weak with the increased concentration of T-AuNCs till the molar ratio of Dzs/T-AuNCs decreased to 0.004. These results demonstrated that Dzs had successfully been absorbed by the positive T-AuNCs.The primary design of AuNCs/Dzs nanoparticles is that the aggregated nanoparticles would be responsively activated and disaggregated for imaging and therapy by the overexpressed protease in the cancer cells. As a consequence, we further explored the activation and disaggregation feasibility of T-AuNCs/Dzs nanoparticles in the presence of a model enzyme. As shown in Fig. 2D, the TEM image showed that the particle size was decreased to 2.98 ± 0.5 nm after incubating T-AuNCs/Dzs with trypsin in PBS for 1 h, implying that Dzs had almost fallen off from T-AuNCs. Moreover, the NSET effect of AuNCs inspired us to verify the detachment of T-AuNCs and Dzs by monitoring the fluorescence intensity change of FAM labeled Dzs (Dzs-FAM) in the reaction system with or without the addition of trypsin. As expected, the T-AuNCs showed almost absolute quenching of Dzs-FAM upon the formation of aggregates, while the quenched Dzs-FAM could be greatly recovered with the aid of trypsin cleavage, accompanied by the fluorescence restoration of T-AuNCs (Fig. 2E). These data clearly suggested that the T-AuNCs/Dzs nanoparticles have the potential to be a protease-responsive nanoplatform for precise imaging and therapy of cancer cells by merely changing the sequence of substrate peptides.
Preparation of legumain-responsive AuNCs (L-AuNCs) and in vitro response of L-AuNCs
After verifying the T-AuNC based aggregation–disaggregation feasibility with the aid of the model enzyme, we synthesized legumain-specific AuNCs (L-AuNCs) and used them as a cationic carrier to absorb the negative therapeutic agent of Dzs. Herein, positive peptide PL1 (CCYAANRRRRRRRRGD), signal peptide PL2 (CCYGGGGAANRK(TRMRA)RRGD) and helper peptide P3 (CCYGGGGRGRKRGD) with a molar ratio of PL1/P3/PL2 at 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 30
30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
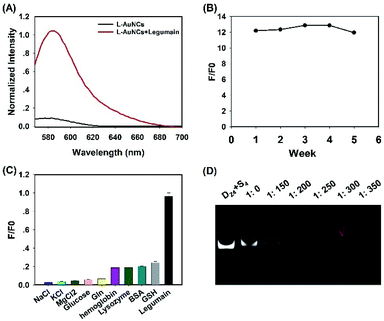
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 were utilized to prepare L-AuNCs. Compared to the ligands of T-AuNCs, a specific recognition site (AAN) of legumain was inserted into the positive peptide and signal peptide of L-AuNCs for the purpose of accelerating the detonation process. The fluorescence response of L-AuNCs to legumain was tested and the S/N ratio was calculated subsequently (Fig. 3A). As anticipated, the L-AuNCs retained the fluorescence off state due to the highly efficient NSET from TAMRA to the AuNC surface, whereas the addition of legumain significantly increased the fluorescence signal by ∼12-fold, suggesting that the legumain specifically hydrolyzed the substrate peptide of PL2 and markedly released TAMRA from L-AuNCs. The stability and selectivity of L-AuNCs were also examined, as shown in Fig. 3B and C, and the favorable results indicated that the L-AuNCs were definitely fit to be applied in the complex intracellular matrix.
1 were utilized to prepare L-AuNCs. Compared to the ligands of T-AuNCs, a specific recognition site (AAN) of legumain was inserted into the positive peptide and signal peptide of L-AuNCs for the purpose of accelerating the detonation process. The fluorescence response of L-AuNCs to legumain was tested and the S/N ratio was calculated subsequently (Fig. 3A). As anticipated, the L-AuNCs retained the fluorescence off state due to the highly efficient NSET from TAMRA to the AuNC surface, whereas the addition of legumain significantly increased the fluorescence signal by ∼12-fold, suggesting that the legumain specifically hydrolyzed the substrate peptide of PL2 and markedly released TAMRA from L-AuNCs. The stability and selectivity of L-AuNCs were also examined, as shown in Fig. 3B and C, and the favorable results indicated that the L-AuNCs were definitely fit to be applied in the complex intracellular matrix.
Aggregation and characterization of L-AuNCs/Dzs nanoparticles
Afterwards, the ability of L-AuNCs to bind DNA was displayed by PAGE. Fig. 3D shows that no free Dzs band could be observed when the molar ratio of Dzs/T-AuNCs decreased to 0.0033, which was chosen as the final ratio for the following experiments. Following the exploratory steps of T-AuNCs, the morphology and zeta potential of pure L-AuNCs and L-AuNCs/Dzs were also obtained (Fig. S5A–C†). The results showed that L-AuNCs were well-dispersed with an average size around 1.81 nm and positive surface charge of 20 mV, while L-AuNCs/Dzs exhibited a nearly neutral potential. The TEM image results of legumain responsive disaggregation of L-AuNCs/Dzs nanoparticles were tested as Fig. S5D,† which was consistent with the results the model enzyme.We then investigated the biological stability of L-AuNCs/Dzs nanoparticles by treating them with FBS and DNase I. PAGE analysis (Fig. S6A†) revealed that the L-AuNCs/Dzs nanoparticles treated with 10% FBS still maintained their integrity without releasing Dzs for 40 h. Besides, DNase I (1.5 U mL−1) was utilized to treat free Dz4-Bhq1-S4-FAM and L-AuNCs/Dzs-FAM, respectively. By monitoring the fluorescence recovery of FAM digested from each of them, it was found that L-AuNCs/Dzs-FAM nanoparticles showed negligible fluorescence recovery compared to free Dzs after incubating with DNase I for 1.4 h (1.5 U mL−1) (Fig. S6B†). To simulate the biological environment in vivo, 100% FBS was used to test the stability of these nanoparticles by fluorescence analysis (Fig. S6C†). The results demonstrated the excellent stability of the formed L-AuNCs/Dzs nanoparticles.
Targeted cellular uptake and in situ activation of L-AuNCs/Dzs for specific cell imaging
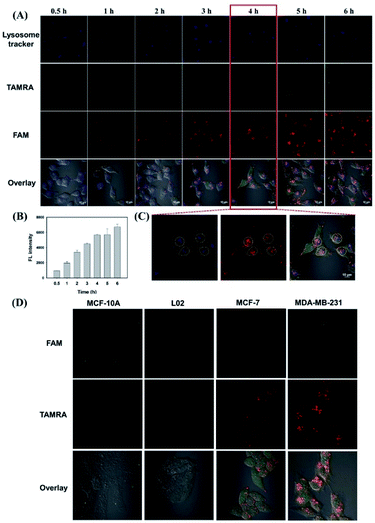
In order to investigate the targeted cellular uptake and in situ activation of L-AuNCs/Dzs for specific cell imaging, Dzs absorbed by the L-AuNCs was labeled with FAM. As we known, TAMRA of L-AuNCs and Dzs-FAM was absolutely quenched by L-AuNCs in the L-AuNCs/Dzs-FAM nanoparticles. First, we monitored the time-dependent cellular internalization of L-AuNCs/Dzs-FAM. The confocal laser scanning microscopy (CLSM) images of MDA-MB-231 cells were recorded after various incubation time-intervals. As shown in Fig. 4A and B, only weak fluorescence could be observed on the cell membranes after 1 h, which might result from the traces of legumain's contribution distributed on the cell surface. Then, the fluorescence intensity of TAMRA and Dzs-FAM in the cells was gradually enhanced with prolonged incubation time till 4 h, indicating the large cleavage of TAMRA labeled substrate peptides by overexpressed legumain and the complete release of Dzs-FAM. As shown in Fig. 4C, by analyzing the co-localization fluorescence, the signal activated by legumain was mainly observed in the lysosome at first and then distributed around the cell, which was consistent with the literature.34 Moreover, as depicted in Fig. S7,† L-AuNCs also served as a control probe to compare the cellular uptake efficiency. The results showed that the internalization efficiency of aggregated AuNCs was significantly enhanced compared to that of the free AuNCs, and thus largely enhanced the imaging contrast. To further demonstrate the specificity of legumain-activated cell imaging, the fluorescence images were tested by CLSM in breast cancer cells (MCF-7 and MDA-MB-231) and normal cells (MCF-10A and L02). As shown in Fig. 4D, the normal cells showed negligible fluorescence recovery after incubation with L-AuNCs/Dzs-FAM for 4 h, while the cancer cells MDA-MB-231 exhibited stronger fluorescence intensity than MCF-7 and much higher than normal cells for their different legumain concentrations and integrin αvβ3 expression levels.34 The control probe of T-AuNCs/Dzs was also utilized to test the specificity of nanoparticle disaggregation, as shown in Fig. S7.† Due to the lack of specific substrates of legumain, the MDA-MB-231 cells treated T-AuNCs/Dzs showed nearly no fluorescence recovery. The above results demonstrated that the aggregation–disaggregation behavior of L-AuNCs/Dzs triggered by legumain significantly improved the imaging contrast due to the activable mechanism and enhanced cellular uptake of AuNCs. In addition, these endogenous stimuli of legumain also specifically initiated the concentrated release of Dzs from the nanoparticles.Investigation of Dox loading and release
To verify the Dox loading capacity of L-AuNCs/Dzs nanoparticles, UV-vis spectra of L-AuNCs (black line), Dzs (red line), Dox (green line), AuNCs + Dzs (yellow line), Dox + Dzs (blue line) and L-AuNCs + Dox + Dzs (purple line) were recorded, as shown in Fig. S2B.† The L-AuNCs showed broad absorption bands from 200 nm to 700 nm with two shoulder peaks at 242 nm and 294 nm, which were consistent with those of T-AuNCs. Dzs exhibited a characteristic peak at 260 nm for purine and pyrimidine bases of DNA; however the peak was apparently weakened and the absorption value in the range of 200 nm to 700 nm showed a general upward after the introduction of L-AuNCs, indicating the aggregation of L-AuNCs and Dzs. Dox showed a typical absorption centered at 480 nm, whereas the peak disappeared upon the addition of Dzs, suggesting a perfect intercalation of Dox into double-stranded GC pairs. Finally, the mixture of L-AuNCs, Dox and Dzs demonstrated stronger absorption signals between 200 nm and 700 nm with the absence of peaks at 480 nm and 260 nm corresponding to free Dox and Dzs respectively, indicating a complete loading of Dox by L-AuNCs/Dzs nanoparticles with a molar ratio of Dzs/Dox/L-AuNCs at 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2.7
2.7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 300, which was selected as the final ratio for the following experiments. To study the legumain-triggered intracellular drug release of L-AuNCs/Dzs-Dox, the fluorescence change of Dox in the MDA-MB-231 cells were monitored by CLSM. After a 4 hour incubation of MDA-MB-231 cells and L-AuNCs/Dzs-Dox, an increasing red fluorescence was detected in the cell cytoplasm and finally in the nucleus (Fig. S8†).
300, which was selected as the final ratio for the following experiments. To study the legumain-triggered intracellular drug release of L-AuNCs/Dzs-Dox, the fluorescence change of Dox in the MDA-MB-231 cells were monitored by CLSM. After a 4 hour incubation of MDA-MB-231 cells and L-AuNCs/Dzs-Dox, an increasing red fluorescence was detected in the cell cytoplasm and finally in the nucleus (Fig. S8†).
Cell cytotoxicity evaluation of L-AuNCs/Dzs-Dox nanoparticles
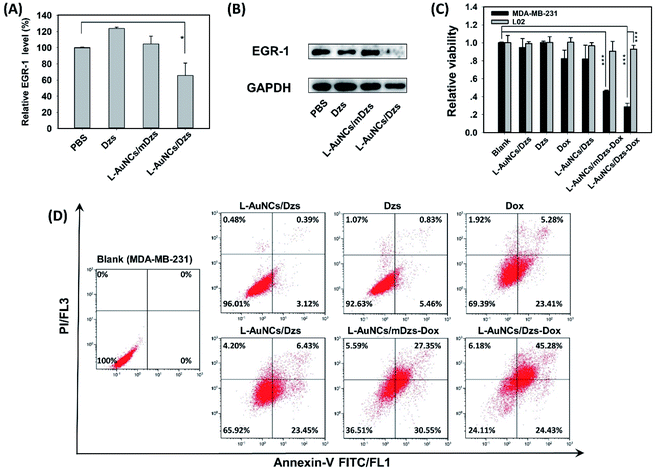
For the purpose of evaluating the cell killing ability of L-AuNCs/Dzs-Dox nanoparticles, the biocompatibility of the delivery system was first tested by using L-AuNCs/mDzs, in which the mutant DNAzyme was introduced to rule out the possibility of gene therapy. In the range of 0–300 nM, L-AuNCs/mDzs exhibited high biocompatibility (Fig. S9†). Afterwards, we further studied the mono-therapy of gene and drug, respectively, in this range. Once L-AuNCs/Dzs was internalized into the cells, the disaggregation triggered by over-expressed legumain could also initiate the cargo release of Dzs. To investigate their cell cytotoxicity, various concentrations of L-AuNCs/Dzs were used to treat MDA-MB-231 cells, as shown in Fig. S10A,† whose cell viability showed a concentration-dependent feature. To further explore the cytotoxicity mechanism, a mutant DNAzyme (mDz) was introduced as the control by substituting one nucleotide of the DNAzyme catalytic core with no effect on its binding affinity. The results showed that the expression level of EGR-1 was apparently down-regulated merely in L-AuNCs/Dzs-treated MDA-MB-231 cells, as shown by qRT-PCR (Fig. 5A) and western blot analysis (Fig. 5B). These data indicated that the detonation strategy based on legumain-triggered disaggregation of AuNCs/Dzs was an effective strategy for the DNAzyme-mediated suppression of EGR-1 of cancer cells.Subsequently, L-AuNCs/mDzs nanoparticles were utilized to load Dox for evaluating chemotherapeutic efficacy. As displayed in Fig. S10B,† the cell viability of MDA-MB-231 cells showed a downward trend in a “fast followed by slow” manner with the increased concentration of L-AuNCs/mDzs-Dox.
Finally, the combinational gene-chemo cancer therapeutic efficacy was studied against MDA-MB-231 and L02 cell lines at a concentration of 200 nM by MTS assay (Fig. 5C). Compared to negligible therapeutic efficacy of free Dzs (0.00%), mono-treatment of L-AuNCs/Dzs (18.2 ± 1.5%) L-AuNCs/mDzs-Dox and AuNCs/mDzs-Dox (54.1 ± 0.1%), gene-chemo combinational treatment using L-AuNCs/Dzs-Dox (71.4 ± 0.4%) obviously exhibited expressively higher therapeutic efficiency to MDA-MB-231 cells in 48 h. In contrast, the distribution of L02 cells treated with varying samples showed a slight distinction due to the low-level of pivotal integrin and legumain. Free Dox presented a similar toxicity to both cell lines due to the lack of selectivity. To further assess the ability of L-AuNCs/Dzs-Dox to trigger tumor cell apoptosis, as shown in Fig. 5D, annexin V-FITC/PI assay by flow cytometry was adopted to analyze the cell viability. As expected, the cell apoptosis percentages of each group were in good accordance and showed an acceptable difference with MTS assays because of distinct postprocessing and instruments. Taken together, these results revealed that AuNC-based stimulus-responsive nanoplatforms are highly efficient for chemo/gene therapy of cancer cells.
In vivo specific response to L-AuNCs/Dzs-Cy7
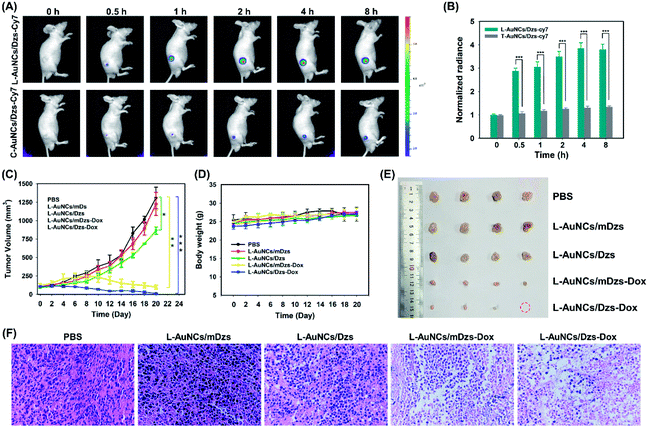
To further explore the feasibility of our L-AuNCs/Dzs system for a more challenging in vivo imaging, the Cy7 labelled L-AuNCs/Dzs system was injected into MDA-MB-231 tumor-bearing mice. The fluorescence of Cy7 of the L-AuNCs/Dzs system was enhanced gradually and reached the maximum value at 2 h and sustained for 8 h at the tumor site after injection (Fig. 6A). To identify the specific response of tumor to the L-AuNCs/Dzs system, T-AuNCs/Dzs, without the legumain specific substrate, was set as another group and the average fluorescence intensity of both groups (L-AuNCs/Dzs-Cy7 and T-AuNCs/Dzs-Cy7) was calculated. As presented in Fig. 6B, the L-AuNCs/Dzs-treated mice displayed 2.7-fold higher intratumoral fluorescence than that treated with the T-AuNCs/Dzs system. Additionally, the fluorescence intensity at the tumor site was found to be about 4-fold higher than that of the normal site in the group of L-AuNCs/Dzs. Therefore, accurate tumor-imaging was achieved in living mice by using the L-AuNCs/Dzs platform.In vivo antitumor efficacy of L-AuNCs/Dzs-Dox
To evaluate the antitumor activities of L-AuNCs/Dzs-Dox, five treatment groups of PBS, L-AuNCs/mDzs, L-AuNCs/Dzs, L-AuNCs/mDzs-Dox and L-AuNCs/Dzs-Dox were intratumorally injected into MDA-MB-231 tumor-bearing mice with an average tumor volume of about 100 mm3. As depicted by the tumor growth curves in Fig. 6C, compared with gene therapy or chemotherapy alone, the combined therapy could significantly suppress tumor growth, demonstrating an improved antitumor treatment. Meanwhile, the safety of this nanocarrier was demonstrated by the general slightly increased body weight observed for most mice and the tumor growth variation of group L-AuNCs/mDzs (Fig. 6D). The therapeutic effect was further illustrated by the photographs of tumors harvested at day 20 post-injection (Fig. 6E), indicating the greatest potency of L-AuNCs/Dzs-Dox for tumor growth inhibition. Hematoxylin–Eosin (H&E) staining revealed the severe structural deformation and more apoptotic cells in the L-AuNCs/Dzs-Dox group, demonstrating the higher antitumor capability of the present chemo/gene therapy (Fig. 6F). These results demonstrated that the multifunctional AuNC-based stimulus-responsive nanoplatform has the potential to be applied for clinical applications as a therapeutic platform to treat cancer in vivo.Conclusions
In summary, we have successfully constructed a multifunctional gold nanocluster-based bomb as a potent and precise co-delivery nanosystem for stimulus-responsive imaging and efficient combination cancer therapy. By utilizing the AuNCs as building blocks of both nanocarrier and reporter, this fabricated nanocluster-bomb demonstrated the following advantages: (1) the endogenous stimuli of legumain were introduced to hydrolyze the bioligand of AuNCs, endowing this nanocluster-bomb system with safe and intelligent performance; (2) based on the highly NSET effect of multifunctional AuNCs and their aggregation–disaggregation mechanism, the targeted cellular uptake efficiency was obviously improved and led to contrast-enhanced cancer imaging; (3) the mechanism of AuNC-based aggregation–disaggregation triggered by legumain not only enriched the payload species, but also realized on-demand and concentrated cargo release. Collectively, the in vitro and in vivo studies revealed that the detonation strategy based on AuNCs/Dzs-Dox readily achieved contrast-enhanced cancer imaging and efficient gene/chemo combination therapy. To our knowledge, this is the first time to report the application of relying on AuNCs' NSET-dependent quenching effect and detonation mechanism for stimulus-responsive cancer imaging and gene/chemo therapy. By replacing the peptide substrate or DNA sequence, the multifunctional AuNCs/Dzs can serve as a versatile platform and can be widely explored for probing complex enzymatic profiles and gene targets, thus achieving precise and efficient cancer theranostics.Experimental
Chemicals and materials
All the peptides used in the experiments were chemically synthesized and purified by a solid phase method from Guoping Pharmaceutical (Hefei, China). The basic information of the synthesized peptides is given in Table S1.† All the oligonucleotides used in this study were synthesized by Sangon Biotechnology (Shanghai, China). Before use, all the oligonucleotides were purified with high performance liquid chromatography (HPLC) and further identified using mass spectrometry. The sequences of oligonucleotides are listed in Table S4.†Hydrogen tetrachloroaurate tetrahydrate (HAuCl4·4H2O, >99%), sodium hydroxide (NaOH), sodium chloride (NaCl), magnesium chloride (MgCl2), and potassium chloride (KCl) were obtained from Sinopharm Chemical Reagent Co. Ltd. (Shanghai, China). Trypsin was purchased from Sigma-Aldrich (St. Louis, MO). Recombinant human legumain protein (Pro form, 10 μg, specific activity >250 pmol min−1 μg−1) was purchased from R & D Systems (Minneapolis, MN). Tris-(2-carboxyethyl) phosphine (TCEP) was purchased from Meilun Biotech (Dalian, China). All other reagents and solvents were of analytical grade and used without further purification. All solutions were prepared using deionized water, obtained from a Milli-Q ultrapure water system (>18.2 MΩ cm resistivity, Billerica, MA).
Synthesis of AuNCs
Synthesis and purification of peptide-capped AuNCs was carried out following the published procedures with appropriate modifications.31 Trypsin was used as a model enzyme in this work for the pretest. We varied the ratio of the positive peptide (PT1), signal peptide (PT2) and helper peptide (P3) in the AuNC synthesis to incorporate functional groups on the AuNCs surface. Briefly, freshly prepared aqueous solution of HAuCl4 (16 μL, 25 mM) was slowly added to 376 μL of aqueous solution of peptides (total final concentration: 1 mM) in an Eppendorf tube with vigorous stirring. Then around 8 μL of 0.5 M NaOH was slowly added to adjust the pH of the solution to about 10. Then the sample was stored and sealed for 13 h at 37 °C undisturbed in the dark to produce the trypsin-responsive AuNCs (T-AuNCs). The obtained T-AuNC solution (400 μL) was concentrated and washed with ultrapure water five times through ultrafiltration using a centrifugal filter unit (Amicon Ultra-0.5 mL, MWCO 10 kDa) so as to remove the unbound peptide and redundant HAuCl4. The concentrated AuNCs were resuspended in deionized water and diluted to 400 μL (1 mM based on the AuNC particle concentration). The synthesis procedure and final optimal peptide ratio were also used to prepare the L-AuNCs (legumain responsive AuNCs) with corresponding functional peptides (PL1, PL2 and P3).NSET investigation of T-AuNCs
T-AuNCs (1 μL, 1 mM) and trypsin (1 μL, 500 U) were introduced into PBS (pH = 7.4, 10 mM) with a total volume of 200 μL. By recording and calculating the fluorescence intensity of T-AuNCs (Noise) and trypsin treated T-AuNCs (Signal) on a PTI QuantaMaster™ 4 (Birmingham, UK) fluorescence spectrophotometer, NSET efficiency (S/N) could be obtained.Gel electrophoresis analysis
12% polyacrylamide gel (PAGE) was employed to test the cleavage activity of 10–23 DNAzyme (DZ) and functional nucleic acid (Dzs) assembly feasibility (Dz4-S4). All samples were prepared in Tris–HCl buffer (pH 7.5, 150 mM NaCl) with a final concentration of nucleic acids at 1 μM and Mg2+ at 15 mM. The electrophoresis analysis was conducted at 100 V for 2 h and recorded by using an Azure C600 Imaging Biosystems (California, USA). Electrophoretic mobility shift assay with 8% PAGE was used to evaluate the aggregation of AuNCs and Dzs.Preparation and characterization of AuNCs/Dzs nanoparticles
To simulate the physiological environment, phosphate buffered solution (PBS, pH = 7.4, 10 mM) was used to assemble the positive AuNCs and negative nucleic acids. For morphology characterization, the AuNCs/Dzs was aggregated in Tris–HCl (pH = 7.4) and examined by transmission electron microscopy (TEM, Tecnai G2 F20 S-TWIN). The surface charge of the prepared nanomaterials was determined using dynamic light scattering (NanoZS, Malvern).Specific legumain responsiveness of L-AuNCs
To determine the specific legumain-responsiveness of L-AuNCs, a hydrolysis assay was used for the evaluation. In a typical activation procedure, pro-legumain protease was diluted in the activation buffer (pH = 4.0, 50 mM CH3CO2Na, and 100 mM NaCl) and incubated at 37 °C for 2 h. Then, the activated legumain was diluted to 10 μg mL−1 and reacted with 5 mM L-AuNCs in the assay buffer (pH = 5.0, 50 mM MES, and 250 mM NaCl) at a total volume of 200 μL. Under the same conditions, various reactants including Na+ (150 mM), K+ (5 mM), Mg2+ (15 mM), glucose (10 mM), glutamine (1 mM), hemoglobin (200 nM), lysozyme (200 nM), BSA (1 mM) and GSH (5 mM) were used to test the selectivity of L-AuNCs.Studies of stability and digestion resistant ability of L-AuNCs/Dzs
To validate the stability and anti-inference ability of the L-AuNC-based nanocarrier system, 8% PAGE was performed after L-AuNCs/Dzs was incubated in 10% FBS at 37 °C for different times. Digestion resistant ability of L-AuNCs/Dzs was also investigated by comparing the fluorescence spectra of Dz4-Bhq1-S4-FAM and L-AuNCs/Dz-S4-FAM after being treated with DNase I. The fluorescence intensity of L-AuNCs/Dzs-FAM, Dzs-FAM, L-AuNCs and L-AuNCs/Dzs-FAM + leguamain was monitored after being incubated with 100% FBS for 120 min.Cell culture
MDA-MB-231 cells (human breast carcinoma) and MCF-7 cells (human breast carcinoma) were obtained from the cell bank of Central Laboratory at Xiangya Hospital (Changsha, China), and cultured in Dulbecco's Modified Eagle Medium (DMEM) supplemented with 10% (v/v) fetal bovine serum (obtained from Invitrogen) and 100 IU mL−1 penicillin–streptomycin. L02 cells (normal human hepatic cell line) were cultured in 1640 medium and MCF-10A cells (normal human mammary epithelial cell line) were grown in DMEM/F12 medium (Procell Life Science & Technology Co., Ltd.) supplemented with 5% HS, 20 ng mL−1 EGF, 10 μg mL−1 insulin, 0.5 μg mL−1 hydrocortisone, 1% NEAA, and 1% P/S solution. All cells were maintained in a 5% CO2 humidified incubator at 37 °C.Confocal laser scanning microscopy (CLSM) experiments
All cells were seeded on 35 mm glass bottom dishes and maintained for 36 h. Afterwards, the medium in the dishes was removed and cells were washed twice with phosphate buffered saline (PBS, pH 7.40, Ca2+ and Mg2+ free). For the time-dependent cellular uptake of L-AuNCs/Dzs and co-localization of lysosomes, L-AuNCs/Dzs-FAM (15 μM![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50 nM) resuspended in PBS (5% FBS) was used to incubate with MDA-MB-231 cells for imaging at different points in time. The cells were introduced with a lysosome tracker for 10 min and washed 3 times with PBS before imaging. For the specific cell imaging investigation, L-AuNCs/Dzs-FAM (22.5 μM
50 nM) resuspended in PBS (5% FBS) was used to incubate with MDA-MB-231 cells for imaging at different points in time. The cells were introduced with a lysosome tracker for 10 min and washed 3 times with PBS before imaging. For the specific cell imaging investigation, L-AuNCs/Dzs-FAM (22.5 μM![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 75 nM) resuspended in PBS (5% FBS) was used to treat the four kinds of cells separately for 4 h. The lasers and emission filters were described as follows. Lysosome tracker/hoechst 33
75 nM) resuspended in PBS (5% FBS) was used to treat the four kinds of cells separately for 4 h. The lasers and emission filters were described as follows. Lysosome tracker/hoechst 33![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 342: Ex = 405 nm, Em = 425–475 nm, FAM: Ex = 488 nm, Em = 500–550 nm, TAMRA: Ex = 561 nm, and Em = 570–620 nm.
342: Ex = 405 nm, Em = 425–475 nm, FAM: Ex = 488 nm, Em = 500–550 nm, TAMRA: Ex = 561 nm, and Em = 570–620 nm.
Dox loading and release investigation
Dox solution (1 μM) was incubated with various concentrations of Dzs at room temperature for 30 min. Then the drug loading capacity of Dzs was quantified by recording the fluorescence of Dox with a PTI QuantaMaster™ 4 fluorescence spectrophotometer. A UV-vis spectrophotometer was used for the quantification of drug loading capacity of L-AuNCs/Dzs and for the purpose of ensuring the absolute absorption of Dzs by L-AuNCs simultaneously. To determine the legumain triggered Dox release behavior of MDA-MB-231 cells, 2 h, 3 h, 4 h and 5 h were set to monitor the fluorescence of Dox, respectively. Dox: Ex = 488 nm and Em = 570–620 nm.Cytotoxicity assays
The cell viability was determined using Cell Titer 96 cell proliferation assay. Briefly, cells (4 × 104 cells per well) were treated with different probes suspended in PBS (37 °C, 5% CO2) for 4 h, respectively. Then, the solution was removed and replenished with fresh medium (10% FBS) for further cell growth (48 h). Subsequently, the MTS reagent (20 μL) and fresh culture medium (100 μL) were added to each well and incubated for 1–2 h at 37 °C after removal of the old medium. The absorbance (490 nm) was recorded using a Bio-RAD (Benchmark, USA). Cell viability was calculated as described by the manufacturer.Quantitative reverse polymerase chain reaction (qRT-PCR) analysis of EGR-1
To quantify the relative expression of EGR-1 in MDA-MB-231 cells, the cells with different treatments were prepared for qRT-PCR. Total cellular RNA was extracted from the cells using TRIzol reagent following the manufacturer's instructions. Afterward, the products were stored at −80 °C for the qRT-PCR analysis (Sangon Co. Ltd., Shanghai, China). The sequences of PCR primers are listed as follows.EGR-1 forward primer: 5′-TGACCGCAGAGTCTTTTCCT-3′
EGR-1 reverse primer: 5′-TGGGTTGGTCATGCTCACTA-3′
GAPDH forward primer: 5′-TGGGTGTGAACCATGAGAAGT-3′
GAPDH reverse primer: 5′-TGAGTCCTTCCACGATACCAA-3′
Western blot analysis of EGR-1
MDA-MB-231 cells were treated with PBS, Dzs, L-AuNCs/mDzs, and L-AuNCs/Dzs for 4 h, respectively. Afterward, the solution was removed and replenished with fresh medium (10% FBS) for further cell growth (48 h). MDA-MB-231 cells were washed with PBS (10 mM, pH = 7.4) and then 100 μL of RIPA lysis buffer supplemented with a protease inhibitor mixture was added and kept on ice for 2 h, and centrifuged at 4 °C. The lysates were placed on ice for 30 min and then centrifuged for 15 min at the speed of 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm to remove cell debris. Subsequently, the protein concentration was determined by BCA (bicinchoninic acid protein assay). Total cellular proteins were resolved on a 12% SDS-PAGE, transferred to a PVDF membrane, and blocked with 5% skimmed milk in PBS. Then, the rabbit polyclonal antibody to EGR-1 (Sangon Biotech) was used as the primary antibody and incubated on the membrane at 4 °C for 12 h. Subsequently, the membrane was washed with PBS containing Tween-20 (0.1%) 3 times, followed by incubation with horseradish peroxidase-conjugated secondary antibody diluted to 1
000 rpm to remove cell debris. Subsequently, the protein concentration was determined by BCA (bicinchoninic acid protein assay). Total cellular proteins were resolved on a 12% SDS-PAGE, transferred to a PVDF membrane, and blocked with 5% skimmed milk in PBS. Then, the rabbit polyclonal antibody to EGR-1 (Sangon Biotech) was used as the primary antibody and incubated on the membrane at 4 °C for 12 h. Subsequently, the membrane was washed with PBS containing Tween-20 (0.1%) 3 times, followed by incubation with horseradish peroxidase-conjugated secondary antibody diluted to 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2000. Finally, the membrane was washed 3 times and protein bands were visualized with the SuperSignal West Pico PLUS chemiluminescent substrate (Thermo Fisher Scientific). The analysis of reference protein GAPDH was the same as above.
2000. Finally, the membrane was washed 3 times and protein bands were visualized with the SuperSignal West Pico PLUS chemiluminescent substrate (Thermo Fisher Scientific). The analysis of reference protein GAPDH was the same as above.
Apoptosis assays
Cell apoptosis was tested by the annexin V-FITC/PI assay. Briefly, the MDA-MB-231 cells were seeded in 1 mL of media in each well of a 12-well plate for 12 h, respectively. After removing the old solution, the cells were then treated with different samples and incubated for another 48 h. Next, the cells were harvested and treated with annexin V-FITC/PI according to the manufacturer's instructions and then examined by flow cytometry.In vivo fluorescence imaging
Male BALB/c mice were obtained from Hunan SJA Laboratory Animal Co. Ltd. They were 4–6 weeks old at the start of each experiment and allowed to acclimate in the laboratory for 1 week. All animal operations were in accord with the institutional animal use and care regulations, according to protocol No. SCXK (Xiang) 2013-0001, approved by the Laboratory Animal Center of Hunan Province. 2 × 106 MDA-MB-231 cells (100 μL) were slowly injected into the thigh subcutaneous region of BALb/c nude mice. Tumors were then allowed to grow for 4–5 weeks to reach about 200 mm3. Before imaging, BALB/c nude mice were anesthetized to be motionless with anesthetics. The nude mice were then intratumorally injected with 30 μL L-AuNCs/Dzs-Cy7 and T-AuNCs/Dzs-Cy7 (the concentration of Dzs was 1.5 μM) and time-lapse fluorescence imaging was carried out by using an IVIS Lumina II in vivo imaging system (Caliper Life Sicence, USA).In vivo anti-tumor efficacy evaluation
The anti-tumor efficacy of L-AuNCs/Dzs-Dox was evaluated using MDA-MB-231 tumor-bearing BALB/c nude mice. When the tumors exhibited a volume of ∼100 mm3, these mice were randomly divided into five groups (4 mice per sample) and injected with PBS, L-AuNCs/mDzs, L-AuNCs/Dzs, L-AuNCs/mDzs-Dox, and L-and AuNCs/Dzs-Dox, respectively. The dosage of Dox in the groups was 0.5 mg kg−1. Drugs were intratumorally injected every other day, and the body weight and tumor volume of each mouse were recorded with calipers every other day. The tumor volume was calculated using the following equation:| Tumor volume = Length × Width 2/2 |
On the 20th day, all mice were sacrificed and the collected tumors were used for H&E staining. The slices were imaged under a fluorescent inverted microscope.
Statistical analysis
All experiments were conducted at least in triplicate. All data were expressed as the mean ± standard deviation and analyzed using one-way analysis of variance (ANOVA) following Tukey's post-test. Differences with *P < 0.05, **P < 0.01, and ***P < 0.001 were noted as statistically significant.Data availability
All experimental procedures, characterization data supporting this article have been described in the manuscript and uploaded as an ESI.† Original data shown in this paper are available from the corresponding author upon reasonable request.Author contributions
X. X. He and K. M. Wang conceived and supervised the project. H. H. Sun and W. J. Ma designed this experiments, performed the experiments and analyzed the data. S. D. Duan performed the western blot experiment. J. Huang, R. C. Jia, H. Cheng and B. Chen provided experimental advice. All authors contributed to ideation, discussion of results, and writing of the paper.Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
This work was supported by the Natural Science Foundation of China (21775036, 21675046, 21735002, 21521063, 21806186 and 21874035).Notes and references
- X. H. Wang, X. Y. Wang, S. X. Jin, M. Nafees and Z. J. Guo, Chem. Rev., 2019, 119, 1138–1192 CrossRef CAS PubMed.
- Y. C. Wang, M. S. Shim, N. S. Levinson, H. W. Sung and Y. N. Xia, Adv. Funct. Mater., 2014, 24, 4206–4220 CrossRef CAS PubMed.
- S. Ahmadi, N. Rabiee, M. Bagherzadeh, F. Elmi, Y. Fatahi, F. Farjadian, N. Baheiraei, B. Nasseri, M. Rabiee, N. T. Dastjerd, A. Valibeik, M. Karimi and M. B. Hamblin, Nano Today, 2020, 34, 100914 CrossRef CAS PubMed.
- Z. H. Qing, J. Y. Xu, J. Zheng, L. He, Z. Zou, S. Yang, W. H. Tan and R. H. Yang, Angew. Chem., Int. Ed., 2019, 58, 11574–11585 CrossRef CAS PubMed.
- M. Peng, Theranostics, 2020, 10, 4557–4588 CrossRef PubMed.
- A. P. Blum, J. K. Kammeyer, A. M. Rush, C. E. Callmann and M. E. Hahn, J. Am. Chem. Soc., 2020, 137, 2140–2154 CrossRef PubMed.
- Y. F. Wang, W. Du, T. Zhang, Y. Zhu, Y. H. Ni, C. C. Wang, R. F. M. Sierra, L. W. Zou, L. S. Wang and G. L. Liang, ACS Nano, 2020, 14, 9585–9593 CrossRef CAS PubMed.
- Z. X. Jiang, B. Feng, J. Xu, T. P. Qing, P. Zhang and Z. H. Qing, Biosens. Bioelectron., 2020, 166, 112471 CrossRef CAS PubMed.
- H. M. Wang, Y. Q. Chen, H. Wang, X. Q. Liu, X. Zhou and F. A. Wang, Angew. Chem., Int. Ed., 2019, 58, 7380–7384 CrossRef CAS PubMed.
- Y. J. Liu, Z. Yan, F. Chan, A. Lee, J. L. Yin, R. Chung, J. B. Park, H. Rizos, W. Tao, M. M. Zheng, O. C. Farokhzad and B. Y. Shi, Nano Lett., 2020, 20, 1637–1646 CrossRef CAS PubMed.
- Q. Lei, S. B. Wang, J. J. Hu, Y. X. Lin, C. H. Zhu, L. Rong and X. Z. Zhang, ACS Nano, 2017, 11, 7201–7214 CrossRef CAS PubMed.
- W. Hai, P. Agarwal, S. T. Zhao, J. H. Yu, X. B. Lu and X. M. He, Adv. Mater., 2016, 28, 347–355 CrossRef PubMed.
- L. J. Zhang, Y. Q. Qi, H. Min, C. Ni, F. Wang, B. Wang, H. Qin, Y. L. Zhang, G. N. Liu, Y. Qin, X. X. Duan, F. Li, X. X. Han, N. Tao, L. R. Zhang, Z. H. Qin, Y. Zhao and G. J. Nie, ACS Nano, 2019, 13, 10852 CrossRef CAS PubMed.
- J. C. Yang, Y. Shang, Y. H. Li, Y. Cui and X. B. Yin, Chem. Sci., 2018, 9, 7210–7217 RSC.
- L. Yang, H. Sun, Y. Liu, W. J. Hou, Y. Yang, R. Cai, C. Cui, P. H. Zhang, X. S. Pan, X. W. Li and L. Li, Angew. Chem., Int. Ed., 2018, 57, 17048–17052 CrossRef CAS PubMed.
- D. Huo, S. Liu, C. Zhang, J. He, Z. Y. Zhou, H. Zhang and Y. Hu, ACS Nano, 2017, 11, 10159–10174 CrossRef CAS PubMed.
- Y. F. Lei, L. X. Tang, Y. Z. Y. Xie, Y. L. Xianyu, L. M. Zhang, P. Wang, Y. Hamada, K. Jiang, W. F. Zheng and X. Y. Jiang, Nat. Commun., 2017, 8, 15130 CrossRef PubMed.
- K. J. Chen, E. Y. Chaung, S. P. Wey, K. J. Lin, F. Cheng, C. C. Lin, H. L. Liu, H. W. Tseng, C. P. Liu, M. C. Wei, C. M. Liu and H. W. Sung, ACS Nano, 2014, 8, 5105–5115 CrossRef CAS PubMed.
- C. N. Loynachan, A. P. Soleimany, J. S. Dudani, Y. Y. Lin, A. Najer, A. Bekdemir and Q. Chen, Nat. Nanotechnol., 2019, 14, 883–890 CrossRef CAS PubMed.
- U. Kauscher, M. N. Holm, M. Björnmalm and M. M. Stevens, Adv. Drug Delivery Rev., 2019, 138, 259–275 CrossRef CAS PubMed.
- B. H. Zhu, L. Y. Wang, J. B. Huang, X. Xiang, Y. J. Tang, C. Cheng, Y. Feng, L. Ma and L. Qiu, J. Mater. Chem. B, 2019, 7, 4581–4591 RSC.
- C. Y. Liu, K. K. Ewert, N. Wang, Y. L. Li, C. R. Safinya and W. H. Qiao, Biomaterials, 2019, 221, 119412 CrossRef CAS PubMed.
- Y. Z. Y. Xie, Y. L. Xianyu, N. X. Wang, Z. Y. Yan, L. Yong, Z. Zhu, N. S. Hatzakis and X. Y. Jiang, Adv. Funct. Mater., 2018, 28, 1702026.1–1702026.7 Search PubMed.
- Y. Li, P. Li, R. Zhu, C. Luo, H. Li, S. F. Hu, Z. Nie, Y. Huang and S. Z. Yao, Anal. Chem., 2016, 88, 11184–11192 CrossRef CAS PubMed.
- T. T. Jia, G. Yang, S. J. Mo, Z. Y. Wang, B. J. Li, W. Ma, Y. X. Guo, X. Y. Chen, X. L. Zhao, J. Q. Liu and S. Q. Zang, ACS Omega, 2020, 5, 22702 CrossRef PubMed.
- H. L. Liu, G. S. Hong, Z. T. Luo, J. C. Chen, J. L. Chang, M. Gong, H. He, J. Yang, X. Yuan, L. L. Li, X. Y. Mu, J. Y. Wang, W. B. Mi, J. Luo, J. P. Xie and X. D. Zhang, Adv. Mater., 2019, 31, 1901015 CrossRef CAS PubMed.
- L. N. Yang, H. L. Wang, D. Y. Li, L. Li, X. F. Lou and H. L. Liu, Chem. Mater., 2018, 30(15), 5507–5515 CrossRef CAS.
- X. Yin, B. Yang, B. B. Chen, M. He and B. Hu, Anal. Chem., 2019, 91, 10596–10603 CrossRef CAS PubMed.
- Y. L. Wang, C. Xu, J. Zhai, F. P. Gao, R. Liu, L. Gao, Y. L. Zhao, Z. F. Chai and X. Y. Gao, Anal. Chem., 2015, 87, 343–345 CrossRef CAS PubMed.
- N. Goswami, Z. T. Luo, X. Yuan, D. T. Leong and J. P. Xie, Mater. Horiz., 2017, 4, 817 RSC.
- C. S. Yun, A. Javier, T. Jennings, M. Fisher, S. Hira, S. Peterson, B. Hopkins, N. O. Reich and G. F. Strouse, J. Am. Chem. Soc., 2005, 127, 3115–3119 CrossRef CAS PubMed.
- T. L. Jennings, M. P. Singh and G. F. Strouse, J. Am. Chem. Soc., 2006, 128, 5462–5467 CrossRef CAS PubMed.
- Y. Z. Lu and W. Chen, Chem. Soc. Rev., 2012, 41, 3594–3623 RSC.
- T. Chen, R. R. Nasaruddin, J. Xie and N. Yan, Coord. Chem. Rev., 2018, 368, 60–79 CrossRef.
- G. H. Liang, X. D. Jin, S. X. Zhang and D. Xing, Biomaterials, 2017, 144, 95–104 CrossRef CAS PubMed.
- Y. L. Wang, Y. Y. Cui, Y. L. Zhao, R. Liu, Z. P. Sun, W. Li and X. Y. Gao, Chem. Commun., 2012, 48, 871–873 RSC.
- H. H. Sun, T. P. Qing, X. X. He, J. F. Shangguan, R. C. Jia, H. C. Bu, J. Huang and K. M. Wang, Anal. Chim. Acta, 2019, 1070, 88–96 CrossRef CAS PubMed.
- C. P. Liu, T. H. Wu, Y. L. Lin, C. Y. Liu, S. Wang and S. Y. Lin, Small, 2016, 12, 4127–4135 CrossRef CAS PubMed.
- C. Liu, C. Sun, H. Huang, K. Janda and T. Edgington, Cancer Res., 2003, 63, 2957–2964 CAS.
- Y. J. Chen, S. C. Wu, C. Y. Chen, S. C. Tzou, T. L. Cheng, Y. F. Huang, S. S. Yuan and Y. M. Wang, Biomaterials, 2014, 35, 304–315 CrossRef CAS PubMed.
- L. E. Edgington, M. Verdoes, A. Ortega, N. P. Withana, J. Lee, S. Syed, M. H. Bachmann, G. Blum and M. Bogyo, J. Am. Chem. Soc., 2016, 135, 174–182 CrossRef PubMed.
- Z. Liu, M. Xiong, J. B. Gong, Y. Zhang, N. Bai, Y. P. Luo, L. Y. Li, Y. Q. Wei, Y. H. Liu, X. Y. Tan and R. Xiang, Nat. Commun., 2014, 5, 4280 CrossRef CAS PubMed.
- Z. Liu, L. S. Xiong, T. Li, P. L. Xie, Q. Li, B. L. Wang, L. Wang, L. L. Li, Y. Q. Wang, H. Chen and K. H. Nan, Nanoscale, 2016, 8, 18400–18411 RSC.
- Y. Zhao, Z. J. Hai, H. Y. Wang, L. H. Su and G. L. Liang, Anal. Chem., 2018, 90, 8732–8735 CrossRef CAS PubMed.
- L. J. Zhang, Y. Q. Qi, H. Min, C. Ni, F. Wang, B. Wang, H. Qin, Y. L. Zhang, G. N. Liu, Y. Qin, X. X. Duan, F. Li, X. X. Han, N. Tao, L. R. Zhang, Z. H. Qin, Y. Zhao and G. J. Nie, ACS Nano, 2019, 13, 5091–5102 CrossRef CAS PubMed.
- G. Gasparini, P. C. Brooks, E. Biganzoli, P. B. Vermeulen and D. A. Cheresh, Clin. Cancer Res., 1998, 4, 2625–2634 CAS.
- R. E. Holiday, Biochem. J., 1936, 30, 1795–1803 CrossRef PubMed.
- F. S. Santiago, H. C. Lowe, M. M. Kavurma, C. N. Chesterman, A. Baker, D. G. Atkins and L. M. Khachigian, Nat. Med., 1999, 5, 1438 CrossRef CAS PubMed.
- G. Z. Zhu, J. Zheng, E. Q. Song, M. Donovan, K. J. Zhang, C. Liu and W. H. Tan, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 7998–8003 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Peptide and oligonucleotide sequences, specific cleavage activity of DNAzyme, TEM images, zeta size and potential, stability investigation, real-time imaging of Dox release, cytotoxicity assays of MDA-MB-231 cells, etc. See DOI: 10.1039/d1sc03847h |
| This journal is © The Royal Society of Chemistry 2021 |