Open Access Article
Open Access ArticleMulticolor fluorescence encoding of different microRNAs in lung cancer tissues at the single-molecule level†
Chen-chen Li‡
ab,
Hui-yan Chen‡a,
Xiliang Luo *b,
Juan Hu
*b,
Juan Hu *c and
Chun-yang Zhang
*c and
Chun-yang Zhang *a
*a
aCollege of Chemistry, Chemical Engineering and Materials Science, Collaborative Innovation Center of Functionalized Probes for Chemical Imaging in Universities of Shandong, Key Laboratory of Molecular and Nano Probes, Ministry of Education, Shandong Provincial Key Laboratory of Clean Production of Fine Chemicals, Shandong Normal University, Jinan 250014, China. E-mail: cyzhang@sdnu.edu.cn
bKey Laboratory of Optic-electric Sensing and Analytical Chemistry for Life Science, MOE, Shandong Key Laboratory of Biochemical Analysis, College of Chemistry and Molecular Engineering, Qingdao University of Science and Technology, Qingdao 266042, China. E-mail: xiliangluo@qust.edu.cn
cSchool of Chemistry and Chemical Engineering, Southeast University, Nanjing 211189, China. E-mail: hujuan@seu.edu.cn
First published on 16th August 2021
Abstract
The simultaneous detection of multiple microRNAs (miRNAs) will facilitate early clinical diagnosis. Herein, we demonstrate the integration of multicolor fluorophore-encoded cascade signal amplification with single-molecule detection for simultaneous measurement of different miRNAs in lung cancer tissues. This assay involves two linear templates and two circular templates without the requirement of any fluorescent-labeled probes. The binding of target miRNAs to their corresponding linear templates initiates the cyclic strand displacement amplification, generating many triggers which can specifically hybridize with the corresponding biotin-labeled AP probes to initiate the apurinic/apyrimidic endonuclease 1-assisted cyclic cleavage reaction for the production of more biotin-labeled primers for each miRNA. The resultant two primers can react with their corresponding circular templates to initiate rolling circle amplification which enables the incorporation of Cy5-dCTP/Cy3-dGTP nucleotides, resulting in the simultaneous production of abundant biotin-/multiple Cy5/Cy3-labeled DNA products. After magnetic separation and exonuclease cleavage, the amplified products release abundant Cy5 and Cy3 fluorescent molecules which can be simply monitored by single-molecule detection, with Cy3 indicating miR-21 and Cy5 indicating miR-155. This assay involves three consecutive amplification reactions, enabling the conversion of extremely low abundant target miRNAs into large numbers of Cy5/Cy3 fluorophore-encoded DNA products which can release abundant fluorescent molecules for the generation of amplified signals. This assay exhibits high sensitivity, good selectivity, and the capability of multiplexed assay. This method can simultaneously quantify miR-155 and miR-21 in living cells and in lung cancer tissues, and it can distinguish the expression of miRNAs between non-small cell lung cancer patients and healthy persons. The accuracy and reliability of the proposed method are further validated by quantitative reverse transcription polymerase chain reaction.
Introduction
Lung cancer is the leading cause of cancer death and accounts for 18.0% of the total cancer deaths. The survival of lung cancer patients in 5 years after diagnosis is only 10%–20%.1 Imaging and tissue biopsy are the standard methods for lung cancer diagnosis, but they suffer from ionizing radiation exposure and invasiveness.2 Traditional tumor diagnostic markers include carcinoembryonic antigen (CEA) and carcinoma antigen 125 (CA125), and they usually exhibit low sensitivity.3 Due to their simpler structure and less post-processing than DNA and protein, microRNAs (miRNAs) can function as potential non-invasive biomarkers for early diagnosis and prognosis of lung cancer.4–9 miRNAs are short endogenous non-coding RNAs with a length of ∼22 nucleotides (nt). miRNAs can regulate the expression of specific genes by binding to complementary regions of messenger RNA (mRNA), and the resulting duplex may inhibit translation by either enhancing the degradation or blocking the initiation of mRNA.10 More than 60% of human protein-coding genes harbor the predicted miRNA target sites. miRNAs play crucial regulatory roles in animal development, cell differentiation, apoptosis, proliferation and immunity,11–13 while dysregulations of miRNAs are frequently observed in a variety of cancers,14 such as the overexpression of miR-155 in lung cancer15–17 and the upregulation of miR-21 in lung cancer.18–20 Consequently, the detection of multiple miRNAs is essential for early diagnosis of lung cancer.Due to the properties of small size, low abundance in real biological tissues and short survival time in vitro, sensitive detection of miRNAs is very challenging.21 Traditional methods include polymerase chain reaction (PCR),22 microarray analysis,23 and northern blotting.24 PCR-based methods exhibit high sensitivity, but they involve the specially designed primers and precise temperature control.22 Although microarray analysis is suitable for multiple miRNA assay, it suffers from high cost, narrow dynamic range, low sensitivity, and poor repeatability.23 Northern blot is a common method for miRNA assay, but it suffers from long analysis time, low sensitivity, and large sample consumption.24 Alternatively, a series of new methods have been developed for miRNA assay based on electrochemical,25 colorimetric,26 Raman spectrum,27,28 and fluorescence measurements.29–31 However, their sensitivities are relatively low without the involvement of signal amplification.25–31 To improve the sensitivity, several signal amplification strategies such as hybridization chain reaction,32,33 strand displacement amplification,34,35 rolling circle amplification,36 and DNA hydrogelation-based signal amplification37 have now been introduced for the detection of a single type of miRNA.33–37 Notably, one disease may correlate with the mis-expression of different types of miRNAs,38 and the combination of multiple miRNAs may greatly improve the diagnostic accuracy.39,40 Therefore, new methods with the capability of sensitive detection of multiple miRNAs are highly required.
Herein, we demonstrate the integration of multicolor fluorophore-encoded cascade signal amplification with single-molecule detection for simultaneous detection of miRNAs in lung cancer tissues. Single-molecule detection has gained overwhelming attention due to the high signal-to-noise ratio, low sample consumption, and ultrahigh sensitivity.41–46 We used two lung cancer biomarkers miR-155 and miR-21 as the targets. The presence of miR-155 and miR-21 can initiate three consecutive amplification reactions (i.e., cyclic strand displacement amplification (SDA), APE1-assisted cyclic cleavage, and rolling circle amplification (RCA)), converting extremely low abundant target miRNAs to abundant Cy5/Cy3 fluorophore-encoded DNA products whose digestion releases larger numbers of fluorescent molecules. The subsequent single-molecule detection enables the identification of miR-155 with Cy5 and miR-21 with Cy3. Due to the high efficiency of the cascade signal amplification and high signal-to-noise ratio of single-molecule detection, this method can measure miRNAs with a detection limit of 25.7 aM for miR-155 and 45.7 aM for miR-21. Moreover, this assay can simultaneously quantify the expression levels of miR-21 and miR-155 in living cells and lung cancer tissues at the single-molecule level, and it can distinguish non-small cell lung cancer (NSCLC) patients from healthy persons as well.
Results and discussion
Principle of multiple miRNA assay
The design for simultaneous detection of multiple miRNAs is shown in Scheme 1. We designed a linear template, a biotinylated AP probe, and a circular template for each miRNA. The 3′ ends of each linear template and each AP probe are modified with NH2 to prevent nonspecific amplification.47 The linear template-155 consists of a miR-155-binding region, a recognition site for the Nt.BstNBI nicking enzyme, and an amplification region for miR-155-induced cyclic SDA. The linear template-21 consists of a miR-21-binding region, a recognition site for Nt.BstNBI, and an amplification region for miR-21-induced cyclic SDA. The biotinylated AP probe-155 consists of a binding region for trigger-155 generated from the first cyclic SDA and an AP site as the recognition domain for APE1-assisted cyclic cleavage. The biotinylated AP probe-21 consists of a binding region for trigger-21 generated from the first cyclic SDA and an AP site. The circular template-155 consists of a special binding domain for primer-155 released from the second APE1-assisted cyclic cleavage and an amplification domain for the RCA reaction. The circular template-21 consists of a special binding domain for primer-21 released from the second APE1-assisted cyclic cleavage and an amplification domain for the RCA reaction.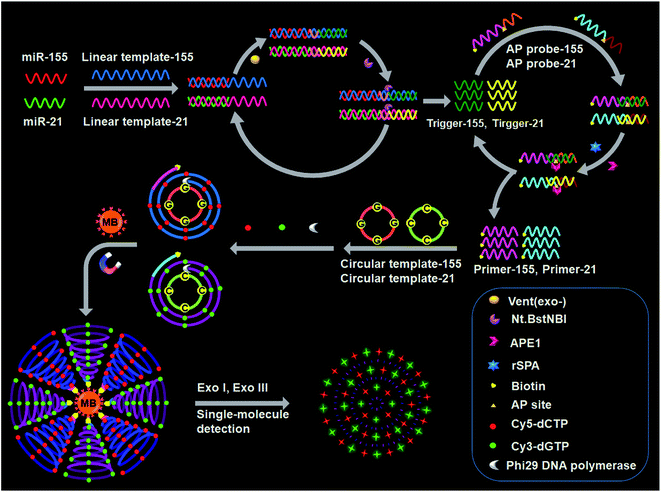 | ||
| Scheme 1 Principle of simultaneous detection of multiple miRNAs based on the integration of multicolor fluorophore-encoded cascade signal amplification with single-molecule detection. | ||
This assay contains the following steps: (1) target miRNA-induced cyclic SDA, (2) APE1-assisted cyclic cleavage, (3) RCA reaction, (4) magnetic separation and the cleavage of amplified products by Exo I/Exo III, and (5) single-molecule detection. In the presence of miR-155 (red color, Scheme 1), the binding of linear template-155 to miR-155 forms a partial duplex, and subsequently miR-155 can be extended along linear template-155 to generate a dsDNA in the presence of Vent (exo-) DNA polymerase and dNTPs. The resultant dsDNA can be cut by Nt.BstNBI at the cleavage recognition domain, generating trigger-155 and a new replication site for DNA polymerase. Consequently, abundant trigger-155 can be produced through repeated extension, cleavage and trigger generation (the first signal amplification). The resultant trigger-155 can bind to AP probe-155 to obtain the partial dsDNA with an AP site. Upon the addition of APE1, the partial dsDNA is cleaved at the AP site, generating 5′ biotinylated primer-155 and simultaneously releasing trigger-155. The free trigger-155 can further hybridize with new AP probe-155 repeatedly, initiating the APE1-assisted cyclic cleavage of biotinylated AP probe-155 through repeated hybridization, cleavage and dissociation, generating abundant 5′ biotinylated primer-155 (the second signal amplification). In the meantime, shrimp alkaline phosphatase (rSAP) is added into the reaction solution to hydrolyse the excess dNTPs from the first step. Subsequently, the resultant primer-155 binds to circular template-155 to initiate the RCA reaction in the presence of phi29 polymerase, dATP, dTTP, Cy5-dCTP and Cy3-dGTP. The circular template-155 containing only three types of bases (i.e., A, T, and G) can generate the RCA product with only three types of bases (i.e., T, A, and C), and the introduction of Cy5-modified dCTP enables the incorporation of abundant Cy5 fluorescent molecules into the RCA product (the third signal amplification). Notably, target miR-155 induces the first cyclic SDA reaction to yield many trigger-155 which can subsequently initiate the second APE1-assisted cyclic cleavage, producing more 5′ biotinylated primer-155. Each resultant 5′ biotinylated primer-155 may produce a biotin-/multiple Cy5-labeled ssDNA through the third RCA reaction, resulting in the generation of abundant biotin-/multiple Cy5-labeled ssDNAs. The ssDNA products can be captured by streptavidin-coated magnetic beads (MBs) and separated from the reaction solution through magnetic separation. Then the product is digested into single nucleotides by Exo I/Exo III to release large numbers of Cy5 fluorescent molecules, which can be simply measured by single-molecule detection.
When miR-21 is present (green color, Scheme 1), it can partially bind to linear template-21 to initiate the cyclic SDA reaction, producing abundant trigger-21. The resultant trigger-21 binds to AP probe-21 to initiate the APE1-assisted cyclic cleavage, generating large numbers of 5′ biotinylated primer-21. Subsequently, the resultant primer-21 can react with circular template-21 to initiate the RCA reaction, and the introduction of Cy3-modified dGTP enables the incorporation of abundant Cy3 fluorescent molecules into the RCA product. When miR-155 and miR-21 coexist, they induce three consecutive amplification events (i.e., target miRNA-induced cyclic SDA, APE1-assisted cyclic cleavage, and RCA reaction), generating abundant biotin-/multiple Cy5-labeled ssDNA products for miR-155 and biotin-/multiple Cy3-labeled ssDNA products for miR-21. The subsequent digestion of amplification products by Exo I/Exo III releases large numbers of fluorescent molecules which can be simultaneously quantified by single-molecule detection, with Cy5 indicating miR-155 and Cy3 indicating miR-21. However, in the absence of miRNAs, three consecutive amplification events cannot occur, and neither Cy5 nor Cy3 signals can be observed.
Validation of the assay
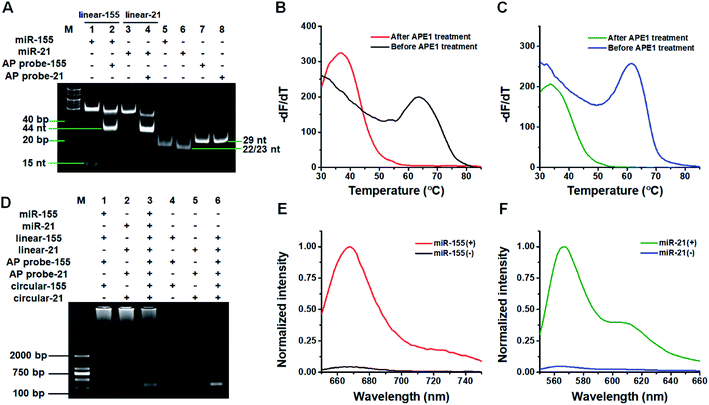
We used 14% native polyacrylamide gel electrophoresis (PAGE) to analyze the products of the target miRNA-induced cyclic SDA reaction (Fig. 1A). Only a 23 nt band is observed for miR-155 alone (Fig. 1A, lane 5). When miR-155, linear template-155, Vent (exo-) DNA polymerase, dNTPs and Nt.BstNBI are present, two significant bands of 53 bp and 15 nt are observed (Fig. 1A, lane 1), with the 53-bp band corresponding to the dsDNA intermediates and the 15 nt band corresponding to trigger-155 generated by the cyclic SDA reaction. The subsequent addition of AP probe-155 (Fig. 1A, lane 2) induces the disappearance of the 15 nt band (i.e., trigger-155) and the appearance of the 44 nt band that is larger than the size of AP probe-155 (Fig. 1A, lane 7, 29 nt), indicating that the resultant trigger-155 (Fig. 1A, lane 1, 15 nt band) can bind to AP probe-155 to form the partial dsDNA (44 nt). These results confirm the occurrence of the target miR-155-induced cyclic SDA reaction. Similarly, only a 22 nt band is observed for miR-21 alone (Fig. 1A, lane 6). In the presence of miR-21 + linear template-21, two significant bands of 52 bp and 15 nt are observed (Fig. 1A, lane 3), with the 52-bp band corresponding to the dsDNA intermediate and the 15 nt band corresponding to trigger-21 generated by the cyclic SDA reaction. The subsequent addition of AP probe-21 (Fig. 1A, lane 4) induces the disappearance of the 15 nt band (i.e., trigger-21) and the appearance of the 44 nt band which corresponds to the partial dsDNA product generated by the hybridization of trigger-21 (Fig. 1A, lane 3, 15 nt band) with AP probe-21 (Fig. 1A, lane 8, 29 nt). These results confirm the occurrence of the target miR-21-induced cyclic SDA reaction.To verify if the binding of the triggers to the AP probes leads to the APE1-assisted cyclic cleavage, we measured the melting curves of the reaction products (Fig. 1B and C). For the trigger-155/AP probe-155 duplex, the melting temperature of the product is 36.5 °C after APE1 treatment (Fig. 1B, red line), much lower than that before APE1 treatment (63.5 °C; Fig. 1B, black line), indicating the occurrence of the APE1-assisted cyclic cleavage reaction. Similarly, for the trigger-21/AP probe-21 duplex, the melting temperature of the product is 33.5 °C after APE1 treatment (Fig. 1C, olive line), much lower than that before APE1 treatment (61.5; Fig. 1C, blue line).
We used agarose gel electrophoresis to confirm the target miRNA-induced RCA reaction (Fig. 1D). No amplification is observed when miR-21 and miR-155 are absent (Fig. 1D, lanes 4–6). However in the presence of miR-155, the bands of amplified DNAs are observed (Fig. 1D, lane 1), suggesting the occurrence of three consecutive amplification events including the miR-155-induced cyclic SDA, APE1-assisted cyclic cleavage, and the subsequent RCA reaction. Similarly, the bands of amplified DNAs are observed in response to miR-21 (Fig. 1D, lane 2), suggesting the occurrence of miR-21-induced three consecutive amplification events. Moreover, the bands of amplified DNAs are observed when both miR-155 and miR-21 are present (Fig. 1D, lane 3).
We further used fluorescence measurement to demonstrate the target miRNA-induced RCA and the incorporation of fluorescent molecules into RCA products (Fig. 1E and F). When miR-155 is present, a distinct Cy5 fluorescence signal is detected (Fig. 1E, red line). Similarly, a distinct Cy3 fluorescence signal is detected when miR-21 is present (Fig. 1F, olive line). However, no distinct fluorescence signal can be detected in the absence of miR-155 (Fig. 1E, black line) and miR-21 (Fig. 1F, blue line). The results demonstrate that the miRNA-induced RCA reaction can only be triggered by targets miR-155 and miR-21 in the presence of specific linear templates, AP probes, and circular templates.
Under the optimal conditions (Fig. S1–S4†), we measured fluorescence emission spectra generated by different concentrations of target miRNAs. For miR-155 assay, the Cy5 fluorescence signal gradually enhances with the increasing concentration of miR-155 from 0 to 3 × 10−8 M (Fig. S5A†), and the fluorescence intensity (F) shows a linear correlation with the logarithm of miR-155 concentration (C) in the range of 1 × 10−15–1 × 10−10 M (Fig. S5B†). The correlation equation is F = 402![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 447 + 25
447 + 25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 902
902![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log10
log10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) C (R2 = 0.9920). The limit of detection (LOD) is 9.83 × 10−16 M by extrapolating the concentration from the signal equal to the mean control signal plus three standard deviations of the control signal. For miR-21 assay, the Cy3 fluorescence signal improves with the increasing concentration of miR-21 from 0 to 3 × 10−8 M (Fig. S5C†), and the fluorescence intensity (F) shows a linear correlation with the logarithm of miR-21 concentration (C) in the range of 1 × 10−14–1 × 10−10 M (Fig. S5D†). The correlation equation is F = 326
C (R2 = 0.9920). The limit of detection (LOD) is 9.83 × 10−16 M by extrapolating the concentration from the signal equal to the mean control signal plus three standard deviations of the control signal. For miR-21 assay, the Cy3 fluorescence signal improves with the increasing concentration of miR-21 from 0 to 3 × 10−8 M (Fig. S5C†), and the fluorescence intensity (F) shows a linear correlation with the logarithm of miR-21 concentration (C) in the range of 1 × 10−14–1 × 10−10 M (Fig. S5D†). The correlation equation is F = 326![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 419 + 21
419 + 21![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 874
874![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log10
log10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) C (R2 = 0.9948). The LOD is evaluated to be 5.01 × 10−15 M.
C (R2 = 0.9948). The LOD is evaluated to be 5.01 × 10−15 M.
Single-molecule/particle detection
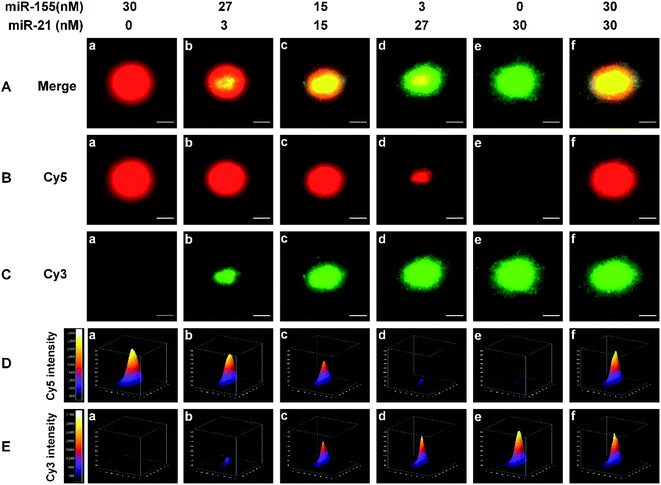
We employed single-molecule/particle imaging to measure different target miRNA-induced fluorescence encoding. After the magnetic beads were incubated with the RCA reaction products, the product-coupled magnetic beads displayed distinct fluorescence images in the red and green channels under the excitation of 640 nm and 561- nm lasers, respectively. When miR-155 is present alone, only the Cy5 signal can be detected (Fig. 2B(a), red color) and no Cy3 signal is observed (Fig. 2C(a)). When miR-21 is present alone, only the Cy3 signal can be detected (Fig. 2C(e), green color), and no Cy5 signal can be detected (Fig. 2B(e)). When both miR-155 and miR-21 are present, both Cy5 (Fig. 2B(b–d and f), red color) and Cy3 (Fig. 2C(b–d and f), green color) signals are simultaneously detected. Moreover, the colocalization of Cy5 and Cy3 fluorescence signals leads to the appearance of yellow color signals (Fig. 2A(b–d and f)).For the optical coding, two different RCA reaction products with distinctively different fluorescence emission spectra (i.e., Cy5 and Cy3) are used to modify the magnetic beads. Five types of color-encoded magnetic beads are identified at the single-particle level. There are red (Fig. 2A(a)), yellow (Fig. 2A(c and f)), and green color alone (Fig. 2A(e)), a red region appearing in the periphery of a yellow region (Fig. 2A(b)), and a green region appearing in the periphery of a yellow region (Fig. 2A(d)). The size of the region around each spot is primarily associated with the concentration of target miRNAs. Notably, the increase of miRNA concentration can induce the formation of larger size product-coupled magnetic beads, generating red/green monomers with tunable size for each spot. The red-green heterodimers display yellow colocalized signals. Thus, a colorimetric signal change at the single-particle level can be quantitatively analyzed with the color profiling method.
Multi-signal magnetic beads can be rationally designed and exploited for the simultaneous detection of miRNAs. We prepared a series of artificial mixtures by mixing miR-155 and miR-21. Fig. 2B(a–e) shows that the increase of miR-155 concentration can induce the formation of larger size product-coupled magnetic beads in the red channel. As expected, the largest size is detected in response to 30 nM miR-155 (Fig. 2B(a), red color), whose size is consistent with the size in response to 30 nM miR-155 + 30 nM miR-21 (Fig. 2B(f), red color). These results indicate that this assay is capable of detecting miR-155 even in the presence of miR-21. Similarly, in the presence of increasing concentration of miR-21, product-coupled magnetic beads exhibit increased size in the green channel (Fig. 2C(a–e)). The largest size is detected in the presence of 30 nM miR-21 (Fig. 2C(e), green color), whose size is consistent with the size in the presence of 30 nM miR-155 + 30 nM miR-21 (Fig. 2C(f), green color). Thus, this method is suitable for the visualization of miRNA concentration changes.
The distribution of the fluorescence intensity of the spot was further analyzed. Upon the addition of increased concentration of miR-155 (0–30 nM), the Cy5 fluorescence intensity is significantly enhanced (Fig. 2D(a–e)). When the miR-155 concentration is 30 nM, the fluorescence intensity reaches the highest value (Fig. 2D(a)). The mixture of miR-155 and miR-21 gives rise to an identical Cy5 intensity (Fig. 2D(f)). When an increased concentration of miR-21 is added to the mixture, the Cy3 intensity is gradually enhanced (Fig. 2E(a–e)), and the highest value is obtained at a concentration of 30 nM (Fig. 2E(e)). There is no significant difference between miR-21 alone and the mixture (Fig. 2E(e and f)). The results demonstrate that this method can be used for monitoring the concentration changes of both miR-21 and miR-155 at the single-particle level.
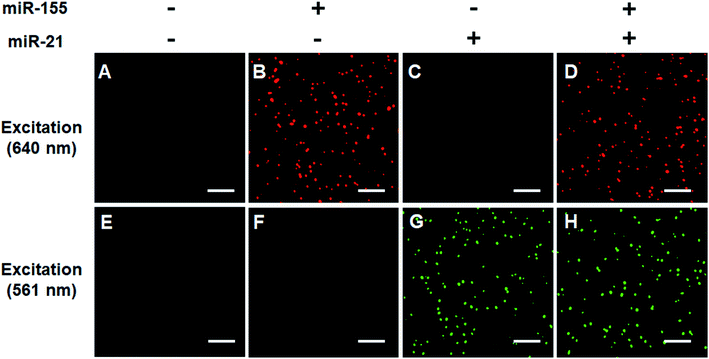
We verified the simultaneous detection of miR-21 and miR-155 at the single-molecule level. When miR-21 and miR-155 are absent, neither Cy5 (Fig. 3A) nor Cy3 fluorescence signals (Fig. 3E) can be observed. When miR-155 is present, only a significant Cy5 signal can be detected (Fig. 3B), but no Cy3 signal is observed (Fig. 3F). When miR-21 is present, only a significant Cy3 signal can be detected (Fig. 3G), but no Cy5 signal is observed (Fig. 3C). When miR-21 and miR-155 coexist, both Cy5 (Fig. 3D) and Cy3 (Fig. 3H) fluorescence signals are simultaneously detected.
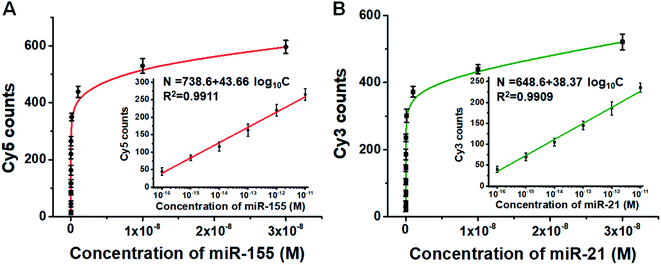
We further employed this method to measure different concentrations of miR-155 and miR-21. The numbers of Cy5 (Fig. S6A†) and Cy3 fluorescence signals (Fig. S6B†) enhance with the increasing concentrations of miR-155 and miR-21, respectively. Moreover, the Cy5 counts (Fig. 4A) and Cy3 counts (Fig. 4B) improve with the increasing concentration of miR-155 and miR-21 in the range from 0 M to 3 × 10−8 M, respectively, and the number of fluorescent molecules (N) exhibits a linear correlation with the logarithm of miRNA concentration (C) in the range of 1 × 10−16–1 × 10−11 M (inset of Fig. 4A and B). For miR-155 assay, the correlation equation is N = 738.6 + 43.66![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log10
log10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) C (R2 = 0.9911), and the LOD is measured to be 2.57 × 10−17 M. The sensitivity of this method has been improved by 38-fold compared with that of ensemble fluorescence measurement (Fig. S5B†). For miR-21 assay, the correlation equation is N = 648.6 + 38.37
C (R2 = 0.9911), and the LOD is measured to be 2.57 × 10−17 M. The sensitivity of this method has been improved by 38-fold compared with that of ensemble fluorescence measurement (Fig. S5B†). For miR-21 assay, the correlation equation is N = 648.6 + 38.37![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log10
log10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) C (R2 = 0.9909), and the LOD is measured to be 4.57 × 10−17 M. The sensitivity of this method has been improved by 110-fold compared with that of ensemble fluorescence measurement (Fig. S5D†). Notably, the proposed method enables simultaneous and sensitive detection of miR-155 and miR-21 with a linear range of 5 orders of magnitude, superior to the reported methods for miRNA assays (Table S1†). The sensitivity of this method has improved by as much as 4 orders of magnitude compared with that of quantum dots and graphene oxide nano-photon switch-based fluorescent assay,48 2 orders of magnitude compared with the peptide nucleic acid-based electrochemical biosensor,49 and 2 orders of magnitude compared with the bi-directional DNA walking machine-based electrochemiluminescent biosensor.50
C (R2 = 0.9909), and the LOD is measured to be 4.57 × 10−17 M. The sensitivity of this method has been improved by 110-fold compared with that of ensemble fluorescence measurement (Fig. S5D†). Notably, the proposed method enables simultaneous and sensitive detection of miR-155 and miR-21 with a linear range of 5 orders of magnitude, superior to the reported methods for miRNA assays (Table S1†). The sensitivity of this method has improved by as much as 4 orders of magnitude compared with that of quantum dots and graphene oxide nano-photon switch-based fluorescent assay,48 2 orders of magnitude compared with the peptide nucleic acid-based electrochemical biosensor,49 and 2 orders of magnitude compared with the bi-directional DNA walking machine-based electrochemiluminescent biosensor.50
Detection specificity
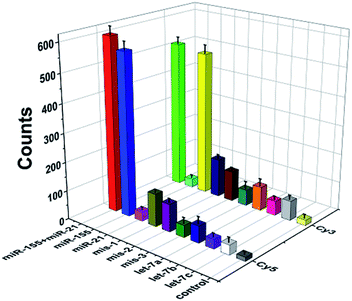
To evaluate the specificity of this method, we used three irrelevant miRNAs, one-base mismatched RNAs (i.e., mis-1-155 and mis-1-21), two-base mismatched RNAs (i.e., mis-2-155 and mis-2-21) and three-base mismatched RNAs (i.e., mis-3-155 and mis-3-21) as the interferents. As shown in Fig. 5, when miR-155 and miR-21 coexist, high Cy5 (Fig. 5, red column) and Cy3 (Fig. 5, green column) fluorescence signals are detected simultaneously. When miR-155 is present, only a high Cy5 fluorescence signal can be detected (Fig. 5, blue column). When miR-21 is present, only a high Cy3 signal can be detected (Fig. 5, yellow column). In contrast, neither significant Cy5 nor significant Cy3 signals can be observed in response to irrelevant miRNAs. Moreover, the Cy5 fluorescence signal generated by target miR-155 is 17-fold, 10-fold, 17-fold and 16-fold higher than those generated by miR-21 (Fig. 5, magenta column), let-7a (Fig. 5, royal blue column), let-7b (Fig. 5, violet column) and let-7c (Fig. 5, white column), respectively, and the Cy3 fluorescence signal generated by target miR-21 is 17-fold, 6-fold, 10-fold and 7-fold higher than those generated by miR-155 (Fig. 5, cyan column), let-7a (Fig. 5, orange column), let-7b (Fig. 5, pink column) and let-7c (Fig. 5, light gray column), respectively. Notably, this assay possesses good specificity and it can differentiate miR-155 from the single-base mismatched miRNAs. The Cy5 fluorescence signal generated by target miR-155 is 5-fold, 6-fold and 14-fold higher than those generated by mis-1-155 (Fig. 5, dark yellow column), mis-2-155 (Fig. 5, purple column), and mis-3-155 (Fig. 5, olive column), respectively. The Cy3 fluorescence signal generated by target miR-21 is 4-fold, 5-fold and 9-fold higher than those generated by mis-1-21 (Fig. 5, navy column), mis-2-21 (Fig. 5, wine column), and mis-3-21 (Fig. 5, dark cyan column), respectively.Profiling of miRNAs in various cell lines
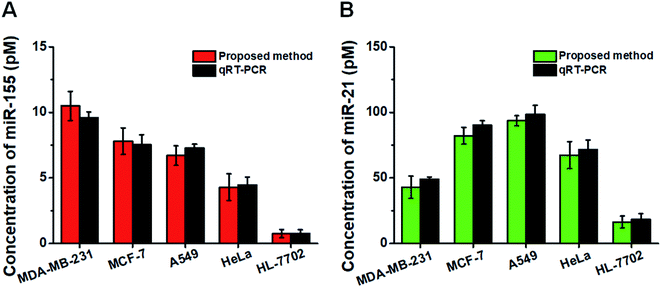
This assay allows for measuring the expression levels of miR-155 and miR-21 in breast cancer cells (MCF-7 cells and MDA-MB-231 cells), lung adenocarcinoma cells (A549 cells), cervical carcinoma cells (HeLa cells), and hepatocyte cells (HL-7702 cells), respectively. In comparison with the negligible signal of the control cell line (HL-7702 cells), a high expression level of miR-155 (Fig. 6A, red column) is observed in four cancer cells (MCF-7 cells, MDA-MB-231 cells, A549 cells, and HeLa cells). The miR-155 concentration decreases in the order of MDA-MB-231 cells > MCF-7 cells > A549 cells > HeLa cells > HL-7702 cells, consistent with previous research studies.48,49 Moreover, the expression level of miR-21 (Fig. 6B, green column) in HL-7702 cells is much lower than those in four cancer cells. The miR-21 concentration decreases in the order of A549 cells > MCF-7 cells > HeLa cells > MDA-MB-231 cells > HL-7702 cells, consistent with previous research studies.51,52 We further validated our results using quantitative-reverse transcription PCR (qRT-PCR) (Fig. S7† and 6, dark gray column). Our results are consistent with those obtained by qRT-PCR (Fig. 6).Measurement of miRNAs in clinical tissue samples
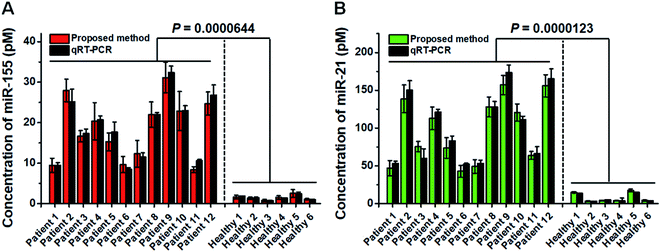
Previous research studies demonstrate the high expression levels of miR-21 and miR-155 in patients with non-small cell lung cancer (NSCLC).53,54 To confirm the proof-of-concept of this assay for real clinical sample analysis, we analyzed human tissue samples from twelve diagnosed NSCLC patients and six healthy persons. As shown in Fig. 7, the miR-155 concentration in NSCLC patients is significantly higher than that in healthy persons (t test, P < 0.0001), with a median concentration of 18.5 pM for NSCLC patients and 1.42 pM for healthy persons. Moreover, the miR-21 concentration in NSCLC patients is significantly higher than that in healthy persons (t test, P < 0.0001), with a median concentration of 94.2 pM for NSCLC patients and 4.08 pM for healthy persons. We further validated the results of the proposed method using qRT-PCR (Fig. 7, dark gray column). The results obtained by the proposed method are in good agreement with those obtained by qRT-PCR for each sample (Fig. 7). The results suggest that miR-155 and miR-21 can function as tumor biomarkers for early diagnosis of NSCLC.Conclusions
In summary, we demonstrate the integration of multicolor fluorophore-encoded cascade signal amplification with single-molecule detection for the simultaneous measurement of miRNAs in lung cancer tissue samples. This assay involves three consecutive amplification reactions which enable the conversion of extremely low abundant target miRNAs into large numbers of Cy5/Cy3 fluorophore-encoded DNA products which can release large numbers of fluorescent molecules for the generation of amplified signals. Unlike the reported methods for miRNA assay with the requirement of carefully designed probes (e.g., hairpin probes labelled with different-color quantum dots,48 signal probes labelled with Cy3/Cy5,55 hairpin probes labelled with ferrocene/methylene blue,49 and the magnetic DNA nanosphere-encoded probes labelled with ferrocene/methylene blue56), this assay enables the simultaneous detection of miRNAs without the need for either miRNA purification or multiple fluorescent-labelled probes. Due to the high efficiency of the cascade signal amplification and high signal-to-noise ratio of single-molecule detection, this assay exhibits high sensitivity, good selectivity, and the capability of multiplexed assay. This method can simultaneously quantify miR-155 and miR-21 in living cells and lung cancer tissues, and it can distinguish the expression of miRNAs between NSCLC patients and healthy persons. We further validate the results of the proposed method using qRT-PCR. Importantly, the proposed method can be extended to detect other cancer-related miRNAs by simply changing the sequence of linear templates, holding great potential in multiplexed detection and clinical diagnosis.Experimental
Chemicals and materials
All oligonucleotides (Table 1) were synthesized and HPLC-purified by TaKaRa Biotechnology Co., Ltd (Dalian, China). The two circular templates were prepared by TaKaRa Biotechnology Co., Ltd (Dalian, China). Nt.BstNBI, Vent (exo-) DNA polymerase, apurinic/apyrimidinic endonuclease 1 (APE1), shrimp alkaline phosphatase (rSAP), phi29 DNA polymerase, exonuclease I (Exo I), exonuclease III (Exo III), 10× ThermoPol buffer, 10× Nt.BstNBI buffer, 10× NEBuffer 4, 10× phi29 DNA polymerase reaction buffer, 10× NEBuffer 1, and 10× exonuclease I reaction buffer were purchased from New England Biolabs (Ipswich, MA, USA). RNase inhibitor, RNase-free water, dATP and dTTP were purchased from TaKaRa Biotechnology Co. Ltd (Dalian, China). Cy5-dCTP and Cy3-dGTP were obtained from PerkinElmer (Foster City, CA, USA). Streptavidin-coated magnetic beads (MBs) were purchased from Invitrogen (California, CA, USA). The miRNeasy Mini Kit and miRNeasy FFPE kit were obtained from Qiagen (Hilden, Germany). All other reagents were of analytical grade and used just as received without further purification. Human breast cancer cell lines (MCF-7 and MDA-MB-231 cells), human lung adenocarcinoma cell line (A549 cells), human cervical carcinoma cell line (HeLa cells) and human hepatocyte cell line (HL-7702 cells) were purchased from the Cell Bank of Chinese Academy of Sciences (Shanghai, China). The tissue samples of non-small cell lung cancer (NSCLC) patients and healthy persons were obtained from the Affiliated Hospital of Guangdong Medical University (Zhanjiang, Guangdong, China), and this research was approved by the ethics committee of the Affiliated Hospital of Guangdong Medical University.| Oligonucleotides | Sequence (5′–3′) |
|---|---|
| a In the linear templates, the recognition sites of Nt.BstNBI are shown in bold. In the AP probes, the letter “X” represents an AP site. In the mismatched RNAs, the underlined bases indicate the mismatched bases. | |
| miR-155 | UUA AUG CUA AUC GUG AUA GGG GU |
| miR-21 | UAG CUU AUC AGA CUG AUG UUG A |
| Linear template-155 | TCA CTA ACT GGC TCT GTA CGA CTC AAA TTT ACC CCT ATC ACG ATT AGC ATT AA – NH2 |
| Linear template-21 | AGT GTA TGT CCT GGA AGA CGA CTC AAA TTT TCA ACA TCA GTC TGA TAA GCT A – NH2 |
| AP probe-155 | Biotin – AAC ATC CCT AAT TTC TCA CTA X CTG GCT CT – NH2 |
| AP probe-21 | Biotin – AAG ATG GGT AAT TAG AGT GTA X GTC CTG GA – NH2 |
| Circular template-155 | TAG TGA GAA ATT AGG GAT GTT AAG TAG GAT GTT GAG TAA AGT TGA AGA ATG GTG A |
| Circular template-21 | TAC ACT CTA ATT ACC CAT CTT AAC TAC CAT CTT CAC TAT TCA ACT TCA ATC CTC A |
| Trigger-155 | AGA GCC AGT TAG TGA |
| Trigger-21 | TCC AGG ACA TAC ACT |
| let-7a | UGA GGU AGU AGG UUG UAU AGU U |
| let-7b | UGA GGU AGU AGG UUG UGU GGU U |
| let-7c | UGA GGU AGU AGG UUG UAU GGU U |
| mis-1-155 | TTA ATG CTA ATC GTG ATA GGG ![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) T T |
| mis-2-155 | TTA ATG CTA ATC GTG ATA ![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) GG GG ![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) T T |
| mis-3-155 | TTA ATG CTA ATC GTG ![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif) TA TA ![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) GG GG ![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) T T |
| mis-1-21 | TAG CTT ATC AGA CTG ATG TT![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) A A |
| mis-2-21 | TAG CTT ATC AGA CTG AT![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) TT TT![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) A A |
| mis-3-21 | TAG CTT ATC AGA CT![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) AT AT![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) TT TT![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) A A |
Target miRNA-induced cyclic SDA and APE1-assisted cyclic cleavage reactions
The reaction mixtures for the target miRNA-induced cyclic SDA reaction were prepared separately on ice as part A and part B. Part A consisted of variable concentrations of miR-155, variable concentrations of miR-21, 30 nM linear template-155, 30 nM linear template-21, 1× ThermoPol buffer, and DEPC-treated water. Part A was incubated at 95 °C for 3 min, followed by cooling down to room temperature. Part B consisted of 8 U of RNase inhibitor, 0.5 U of Vent (exo-) DNA polymerase, 2 U of Nt.BstNBI, 0.5× Nt.BstNBI buffer, and 600 μM dNTPs. The reaction of part A with part B was performed in 10 μL of the reaction solution at 55 °C for 1 h to generate trigger-155 and trigger-21. Then the APE1-assisted cyclic cleavage reaction was performed in 15 μL of reaction mixture containing the above reaction solution, 160 nM AP probe-155, 160 nM AP probe-21, 1 U of APE1, 0.2 U of rSAP, 1× NEBuffer 4, and DEPC-treated water, followed by incubation at 37 °C for 1 h to generate primer-155 and primer-21.RCA reaction
The RCA reaction was performed in 25 μL of reaction mixture containing the above reaction solution, 96 nM circular template-155, 96 nM circular template-21, 240 μM dATP, 240 μM dTTP, 8 μM Cy5-dCTP, 8 μM Cy3-dGTP, 5 U of phi29 DNA polymerase, and 0.5× phi29 reaction buffer at 30 °C for 1.5 h in the dark, and the reaction was terminated by incubation at 65 °C for 10 min.Conjugation of RCA products with the streptavidin-coated MBs and exonuclease digestion reaction
Prior to the reaction, 5 μL of streptavidin-coated MB solution (10 mg mL−1) was washed twice with 2× binding and washing (B&W) buffer (10 mM Tris–HCl, pH 7.5, 1 mM EDTA, and 2 M NaCl) and resuspended in 2× B&W buffer with a final concentration of 2 mg mL−1. Then 2 mg mL−1 MB solution was mixed with biotinylated RCA products, followed by incubation in the dark for 15 min on a roller mixer at room temperature. The mixture was then washed three times by magnetic separation using 1× B&W buffer to remove the excess Cy5-dCTP and Cy3-dGTP, and the single-stranded DNA-MB conjugates (MB-ssDNAs) were resuspended in 1× NEBuffer 1. The exonuclease digestion reaction was performed in 20 μL of reaction mixture containing MB-ssDNAs, 10 U of Exo I, 10 U of Exo III, 1× Exo I reaction buffer, and 1× NEBuffer 1 at 37 °C for 30 min. Finally, the streptavidin-coated MBs were separated by magnetic separation for 3 min in the dark, and the supernatant solution was subjected to subsequent detection.Gel electrophoresis and fluorescence measurement
The products of the target miRNA-induced cyclic SDA reaction were analyzed by 14% nondenaturing polyacrylamide gel electrophoresis (PAGE) in TBE buffer (9 mM Tris–HCl, pH 7.9, 9 mM boric acid, and 0.2 mM EDTA) at a constant voltage of 110 V for 50 min at room temperature. Then the gel was stained with SYBR gold and analyzed by using a Bio-Rad ChemiDoc MP imaging system (Hercules, CA, USA). The RCA reaction products stained with SYBR gold were analyzed by 1% agarose gel electrophoresis in TAE buffer (40 mM Tris-acetic acid and 2 mM EDTA) at a constant voltage of 110 V for 60 min. 20 μL of the reaction products were diluted to a final volume of 80 μL with ultrapure water, and subsequently subjected to fluorescence emission spectra measurement by using an FLS1000 (Edinburgh Instruments, UK). Cy5 fluorescence was measured at an excitation wavelength of 635 nm, with the fluorescence intensity at 668 nm being used for the quantitative analysis of miR-155. Cy3 fluorescence was measured at an excitation wavelength of 532 nm, with the fluorescence intensity at 568 nm being used for the quantitative analysis of miR-21.Single-molecule detection and data analysis
For single-molecule measurement, the reaction products were diluted 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000-fold with 1× imaging buffer (3 mM MgCl2, 100 mM Tris–HCl, pH 8.0, and 10 mM (NH4)2SO4). 10 μL of samples were spread on a glass coverslip for imaging. The images were acquired by total internal reflection fluorescence (TIRF) microscopy (Nikon, Ti-E, Japan). 640 nm and 561 nm lasers were used to excite Cy5 and Cy3 fluorescent molecules, respectively. The photons of Cy5 and Cy3 were collected by an oil immersion 100× objective (Nikon, Japan), and were split into the Cy5 channel (661.5–690.5 nm filter) and Cy3 channel (573–613 nm filter) by a dichroic mirror, and were imaged onto a digital CMSO EMCCD camera (Hamamatsu Photonics K. K., Japan) with an exposure time of 500 ms. For data analysis, regions of interest of 600 × 600 pixels were selected for Cy5 and Cy3 molecule counting by using Image J software (NIH, Bethesda, MD, USA). The numbers of Cy5 and Cy3 were the average of ten frames, respectively.
000-fold with 1× imaging buffer (3 mM MgCl2, 100 mM Tris–HCl, pH 8.0, and 10 mM (NH4)2SO4). 10 μL of samples were spread on a glass coverslip for imaging. The images were acquired by total internal reflection fluorescence (TIRF) microscopy (Nikon, Ti-E, Japan). 640 nm and 561 nm lasers were used to excite Cy5 and Cy3 fluorescent molecules, respectively. The photons of Cy5 and Cy3 were collected by an oil immersion 100× objective (Nikon, Japan), and were split into the Cy5 channel (661.5–690.5 nm filter) and Cy3 channel (573–613 nm filter) by a dichroic mirror, and were imaged onto a digital CMSO EMCCD camera (Hamamatsu Photonics K. K., Japan) with an exposure time of 500 ms. For data analysis, regions of interest of 600 × 600 pixels were selected for Cy5 and Cy3 molecule counting by using Image J software (NIH, Bethesda, MD, USA). The numbers of Cy5 and Cy3 were the average of ten frames, respectively.
Real sample analysis
MDA-MB-231 cells, MCF-7 cells, A549 cells, HeLa cells, and HL-7702 cells were cultured with 10% fetal bovine serum (FBS; Gibco, USA) and 1% penicillin–streptomycin (Invitrogen, USA) in Dulbecco's modified Eagle's medium (DMEM). The cells were cultured at 37 °C in a humidified atmosphere containing 5% CO2. The number of cells was counted by using a Countstar cell counter. The RNA extracts were prepared by using the miRNeasy Mini Kit according to the manufacturer's protocol. The extraction of total RNA from lung tissues of NSCLC patients and healthy persons embedded in paraffin was performed by using the miRNeasy FFPE kit according to the manufacturer's protocol. The concentration of total RNA was determined by using a NanoDrop 2000c spectrophotometer (Thermo Scientific, Wilmington, Delaware, USA).Author contributions
Chen-chen Li: conceptualization, methodology, investigation, writing – original draft preparation; Hui-yan Chen: methodology, investigation; Xiliang Luo: conceptualization, writing – review and editing; Juan Hu: conceptualization, writing – review and editing; Chun-yang Zhang: conceptualization, funding acquisition, writing – review and editing.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the National Natural Science Foundation of China (Grant no. 21735003) and the Award for Team Leader Program of Taishan Scholars of Shandong Province, China.Notes and references
- H. Sung, J. Ferlay, R. L. Siegel, M. Laversanne, I. Soerjomataram, A. Jemal and F. Bray, CA Cancer J. Clin., 2021, 71, 209–249 CrossRef.
- J. C. M. Wan, C. Massie, J. Garcia-Corbacho, F. Mouliere, J. D. Brenton, C. Caldas, S. Pacey, R. Baird and N. Rosenfeld, Nat. Rev. Cancer, 2017, 17, 223–238 CrossRef CAS.
- G. Yang, Z. Xiao, C. Tang, Y. Deng, H. Huang and Z. He, Biosens. Bioelectron., 2019, 141, 111416 CrossRef CAS.
- L. F. R. Gebert and I. J. MacRae, Nat. Rev. Mol. Cell Biol., 2019, 20, 21–37 CrossRef CAS.
- X. W. He, T. Zeng, Z. Li, G. L. Wang and N. Ma, Angew. Chem., Int. Ed., 2016, 55, 3073–3076 CrossRef CAS.
- S. Xu, Y. Nie, L. Jiang, J. Wang, G. Xu, W. Wang and X. Luo, Anal. Chem., 2018, 90, 4039–4045 CrossRef CAS PubMed.
- J. Hu, M. H. Liu and C. Y. Zhang, Chem. Sci., 2018, 9, 4258–4267 RSC.
- L. Xian, F. Xu, J. Liu, N. Xu, H. Li, H. Ge, K. Shao, J. Fan, G. Xiao and X. Peng, J. Am. Chem. Soc., 2019, 141, 20490–20497 CrossRef CAS.
- J. Su, F. Wu, H. Xia, Y. Wu and S. Liu, Chem. Sci., 2020, 11, 80–86 RSC.
- D. P. Bartel, Cell, 2009, 136, 215–233 CrossRef CAS.
- L. P. Lim, N. C. Lau, P. Garrett-Engele, A. Grimson, J. M. Schelter, J. Castle, D. P. Bartel, P. S. Linsley and J. M. Johnson, Nature, 2005, 433, 769–773 CrossRef CAS.
- K. K. Farh, A. Grimson, C. Jan, B. P. Lewis, W. K. Johnston, L. P. Lim, C. B. Burge and D. P. Bartel, Science, 2005, 310, 1817–1821 CrossRef CAS.
- W. P. Kloosterman and R. H. Plasterk, Dev. Cell, 2006, 11, 441–450 CrossRef CAS PubMed.
- C. P. Bracken, H. S. Scott and G. J. Goodall, Nat. Rev. Genet., 2016, 17, 719–732 CrossRef CAS.
- F. Gao, J. Chang, H. Wang and G. Zhang, Oncol. Rep., 2014, 31, 351–357 CrossRef CAS PubMed.
- N. Yanaihara, N. Caplen, E. Bowman, M. Seike, K. Kumamoto, M. Yi, R. M. Stephens, A. Okamoto, J. Yokota, T. Tanaka, G. A. Calin, C. G. Liu, C. M. Croce and C. C. Harris, Cancer Cell, 2006, 9, 189–198 CrossRef CAS.
- T. Donnem, K. Eklo, T. Berg, S. W. Sorbye, K. Lonvik, S. Al-Saad, K. Al-Shibli, S. Andersen, H. Stenvold, R. M. Bremnes and L. T. Busund, J. Transl. Med., 2011, 9, 6 CrossRef.
- C. Bica-Pop, R. Cojocneanu-Petric, L. Magdo, L. Raduly, D. Gulei and I. Berindan-Neagoe, Cell. Mol. Life Sci., 2018, 75, 3539–3551 CrossRef CAS PubMed.
- S. Volinia, G. A. Calin, C. G. Liu, S. Ambs, A. Cimmino, F. Petrocca, R. Visone, M. Iorio, C. Roldo, M. Ferracin, R. L. Prueitt, N. Yanaihara, G. Lanza, A. Scarpa, A. Vecchione, M. Negrini, C. C. Harris and C. M. Croce, Proc. Natl. Acad. Sci. U. S. A., 2006, 103, 2257–2261 CrossRef CAS PubMed.
- A. A. Abd-El-Fattah, N. A. Sadik, O. G. Shaker and M. L. Aboulftouh, Cell Biochem. Biophys., 2013, 67, 875–884 CrossRef CAS.
- C. C. Pritchard, H. H. Cheng and M. Tewari, Nat. Rev. Genet., 2012, 13, 358–369 CrossRef CAS PubMed.
- Y. Niu, L. Zhang, H. Qiu, Y. Wu, Z. Wang, Y. Zai, L. Liu, J. Qu, K. Kang and D. Gou, Sci. Rep., 2015, 5, 15100 CrossRef CAS.
- J. M. Lee, H. Cho and Y. Jung, Angew. Chem., Int. Ed., 2010, 49, 8662–8665 CrossRef CAS.
- S. Bagga, J. Bracht, S. Hunter, K. Massirer, J. Holtz, R. Eachus and A. E. Pasquinelli, Cell, 2005, 122, 553–563 CrossRef CAS.
- X. L. Zhang, Z. H. Yang, Y. Y. Chang, D. Liu, Y. R. Li, Y. Q. Chai, Y. Zhuo and R. Yuan, Chem. Sci., 2020, 11, 148–153 RSC.
- H. Lee, J. Lee, S. G. Lee and P. S. Doyle, Anal. Chem., 2020, 92, 5750–5755 CrossRef CAS PubMed.
- W. Ma, P. Fu, M. Sun, L. Xu, H. Kuang and C. Xu, J. Am. Chem. Soc., 2017, 139, 11752–11759 CrossRef CAS PubMed.
- Y. Si, L. Xu, N. Wang, J. Zheng, R. Yang and J. Li, Anal. Chem., 2020, 92, 2649–2655 CrossRef CAS PubMed.
- F. Causa, A. Aliberti, A. M. Cusano, E. Battista and P. A. Netti, J. Am. Chem. Soc., 2015, 137, 1758–1761 CrossRef CAS.
- X. Ou, S. Zhan, C. Sun, Y. Cheng, X. Wang, B. Liu, T. Zhai, X. Lou and F. Xia, Biosens. Bioelectron., 2019, 124–125, 199–204 CrossRef CAS PubMed.
- Z. Zhou, Y. S. Sohn, R. Nechushtai and I. Willner, ACS Nano, 2020, 14, 9021–9031 CrossRef CAS PubMed.
- P. Zhang, Y. Ouyang and I. Willner, Chem. Sci., 2021, 12, 4810–4818 RSC.
- Y. Zhang, Y. Zhang, X. Zhang, Y. Li, Y. He, Y. Liu and H. Ju, Chem. Sci., 2020, 11, 6289–6296 RSC.
- L. J. Wang, M. Ren, H. X. Wang, J. G. Qiu, B. Jiang and C. Y. Zhang, Anal. Chem., 2020, 92, 8546–8552 CrossRef CAS PubMed.
- B. Li, Y. Liu, Y. Liu, T. Tian, B. Yang, X. Huang, J. Liu and B. Liu, ACS Nano, 2020, 14, 8116–8125 CrossRef CAS.
- R. Deng, L. Tang, Q. Tian, Y. Wang, L. Lin and J. Li, Angew. Chem., Int. Ed., 2014, 53, 2389–2393 CrossRef CAS PubMed.
- L. Zou, Z. Wu, X. Liu, Y. Zheng, W. Mei, Q. Wang, X. Yang and K. Wang, Anal. Chem., 2020, 92, 11953–11959 CrossRef CAS.
- K. P. Porkka, M. J. Pfeiffer, K. K. Waltering, R. L. Vessella, T. L. Tammela and T. Visakorpi, Cancer Res., 2007, 67, 6130–6135 CrossRef CAS PubMed.
- T. Erbes, M. Hirschfeld, G. Rücker, M. Jaeger, J. Boas, S. Iborra, S. Mayer, G. Gitsch and E. Stickeler, BMC Cancer, 2015, 15, 193 CrossRef.
- H. Butz, R. Nofech-Mozes, Q. Ding, H. W. Z. Khella, P. M. Szabó, M. Jewett, A. Finelli, J. Lee, M. Ordon, R. Stewart, S. Krylov and G. M. Yousef, Eur. Urol. Focus, 2016, 2, 210–218 CrossRef.
- S. C. Liu, Y. L. Ying, W. H. Li, Y. J. Wan and Y. T. Long, Chem. Sci., 2021, 12, 3282–3289 RSC.
- R. J. Yu, S. M. Lu, S. W. Xu, Y. J. Li, Q. Xu, Y. L. Ying and Y. T. Long, Chem. Sci., 2019, 10, 10728–10732 RSC.
- L. J. Wang, X. Han, J. G. Qiu, B. Jiang and C. Y. Zhang, Chem. Sci., 2020, 11, 9675–9684 RSC.
- Y. Zhang, C. Wang, X. Zou, X. Tian, J. Hu and C. Y. Zhang, Nano Lett., 2021, 21, 4193–4201 CrossRef CAS PubMed.
- L.-M. Needham, J. Weber, J. A. Varela, J. W. B. Fyfe, D. T. Do, C. K. Xu, L. Tutton, R. Cliffe, B. Keenlyside, D. Klenerman, C. M. Dobson, C. A. Hunter, K. H. Müller, K. O'Holleran, S. E. Bohndiek, T. N. Snaddon and S. F. Lee, Chem. Sci., 2020, 11, 4578–4583 RSC.
- G. N. Samaan, M. K. Wyllie, J. M. Cizmic, L.-M. Needham, D. Nobis, K. Ngo, S. Andersen, S. W. Magennis, S. F. Lee and B. W. Purse, Chem. Sci., 2021, 12, 2623–2628 RSC.
- C. C. Li, W. X. Liu, J. Hu and C. Y. Zhang, Chem. Sci., 2019, 10, 8675–8684 RSC.
- R. Huang, Y. H. Liao, X. M. Zhou, Y. Fu and D. Xing, Sens. Actuators, B, 2017, 247, 505–513 CrossRef CAS.
- P. Fu, S. Xing, M. J. Xu, Y. Zhao and C. Zhao, Sens. Actuators, B, 2020, 305, 127545 CrossRef CAS.
- L. Peng, P. Zhang, Y. Chai and R. Yuan, Anal. Chem., 2017, 89, 5036–5042 CrossRef CAS.
- L. Shi, J. Lei, B. Zhang, B. Li, C. J. Yang and Y. Jin, ACS Appl. Mater. Interfaces, 2018, 10, 12526–12533 CrossRef CAS.
- F. Ma, Q. Zhang and C. Y. Zhang, Nano Lett., 2019, 19, 6370–6376 CrossRef CAS PubMed.
- X. Xue, Y. Liu, Y. Wang, M. Meng, K. Wang, X. Zang, S. Zhao, X. Sun, L. Cui, L. Pan and S. Liu, Oncotarget, 2016, 7, 84508–84519 CrossRef PubMed.
- M. Yang, H. Shen, C. Qiu, Y. Ni, L. Wang, W. Dong, Y. Liao and J. Du, Eur. J. Cancer, 2013, 49, 604–615 CrossRef CAS PubMed.
- H. D. Zhang, X. D. Huang, J. W. Liu and B. H. Liu, Chem. Sci., 2020, 11, 3812–3819 RSC.
- Z. Shen, L. He, W. Wang, L. Tan and N. Gan, Biosens. Bioelectron., 2020, 148, 111831 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1sc02982g |
| ‡ These authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2021 |