 Open Access Article
Open Access ArticleA minimal hybridization chain reaction (HCR) system using peptide nucleic acids†
Ki Tae
Kim
,
Simona
Angerani
and
Nicolas
Winssinger
 *
*
University of Geneva, Switzerland. E-mail: nicolas.winssinger@unige.ch
First published on 6th May 2021
Abstract
The HCR represents a powerful tool for amplification in DNA-based circuitry and sensing applications, yet requires the use of long DNA sequences to grant hairpin metastability. Here we describe a minimal HCR system based on peptide nucleic acids (PNAs). A system comprising a 5-mer stem and 5-mer loop/toehold hairpins was found to be suitable to achieve rapid amplification. These hairpins were shown to yield >10-fold amplification in 2 h and be suitable for the detection of a cancer biomarker on live cells. The use of γ-peg-modified PNA was found to be beneficial.
Introduction
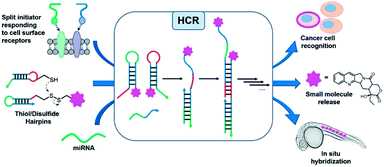
Oligonucleotides are an attractive platform for the design of programmable circuitry, empowering logic gated operations and enzyme-free amplifications. The modular nature of such systems allows a variety of inputs to be integrated into diverse outputs with emerging functions.1 In addition to logic-gated operations, oligonucleotide circuitry lends itself to the amplification mechanism and is being increasingly embraced for sensing applications and smart diagnostics, integrating logic-based operations with an amplification.2–5 A particularly important advent for amplification is the development of the hybridization chain reaction (HCR), where a single strand oligonucleotide initiator opens a metastable hairpin (kinetically trapped) freeing a single strand domain capable of opening a second hairpin to repeat the process in a chain reaction (Fig. 1).6–8 This isothermal polymerization process can be used to amplify the detection of nucleic acids and other biomolecules.9,10 The HCR has also been shown to proceed in live cells.11–13 In addition to a simple amplification, the polymerization reaction can be used to turn on diverse outputs such as fluorescence signals (such as an excimer),14 DNAzyme activity,15 and pro-drug activation,16 and deliver drugs for photodynamic therapy.17 HCRs have been used to detect cell-surface receptors on live cells.18 Furthermore, the ability to initiate an HCR by the proximity of analytes (AND logic gate) has been harnessed to detect specific protein–protein interactions (PPIs), posttranslational modifications19 and specific biomarkers on cancer cells.20 Pierce and coworkers' original report of the HCR made use of hairpins that were 48 nt (18 nt stem and 6 nt loop/toehold).6 Investigations into minimizing this length in order to reduce cost and increase density have been reported with 36–42 nt hairpins.21,22 It has also been demonstrated that HCR systems with hairpins containing shorter stems (12-mers) show faster kinetics (10-fold)21 but the shorter stems compromise the metastability resulting in an untriggered HCR. Surprisingly, the performances of DNA mimics have yet to be explored to further minimize the hairpin size. The unique properties of peptide nucleic acids (PNAs)23–26 are well suited for this goal. PNAs have been shown to form a stable hairpin with just 3 or 4-mer stems with high thermal stability.27 Moreover, modified monomers can further tune the PNA properties. In particular, modification at the gamma position of the PNA backbone28 can be used to tune affinities and its chirality can be exploited for orthogonal hybridizations.29–31 In addition, the enzymatic stability of PNAs against nucleases and proteases would make PNA-based HCRs applicable for in vivo applications.Herein, we report a minimal hybridization chain reaction process enabled by PNAs. Different lengths of hairpin stems and loops were tested, as well as different modifications at the γ-position of PNA monomers (L-serine and mini-peg) in order to identify the minimal structure. The kinetics and extent of amplification were characterized indicating more than 10-fold amplification after two hours of incubation. The promising performance of the PNA-based HCR in vitro was successfully translated in cellulo, where the PNA HCR was applied for the visualization of carbonic anhydrase IX (CA IX) expressed on the surface of cancer cells. Taken together, our results suggest that PNAs are a valid alternative to DNA for the construction of complex networks with a minimal hairpin length.
Results and discussion
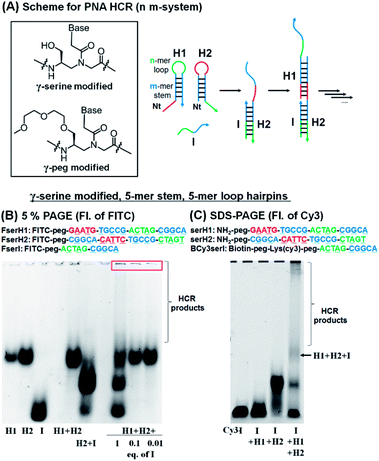
For the HCR to proceed selectively upon addition of a trigger sequence the hairpins must have sufficient metastability (length of the stem). Based on previous results showing that a stem with four G:C pairs can induce stable hairpin structures (Tm = 75 °C),27 we investigated PNA hairpins having a 3-mer loop and 4-mer stem (3,4-system), 4-mer loop and 5-mer stem (4,5-system), and 5-mer loop and 5-mer stem (5,5-system) as HCR scaffolds (Fig. 2A; see Table S1† for full sequences) to gain insight into the minimum length required for a PNA-based HCR. A γ-serine modified PNA backbone was first utilized as it improves the solubility and duplex stability compared to unmodified PNAs.28,32,33 To evaluate the stability of the hairpins (H1 and H2) and the product of the reaction with the initiator strand (I), the PNAs were labelled with fluorescein (FITC), to confer negative charges and thus allow the PNAs to migrate in polyacrylamide gel electrophoresis (PAGE). In a desirable system the initiator strand strongly hybridizes to the first hairpin to form an I:H2 duplex, which in turn opens the second hairpin (H1) for the HCR. Gel analysis conducted on these three systems showed that the 5,5-system gave a >90% hybridization yield for the first reaction step (H2 + I) and that the metastability was sufficient to preclude an untriggered HCR (H1 + H2 does not react, Fig. 2B and S1†). In the case of the 3,4-system, the metastability was sufficient, but the H2 + I product was not observable by gel analysis (Fig. S1†). However, the HCR mixture (H1 + H2 + I) yielded species with a higher molecular weight, suggesting that the duplex H2 + I is present in solution but it is too dynamic to withstand migration through the gel. The 4,5-system showed results that were intermediate between the 3,4- and 5,5-systems, with partial formation of the H2:I duplex observed by PAGE (Fig. S1†). Based on these results, the 5,5-system was adopted as a basic design for further investigations.The addition of 1 equivalent of I (FserI) to H1 and H2 (FserH1 + FserH2) affords a mixture of products as could be anticipated for a statistical reaction, if the rate of initiation is comparable to the rate of the HCR, with higher order HCR oligomers appearing as a smear band. Triggering the HCR with a sub-stoichiometric initiator (0.1 or 0.01 equivalent) led to complete consumption of the initiator but not of H1 and H2 suggesting that the reaction terminates. Moreover, the precipitation of bigger size products was observed in the loading zone of the gel, possibly due to the low solubility of long HCR products (Fig. 2B, red box). These results suggested that the I strand produces desired HCR products; however, the elongation yield of the HCR appeared prematurely terminated (estimated fewer than 10 HCR cycles based on the fact that 0.1 equivalent of initiator did not consume all the hairpins).
To corroborate the results obtained by PAGE, we also sought to analyse the reaction by SDS-PAGE gel electrophoresis and in addition, immobilize the initiator strand on streptavidin beads. In this case, the initiator sequence was labelled with a biotin and Cy3 (BCy3serI). As shown in Fig. 2C, BCy3serI successfully hybridized with the hairpin H2 to produce the H2 + I complex, while showing no interaction with the H1 hairpin, as observed with FserI (Fig. 2B). In the presence of both H1 and H2, the HCR afforded H1 + H2 + I species, together with longer HCR products that showed poor mobility in gel (Fig. 2C, HCR products). Taken together these results suggest that PNAs can be employed as a substrate for HCR systems and that a 5-mer stem is sufficient to provide hairpin metastability. However, the analysis of the HCR was limited, in part by the solubility of HCR products. The quantification of the HCR by fluorescence using BCy3serI immobilized on streptavidin beads and FITC-labelled hairpins indicated that the reaction with 0.1 equivalent of I consumed ca. 50% of the hairpins (Fig. S2†), indicating 5 cycles of the HCR.
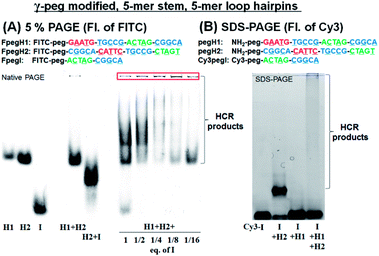
We turned our attention to a γ-peg modified-PNA as such modification was shown to confer high aqueous solubility and low aggregation.34 To allow for direct comparison, the same sequences were used, simply changing the γ-position of the PNA from a serine side chain to polyethylene glycol (peg). Once again, the HCR using FITC-labelled PNAs (FpegH1, FpegH2, and FpegI) was assessed by native PAGE analysis (Fig. 3A). These experiments confirmed the metastability of the hairpins and the efficacy of the initiation. The use of the γ-peg modified-PNA improves the quality of the gels but the initiation of the HCR with a low equivalence of I did not result in the complete consumption of the hairpin after 2 h of reaction (Fig. 3A, 1/8 and 1/16 eq. of I). A similar pattern was observed for SDS-PAGE with γ-peg modified-PNA hairpins (Fig. 3B, 87% consumption of hairpins in the reaction of I + H1 + H2 based on fluorescence integration, and Fig. S3†).
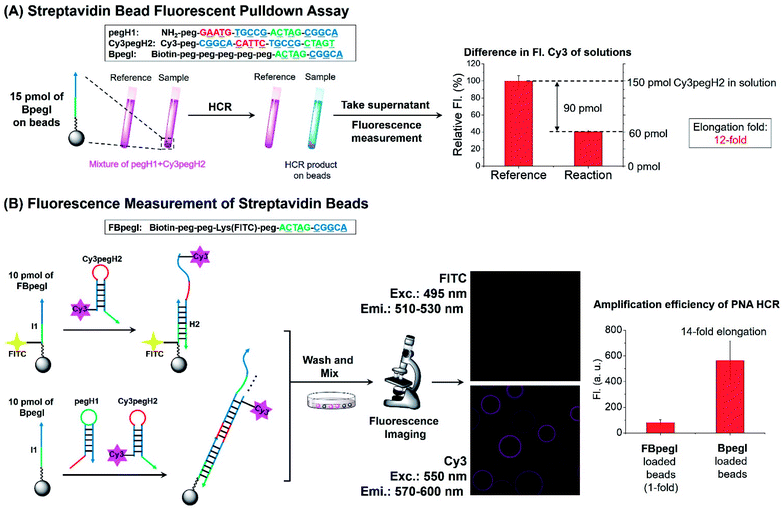
Next, the elongation efficiency of the HCR was evaluated by fluorescence-based techniques. First, a fluorescence pulldown assay was utilized to estimate the number of hairpins that participate in the formation of HCR products. In this experiment, a mixture of pegH1 and Cy3pegH2 was added to streptavidin beads loaded with a biotinylated initiator (BpegI) to trigger the HCR (Fig. 4A). The reaction yield of the HCR was evaluated by the quantification of consumed hairpins upon reaction both in the reaction sample (BpegI + pegH1 + Cy3pegH2) and in a reference sample lacking the initiator strand. Based on this method, we observed that 90 pmol of Cy3pegH2 was consumed by 15 pmol of BpegI, which indicates 12 cycles of the HCR (Fig. 4A). In addition, the reaction was quantified by fluorescence microscopy analysis of streptavidin beads. In this case, two different biotinylated initiators (BpegI and FBpegI) were used; each initiator was loaded on separated beads and the beads containing BpegI were submitted to a regular HCR with pegH1 + Cy3pegH2, while beads with FBpegI (fluorescein labelled) were treated with an excess amount of Cy3-labelled hairpin H2 (Cy3pegH2) but not H1 such that only one cycle of the HCR can take place. After incubation, the beads from each reaction were washed, mixed and screened with a confocal microscope for the quantification of the fluorescence. The beads that underwent the HCR were 7-fold more fluorescent in the Cy3 channel than the beads that were treated with only H2, indicating 14 cycles of the HCR, which is consistent with previous quantification accomplished by fluorescence pulldown assay. The elongation efficiency was confirmed by experiments carried out at different concentrations of reagents and for different reaction times (Fig. S4†). Moreover, size-exclusion chromatography (SEC) experiments further supported the formation of H2 + I, H2 + I + H1 and HCR products with a higher molecular weight (Fig. S5†); the estimated size of the HCR product (57 kDa, ∼8 HCR cycles) is consistent with previous results obtained by fluorescence measurements. Overall, these quantification experiments suggested that the length of HCR products benefited from the use of peg-modified PNA monomers, enabling more than 10 cycles of the HCR to occur (amplification).
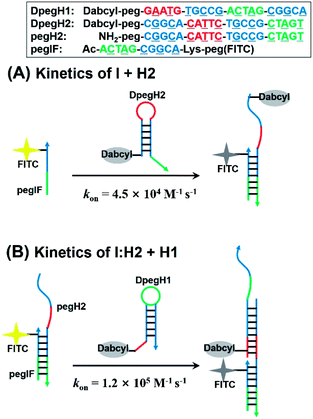
Next, we evaluated the kinetics of hairpin hybridization. FITC or dabcyl quenchers were introduced into PNA strands to monitor the dissociation constants and the rate constants of the first (H2 + I) and second (H2 + I + H1) hybridization steps. The first step was analysed by using a FITC labelled initiator (FpegI) and a dabcyl-modified H2 (DpegH2). Pseudo-first-order kinetics experiments (dabcyl-PNA in excess) revealed a rate constant of 4.5 × 104 M−1 s−1 (kon, Fig. 5A) (half-life = 78 s; kobs = 8.9 × 10−3 s−1, Fig. S6†). Interestingly, the second step (H2:I + H1) showed a 2.6-fold faster reaction rate constant (1.2 × 105 M−1 s−1, Fig. 5B) (half-life = 30 s; kobs = 2.3 × 10−2 s−1, Fig. S6†). These kinetic data are in line with those of previously reported DNA HCR systems.21 The faster reaction for the second may be explained by additional base stacking interaction between pegIF and DpegH1, mediated by the formation of a pegIF:pegH2 duplex.35–37 These results are consistent with previous PAGE results, which showed that some of the initiator remained unreacted when using a stoichiometric ratio of reagents (Fig. 2B, lane H1 + H2 + 1 eq. of I and Fig. 3a), indicating that propagation is faster than initiation.
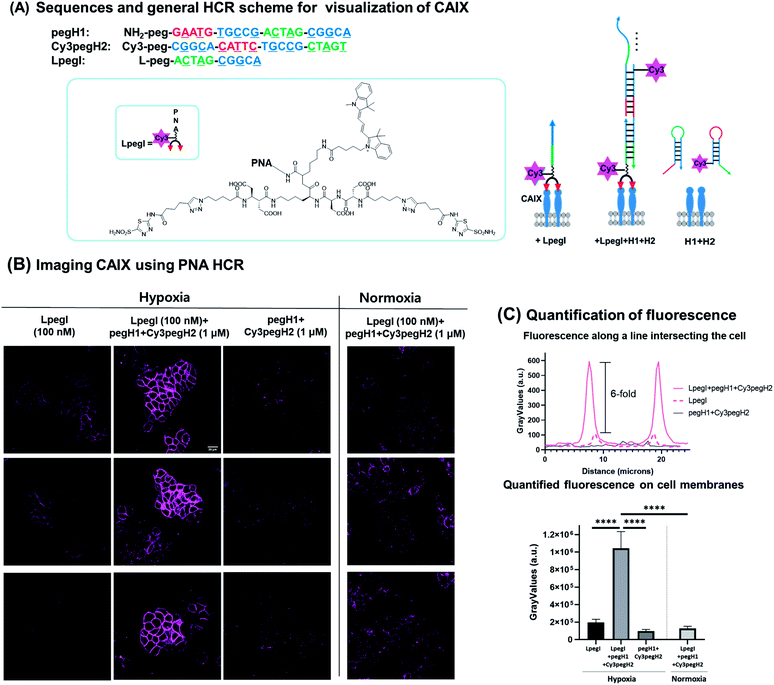
Next, we sought to apply this minimal HCR system for the detection of biomarkers expressed on the surface of cancer cells. We selected carbonic anhydrase IX (CA IX) as the protein of interest based on the fact that it is a marker of hypoxia in various types of solid tumors.38–40 CA IX has been established as a prognostic marker and imaging target for solid tumors.41,42 HT-29, a human colon cancer cell line, was selected as it is known to express CA IX under hypoxic conditions. To ensure anchoring of the initiator strand to the target protein, an acetazolamide-based bivalent ligand43–45 was conjugated to the initiator strand (LpegI) (Fig. 6A). LpegI was also Cy3-labelled to be able to monitor the fluorescence signal arising from direct binding between the ligand initiator strand conjugate and CA IX (Fig. 6A). Upon binding of LpegI (Fig. 6A) to CA IX overexpressing cells (100 nM, 30 min, and hypoxic conditions), treatment with hairpins pegH1 + Cy3pegH2 (1 μM, 1 hour, and hypoxic conditions) triggered the HCR (Fig. 6A and B), resulting in a 6-fold increase in the fluorescent signal (corresponding to 12 elongations) localized on the cell membrane (Fig. 6B and C). As a control, cells treated with hairpins pegH1 + Cy3pegH2 (1 μM, 1 hour, and hypoxic conditions) but lacking the initiator strand showed no fluorescence upon washing (Fig. 6B and C), highlighting the fact that the PNA HCR is suitable for the recognition of biological markers and that the system is specifically responsive to the initiator trigger. Once again, this result was consistent with the previous experiments using loaded beads. Moreover, the system was tested under normoxic conditions (CA IX weakly expressed), showing a dramatically weaker signal (Fig. 6B and C), confirming the specificity of the HCR event as a consequence of the target protein concentration (8-fold difference between hypoxia and normoxia). Accordingly, the results demonstrate that the PNA HCR system can be used to readily visualize CA IX in cellulo. The advantage of an amplified fluorescence signal makes this setup particularly appealing for the imaging of cellular targets with a low expression level. The amplification levels observed are comparable to those of previously reported amplification with a DNA-based HCR.46
Conclusions
In conclusion, we introduced a minimal HCR system based on γ-modified PNAs (including L-serine or peg modification). Among the screened options, a γ-peg-modified 5-mer stem and 5-mer toehold/loop hairpins were found to be better performing, showing the metastability of hairpins together with fast kinetics (1.2 × 105 M−1 s−1) in the chain reaction. The elongation efficiency for the PNA HCR was improved to >10-fold by using the γ-peg-modified PNA, by virtue of the increased solubility of HCR products. The optimized 5,5-system was applied to signal amplification and distinct visualization of CA IX (a cancer biomarker) in live cells by fluorescence. Classic DNA HCR systems require long sequences to allow for the formation of elongated products without leakage (hairpin metastability is lost with stems below a 12 nucleobase length); this work demonstrates that PNAs represent a valuable alternative for the design of minimal systems. Moreover, our results suggest the suitability of the PNA HCR for the detection of targets in biological environments. The work enables the power of HCR amplifications to be tapped with an alternative platform that has significantly different physiochemical properties than DNA.Data availability
Raw data corresponding to the experiments reported have been deposited at (https://zenodo.org/record/4740000#.YJ-x36j7Q2x with DOI: 10.5281/zenodo.4740000).Author contributions
The work was conceived by K. T. K., S. A. and N. W., K. T. K. synthesized and characterized the HCR system. S. A. performed in cellulo experiments. K. T. K., S. A., N. W., analysed and discussed the results. N. W. directed the project. K. T. K., S. A., N. W. wrote the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors thank the SNSF (grant: 200020_169141 and 200020_188406) and the NCCR Chemical Biology for financial support.Notes and references
- E. E. Watson, S. Angerani, P. M. Sabale and N. Winssinger, J. Am. Chem. Soc., 2021, 143, 4467–4482 CrossRef CAS PubMed.
- P. Yin, H. M. T. Choi, C. R. Calvert and N. A. Pierce, Nature, 2008, 451, 318–322 CrossRef CAS PubMed.
- D. Y. Zhang and G. Seelig, Nat. Chem., 2011, 3, 103–113 CrossRef CAS PubMed.
- C. Jung and A. D. Ellington, Acc. Chem. Res., 2014, 47, 1825–1835 CrossRef CAS PubMed.
- K. Gorska and N. Winssinger, Angew. Chem., Int. Ed., 2013, 52, 6820–6843 CrossRef CAS PubMed.
- R. M. Dirks and N. A. Pierce, Proc. Natl. Acad. Sci. U. S. A., 2004, 101, 15275–15278 CrossRef CAS PubMed.
- H. M. Choi, V. A. Beck and N. A. Pierce, ACS Nano, 2014, 8, 4284–4294 CrossRef CAS PubMed.
- H. M. T. Choi, M. Schwarzkopf, M. E. Fornace, A. Acharya, G. Artavanis, J. Stegmaier, A. Cunha and N. A. Pierce, Development, 2018, 145, dev165753 CrossRef PubMed.
- H. M. Choi, J. Y. Chang, A. Trinh le, J. E. Padilla, S. E. Fraser and N. A. Pierce, Nat. Biotechnol., 2010, 28, 1208–1212 CrossRef CAS PubMed.
- S. Bi, S. Yue and S. Zhang, Chem. Soc. Rev., 2017, 46, 4281–4298 RSC.
- Z. Cheglakov, T. M. Cronin, C. He and Y. Weizmann, J. Am. Chem. Soc., 2015, 137, 6116–6119 CrossRef CAS PubMed.
- Z. Wu, G. Q. Liu, X. L. Yang and J. H. Jiang, J. Am. Chem. Soc., 2015, 137, 6829–6836 CrossRef CAS PubMed.
- L. Li, J. Feng, H. Liu, Q. Li, L. Tong and B. Tang, Chem. Sci., 2016, 7, 1940–1945 RSC.
- J. Huang, Y. Wu, Y. Chen, Z. Zhu, X. Yang, C. J. Yang, K. Wang and W. Tan, Angew. Chem., Int. Ed., 2011, 50, 401–404 CrossRef CAS PubMed.
- F. Wang, J. Elbaz, R. Orbach, N. Magen and I. Willner, J. Am. Chem. Soc., 2011, 133, 17149–17151 CrossRef CAS PubMed.
- A. L. Prinzen, D. Saliba, C. Hennecker, T. Trinh, A. Mittermaier and H. F. Sleiman, Angew. Chem., Int. Ed., 2020, 59, 12900–12908 CrossRef CAS PubMed.
- J. Wang, D. X. Wang, J. Y. Ma, Y. X. Wang and D. M. Kong, Chem. Sci., 2019, 10, 9758–9767 RSC.
- G. Zhu, S. Zhang, E. Song, J. Zheng, R. Hu, X. Fang and W. Tan, Angew. Chem., Int. Ed., 2013, 52, 5490–5496 CrossRef CAS PubMed.
- B. Koos, G. Cane, K. Grannas, L. Lof, L. Arngarden, J. Heldin, C. M. Clausson, A. Klaesson, M. K. Hirvonen, F. M. S. de Oliveira, V. O. Talibov, N. T. Pham, M. Auer, U. H. Danielson, J. Haybaeck, M. Kamali-Moghaddam and O. Soderberg, Nat. Commun., 2015, 6, 7294 CrossRef CAS PubMed.
- X. Chang, C. Zhang, C. Lv, Y. Sun, M. Z. Zhang, Y. M. Zhao, L. L. Yang, D. Han and W. H. Tan, J. Am. Chem. Soc., 2019, 141, 12738–12743 CrossRef CAS PubMed.
- Y. S. Ang and L. Y. L. Yung, Chem. Commun., 2016, 52, 4219–4222 RSC.
- Y. Tsuneoka and H. Funato, Front. Mol. Neurosci., 2020, 13, 75 CrossRef CAS PubMed.
- M. Egholm, O. Buchardt, L. Christensen, C. Behrens, S. M. Freier, D. A. Driver, R. H. Berg, S. K. Kim, B. Norden and P. E. Nielsen, Nature, 1993, 365, 566–568 CrossRef CAS PubMed.
- P. E. Nielsen, M. Egholm, R. H. Berg and O. Buchardt, Science, 1991, 254, 1497–1500 CrossRef CAS PubMed.
- S. Barluenga and N. Winssinger, Acc. Chem. Res., 2015, 48, 1319–1331 CrossRef CAS PubMed.
- J. Saarbach, P. M. Sabale and N. Winssinger, Curr. Opin. Chem. Biol., 2019, 52, 112–124 CrossRef CAS PubMed.
- T. Machida, S. Dutt and N. Winssinger, Angew. Chem., Int. Ed., 2016, 55, 8595–8598 CrossRef CAS PubMed.
- A. Dragulescu-Andrasi, S. Rapireddy, B. M. Frezza, C. Gayathri, R. R. Gil and D. H. Ly, J. Am. Chem. Soc., 2006, 128, 10258–10267 CrossRef CAS PubMed.
- J. Sacui, W. C. Hsieh, A. Manna, B. Sahu and D. H. Ly, J. Am. Chem. Soc., 2015, 137, 8603–8610 CrossRef PubMed.
- D. Chang, E. Lindberg and N. Winssinger, J. Am. Chem. Soc., 2017, 139, 1444–1447 CrossRef CAS PubMed.
- S. A. Thadke, V. M. Hridya, J. D. R. Perera, R. R. Gil, A. Mukherjee and D. H. Ly, Commun. Chem., 2018, 1, 79 CrossRef.
- J. M. Vieville, S. Barluenga, N. Winssinger and M. A. Delsuc, Biophys. Chem., 2016, 210, 9–13 CrossRef CAS PubMed.
- A. Manicardi and R. Corradini, Artificial DNA: PNA & XNA, 2014, 5, e1131801 Search PubMed.
- B. Sahu, I. Sacui, S. Rapireddy, K. J. Zanotti, R. Bahal, B. A. Armitage and D. H. Ly, J. Org. Chem., 2011, 76, 5614–5627 CrossRef CAS PubMed.
- B. F. Yuan, X. Y. Zhuang, Y. H. Hao and Z. Tan, Chem. Commun., 2008, 6600–6602, 10.1039/b812929k.
- K. T. Kim and N. Winssinger, Chem. Sci., 2020, 11, 4150–4157 RSC.
- K. T. Kim, S. Angerani, D. L. Chang and N. Winssinger, J. Am. Chem. Soc., 2019, 141, 16288–16295 CrossRef CAS PubMed.
- K. Maseide, R. A. Kandel, R. S. Bell, C. N. Catton, B. O'Sullivan, J. S. Wunder, M. Pintilie, D. Hedley and R. P. Hill, Clin. Cancer Res., 2004, 10, 4464–4471 CrossRef CAS PubMed.
- S. Pastorekova and R. J. Gillies, Cancer Metastasis Rev., 2019, 38, 65–77 CrossRef CAS PubMed.
- N. K. Tafreshi, M. C. Lloyd, M. M. Bui, R. J. Gillies and D. L. Morse, Subcell. Biochem., 2014, 75, 221–254 CAS.
- B. Muz, P. de la Puente, F. Azab and A. K. Azab, Hypoxia, 2015, 3, 83–92 CrossRef PubMed.
- S. W. Choi, J. Y. Kim, J. Y. Park, I. H. Cha, J. Kim and S. Lee, Hum. Pathol., 2008, 39, 1317–1322 CrossRef CAS PubMed.
- N. Krall, F. Pretto and D. Neri, Chem. Sci., 2014, 5, 3640–3644 RSC.
- M. Wichert and N. Krall, Curr. Opin. Chem. Biol., 2015, 26, 48–54 CrossRef CAS PubMed.
- S. Angerani and N. Winssinger, J. Am. Chem. Soc., 2020, 142, 12333–12340 CrossRef CAS PubMed.
- X. H. Wen, B. Y. Yuan, J. X. Zhang, X. X. Meng, Q. P. Guo, L. Li, Z. H. Li, H. S. Jiang and K. M. Wang, Chem. Commun., 2019, 55, 6114–6117 RSC.
Footnote |
| † Electronic supplementary information (ESI) available: Fig. S1–S10 and Tables S1 and S2; procedures. See DOI: 10.1039/d1sc01269j |
| This journal is © The Royal Society of Chemistry 2021 |

![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
: