Open Access Article
Open Access ArticleConstruction of diverse peptide structural architectures via chemoselective peptide ligation†
Carina Hey Pui
Cheung‡
a,
Jianchao
Xu‡
a,
Chi Lung
Lee
 a,
Yanfeng
Zhang
a,
Ruohan
Wei
a,
Donald
Bierer
b,
Xuhui
Huang
a,
Yanfeng
Zhang
a,
Ruohan
Wei
a,
Donald
Bierer
b,
Xuhui
Huang
 c and
Xuechen
Li
c and
Xuechen
Li
 *a
*a
aDepartment of Chemistry, State Key Lab of Synthetic Chemistry, The University of Hong Kong, Hong Kong. E-mail: xuechenl@hku.hk
bDepartment of Medicinal Chemistry, Bayer AG, Aprather Weg 18A, 42096 Wuppertal, Germany
cDepartment of Biological and Chemical Engineering, The Hong Kong University of Science and Technology, Clear Water Bay, Kowloon, Hong Kong
First published on 13th April 2021
Abstract
Herein, we report the development of a facile synthetic strategy for constructing diverse peptide structural architectures via chemoselective peptide ligation. The key advancement involved is to utilize the benzofuran moiety as the peptide salicylaldehyde ester surrogate, and Dap–Ser/Lys–Ser dipeptide as the hydroxyl amino functionality, which could be successfully introduced at the side chain of peptides enabling peptide ligation. With this method, the side chain-to-side chain cyclic peptide, branched/bridged peptides, tailed cyclic peptides and multi-cyclic peptides have been designed and successfully synthesized with native peptidic linkages at the ligation sites. This strategy has provided an alternative strategic opportunity for synthetic peptide development. It also serves as an inspiration for the structural design of PPI inhibitors with new modalities.
Introduction
Protein–protein interactions (PPIs) are involved in many biological processes, such as antigen–antibody, ligand–GPCRs, and substrate–enzymes.1–3 Abnormal PPIs driving signaling changes can be pathogenic, hence PPIs are considered as the therapeutic targets for various diseases.4–7 Interests in targeting disease-associated PPIs have been growing in both academia and industry. The rationale behind the inhibitor design is to block the PPIs through mimicry of the topologically defined regions. However, molecular targeting in PPIs is extremely challenging due to the flat and shallow binding pocket in proteins.4,8 Hence, PPIs have long been considered to be undruggable using traditional small molecules with molecular weight less than 500 as the disrupters/inhibitors.9 To address this issue, middle-sized modalities, such as peptides and peptidomimetics have been explored.10,11 Synthetic peptides have been reported to have higher potency and specificity in targeting PPIs, not only because they possess the capability in binding to the large grooves on the interacting face, but their residues can also be modified to mimic the conformational features of the protein domain at the binding interface and disrupt the PPIs.8 More importantly, compared with the traditional small molecules-based drugs, peptide drugs being able to bind to the PPI targets more specifically implies less off-target effects.12 Moreover, peptide drugs have also been reported to have less cytotoxicity.13Although the peptides represent a promising class of therapeutic drugs in various therapeutic areas, they suffer from some limitations.6 For example, linear peptides are not stable in vivo and are prone to protease degradation.4,14,15 They also have poor permeability to access desirable intracellular targets.4,8 Furthermore, the binding of peptides to the PPI interface may not be always thermodynamically favorable as they will need to overcome the entropic penalty to reorganize themselves into its constrained bioactive state.4 To overcome the intrinsic limitations of the natural peptides, structural modifications with unnatural elements are being explored. One of the most notable strategies is to develop side chain-to-side chain cyclic peptides from the mimicry of interfacial α-helical domains.16–18 Previous studies have shown that via varying the structural design including the stapled positions, structures, lengths and peptide sequence, one can change the dynamics of the peptide–protein interaction8 thus optimize the engagement of inhibitors at the PPIs interface. Peptide drugs with optimized structural designs were found having higher protease resistance, biological potency and binding affinity.17 Successful examples of stapled peptides offering therapeutic modality include all-hydrocarbon-linked stapled peptide ALRN-6924 which is under clinical development as an anti-cancer drug targeting HDM2/p53.19,20 Other examples include peptides targeting HIV integrase, BCl-2 and β-catenin.21–23 Apart from the stapling chemistry, some other motifs have been explored, including the β-strands mimetics24 and loops motif25 that display more complex topologies. Examples of tertiary mimetics as PPI inhibitors has also been reported, including α- and α/β-peptides derived from the “Z-domain” scaffold.26 As PPIs have pivotal roles in the regulation of biological systems, novel and practical tools for the generation of new peptide architectures and structural complexities will be worth being explored.
Here, we report the development of chemical ligation chemistry for constructing diverse peptide structural motifs, including side chain-to-side chain cyclic peptides, branched and bridged peptides, tailed cyclic peptides and multi-cyclic peptides. We expect these peptides will represent new structural motifs and offer new modalities for developing inhibitors of PPIs with enhanced stability and binding affinity.
Results and discussion
Our design involves using chemoselective peptide ligation to link the side chain unprotected (cyclic)peptide segments for architecture construction. To this end, the reacting groups necessary for executing ligation need be installed at the side chain of the peptide. This is a challenging task and has not been well explored in the literature. The notable native chemical ligation (NCL) requires C-terminal thioesters and N-terminal cysteinyl peptide fragments as the requisite reacting counterparts.27 A side chain NCL was reported between the N-linked glycosyl auxiliary to the thioester side chain of an N-terminal aspartate oligopeptide.28 A drawback is that extra orthogonal protecting groups for Asp/Glu are needed for selective deprotection of the aspartic acid side chain protecting group prior to thioester formation.28 Another limitation of this technique is that the thioester generated will be easily decomposed upon piperidine treatment in Fmoc-solid phase peptide synthesis (SPPS).29Peptide C-terminal salicylaldehyde esters have been developed to ligate side chain unprotected peptides with N-terminal Ser/Thr/Cys/Pen for Ser/Thr ligation (STL)30,31 and Cys/Pen ligation (CPL)32 used in chemical protein synthesis. The chemoselective reaction between the two counterparts affords an acid-cleavable N,O/S-benzylidene acetal intermediate. Followed by acidolysis, the native peptidic linkage is generated at the ligation junction. Apart from linear peptide/protein synthesis, intramolecular Ser/Thr ligation has been developed for peptide cyclization to produce head-to-tail cyclic peptides of various sizes.33–39 In this paper, we ought to further expand the scope of Ser/Thr ligation by solving the problem of introducing the peptide salicylaldehyde ester on the side chain, with which to construct diverse peptide scaffolds.
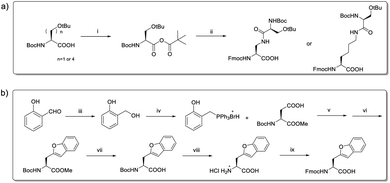
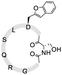
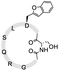
To install these two reactive groups onto the side chain of the unprotected peptides for executing side-chain STL, we first identified the Dap–Ser or Lys–Ser dipeptide as the side chain hydroxyl amino functionality. The Dap–Ser or Lys–Ser dipeptide was successfully synthesized via mixed anhydride derivatives from isobutyl chloroformate (Scheme 1a). Boc-Ser(tBu)-OH was first dissolved in dry CH2Cl2, isobutyl chloroformate and DIEA were then added at 0 °C to afford the mixed anhydride. The amine nucleophile, Fmoc-Dap-OH or Fmoc-Lys-OH, was added after an hour to generate the dipeptide ready for SPPS.
The introduction of the salicylaldehyde ester to the side chain of Asp/Glu of the peptide is very challenging. As priorly mentioned, there is no simple and direct method reported yet for synthesizing peptides with Asp/Glu side chain active esters. The complication occurs as the coupling of Asp (O-salicylaldehyde ester) or any Asp active ester building block will lead to aspartimide formation during SPPS or the condensation step. Moreover, the Asp/Glu active esters cannot survive piperidine treatment during Fmoc-SPPS. All previous strategies for synthesizing peptide C-terminal salicylaldehyde esters are not applicable for introducing salicylaldehyde esters at the peptide side chain.31 To solve this problem, we have come to identify a novel synthetic strategy utilizing the benzofuran moiety as the peptide crypto-salicylaldehyde ester. Upon ozonolysis, the benzofuran moiety could readily become a salicylaldehyde ester for subsequent chemoselective ligation.
Previous synthetic routes to benzofuranylalanine via transition metal-catalyzed or chemoenzymatic synthesis were reported.40,41 In our design, preparation of the benzofuran moiety started from salicylaldehyde. Salicylaldehyde was first treated with reducing agent NaBH4, followed by reflux with triphenylphosphine hydrobromide. The generated 2-hydroxybenzyltriphenylphosphonium bromide was then added to Boc-Asp-OMe and transformed into an activated ester intermediate upon DIC treatment (Scheme 1b). Subsequently, the resulted intermediate was refluxed with triethylamine to offer the benzofuran moiety. Finally, the desired benzofuran building block was obtained by C-terminal methyl ester group deprotection, followed by N-terminal Boc group deprotection and Fmoc group installation. As the Fmoc-Asp(benzofuran)-OH moiety is stable under both acidic and basic conditions during peptide synthesis, it could be directly used in Fmoc-SPPS. We also obtained Fmoc-Glu(benzofuran)-OH in similar route (see ESI†).
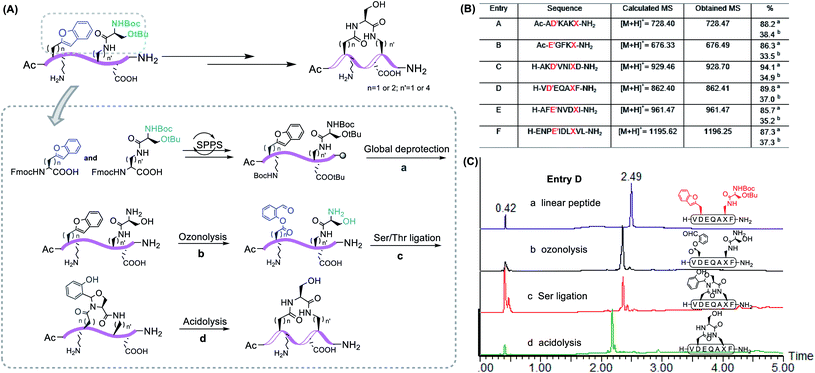
With these building blocks in hand, we continued to incorporate them into the peptide to execute our construction plan. As a proof of concept, the linear peptide with Dap–Ser and Asp(benzofuran) could be readily synthesized under standard Fmoc-SPPS conditions.
After global deprotection, the resultant side chain unprotected peptides were treated with ozonolysis in H2O/ACN (v![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) v = 1
v = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
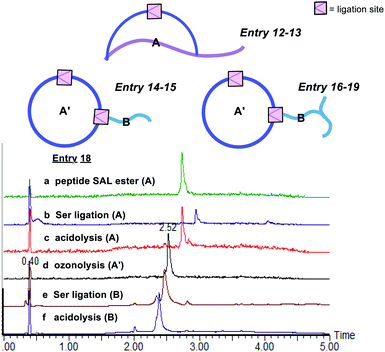
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) with 0.7% TFA under 0 °C for 1 minute, affording the peptide salicylaldehyde ester smoothly. It is noting that oxidation-prone residues are not compatible with the ozone treatment, i.e. Cys and Met react with ozone at the sulfhydryl to give sequential O-atom addition products,42 and Trp becomes kynurenine after ozonolysis.43 Therefore, these amino acid residues are excluded from the chemical space available for this chemistry. Subsequently, side chain-to-side chain Ser/Thr ligation with unprotected peptides was performed. The peptide was dissolved in pyridine acetate buffer at 0.5 mM concentration, which spontaneously proceeded to cyclize, followed by acidolysis to cleave the N,O-benzylidene acetal intermediate to afford the cyclic peptides with an overall yield of ∼35% after one HPLC purification. With this strategy, we were able to generate Asp–Dap/Lys lactam linkage with a hydrophilic hydroxyl group on different peptide sequences (Fig. 2). Notably, the chemoselective cyclization can tolerate most of the side chain functionalities presenting in the peptide except oxidation-prone residues.
1) with 0.7% TFA under 0 °C for 1 minute, affording the peptide salicylaldehyde ester smoothly. It is noting that oxidation-prone residues are not compatible with the ozone treatment, i.e. Cys and Met react with ozone at the sulfhydryl to give sequential O-atom addition products,42 and Trp becomes kynurenine after ozonolysis.43 Therefore, these amino acid residues are excluded from the chemical space available for this chemistry. Subsequently, side chain-to-side chain Ser/Thr ligation with unprotected peptides was performed. The peptide was dissolved in pyridine acetate buffer at 0.5 mM concentration, which spontaneously proceeded to cyclize, followed by acidolysis to cleave the N,O-benzylidene acetal intermediate to afford the cyclic peptides with an overall yield of ∼35% after one HPLC purification. With this strategy, we were able to generate Asp–Dap/Lys lactam linkage with a hydrophilic hydroxyl group on different peptide sequences (Fig. 2). Notably, the chemoselective cyclization can tolerate most of the side chain functionalities presenting in the peptide except oxidation-prone residues.
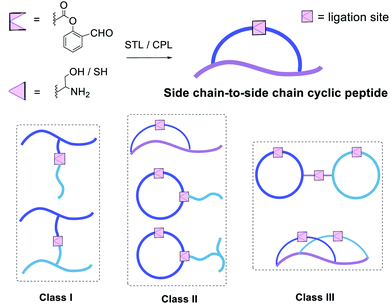
These results were very encouraging and opened up new opportunities to construct novel peptide structures via chemical ligation. We envisaged that our strategy could be flexibly applied to afford innovative peptide modalities via ligating peptides with side chain salicylaldehyde ester with other peptides carrying Dap/Lys–Ser dipeptide or N-terminal Ser/Thr/Cys/Pen. Thus, in addition to the side chain-to-side chain cyclic peptides, we continued to apply this new chemistry to synthesize three classes of structural motifs, as shown in Fig. 1.
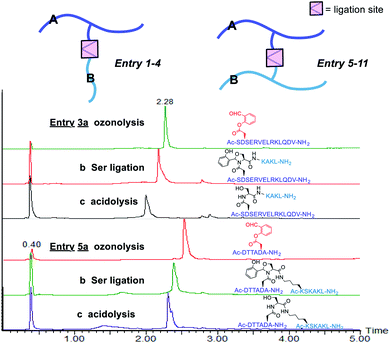
The first class includes branched peptide and bridged peptide (Fig. 3 and Table 1). The peptide with the side chain salicylaldehyde reacted with another peptide carrying N-terminal Ser/Thr under the Ser/Thr ligation conditions to produce the branched peptide (Table 1, entries 1–4). Alternatively, the peptide with side chain salicylaldehyde ester can ligate with another peptide carrying side chain Dap–Ser or Lys–Ser under the Ser/Thr ligation conditions to produce bridged peptides (Table 1, entries 5–11). Such structural motifs will allow combining two functional peptides to generate a hybrid peptide with dual activities.
| Entry | A | B | % |
|---|---|---|---|
| a D′ = Asp(benzofuran), (K–S) = Lys–Ser dipeptide. b Percentage conversion to product based on UPLC-MS analysis of the crude reaction mixture. c Isolated yield based on ligation. | |||
| 1 | Ac-DTTAD′A-NH2 | H-SNVKAQFL-NH2 | 78.2b, 36.6c |
| 2 | Ac-SRQQGESNQERGARD′RL-NH2 | H-SKAKL-NH2 | 80.0b, 39.4c |
| 3 | Ac-D′SERVELRKLQDV-NH2 | H-SKAKL-NH2 | 95.0b, 39.1c |
| 4 | H-VIGGVGD′N-NH2 | H-TLHAPTD-OH | 89.6b, 32.8c |
| 5 | Ac-DTTAD′A-NH2 | Ac-(K–S)SKAKL-NH2 | 90.5b, 48.5c |
| 6 | Ac-SRQQGESNQERGARD′RL-NH2 | Ac-(K–S)SKAKL-NH2 | 91.9b, 53.0c |
| 7 | Ac-SRQQGESNQERGARD′RL-NH2 | Ac-(K–S)SARKYFAGNLPE-NH2 | 95.6b, 37.4c |
| 8 | Ac-D′SERVELRKLQDV-NH2 | Ac-(K–S)SKAKL-NH2 | 88.9b, 33.3c |
| 9 | Ac-D′SERVELRKLQDV-NH2 | H-NIGTYLP(K–S)NVK-NH2 | 93.4b, 31.1c |
| 10 | Ac-D′SERVELRKLQDV-NH2 | Ac-ARE(K–S)TPEP-NH2 | >98b, 38.4c |
| 11 | Ac-LSQD′RG-NH2 | Ac-ARE(K–S)TPEP-NH2 | 91.7b, 34.7c |
The second class includes tailed cyclic peptides (Fig. 4 and Table 2). Due to the compact structures of cyclic peptides and their ultra-stability against enzymatic degradation and physical denaturation, cyclic peptides have attracted a lot of attention in the pharmaceutical industry. In this regard, the development of effective methods for peptide cyclization has been an intensively researched subject. In our design, a peptide with the side chain salicylaldehyde ester and N-terminal Ser/Thr could cyclize under the Ser/Thr ligation condition to generate head-to-side chain cyclic peptides (Table 2, entries 12 and 13). Furthermore, a cyclic peptide with the side chain salicylaldehyde ester could react with another peptide with side chain Lys–Ser to generate cyclic peptide with a branched tail (Table 2, entries 14–19). Our strategy not only provides an effective synthetic method but also generates new structural motifs.
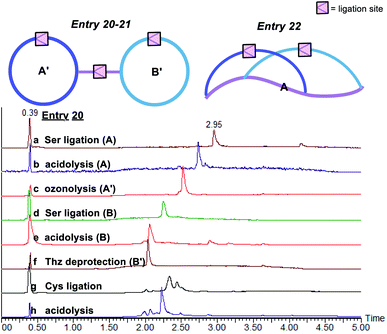
The third class of peptide structural architectures includes the bridged cyclic and embedded bicyclic peptides (Fig. 5 and Table 3). One side chain-to-tail cyclic peptide with free N-terminal Cys could react with another head-to-tail cyclic peptide with side chain salicylaldehyde ester under Cys ligation conditions to generate a bridged cyclic peptide, which will provide a way to conjugate cell-penetrating peptide to a peptide ligand. Alternatively, the embedded bicyclic peptides could be synthesized by performing first the side chain-to-tail Ser/Thr ligation involving the side chain Lys–Ser and C-terminal salicylaldehyde ester, followed by the head-to-side chain Ser/Thr ligation involving the N-terminal Ser/Thr and the side chain salicylaldehyde ester. Based on this synthetic route, we envisioned that cyclic peptides of a higher order would be achievable through the rational design of combining the above bridged cyclization and embedded bicyclization approaches.
| Entry | A | A′ | B | B′ | A′ + B′ | ||
|---|---|---|---|---|---|---|---|
| Sequence | Sequence | % | Sequence | Sequence | % | % | |
| a D′ = Asp(benzofuran), (K–S) = Lys–Ser dipeptide, SAL = salicylaldehyde. b Percentage conversion to the ligation product based on UPLC-MS analysis of the crude reaction mixture. c Isolated yield based on ligation. d Isolated yield over 2 steps (intramolecular STL and one-pot Thz deprotection). e Isolated yield over 2 steps (intramolecular STL and one-pot Fmoc deprotection). | |||||||
| 20 | H-SD′LSQRG-SAL ester |
|
>98a, 48.0b | Boc-(Thz)(K–S)LSQRG-SAL ester |
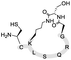
|
88.9b, 37.2d | 84.3b, 50.4c |
| 21 | Boc-(Thz)NFS(K–S)QSNKRFLSKTQG-SAL ester |

|
90.1b, 35.4d | 89.9b, 45.8c | |||
Conclusions
In summary, we have developed a facile synthetic strategy for constructing diverse peptide structural motifs via chemoselective peptide ligation. The key advancement of our synthetic strategy is to utilize the benzofuran moiety as the peptide salicylaldehyde ester surrogate, and Dap–Ser/Lys–Ser dipeptide as the hydroxyl amino functionality. These two building blocks could be incorporated into the peptide side chain under the standard Fmoc-SPPS conditions. After global deprotection, the desired peptide salicylaldehyde ester could be efficiently generated upon ozonolysis. Subsequently, Ser/Thr ligation or Cys/Pen ligation is performed to covalently link up the two reacting counterparts to form different peptide architectures. As a proof of concept, different models of side chain-to-side chain cyclic peptides were successfully synthesized. We then proposed that the application scope of this strategy could be further expanded to afford innovative peptide modalities. In addition to the side chain-to-side chain cyclic peptides, three classes of structural motifs have been designed and successfully synthesized by ligation. Peptides with side chain salicylaldehyde ester have reacted smoothly with different peptide counterparts carrying Dap/Lys–Ser dipeptide or N-terminal Ser/Thr/Cys/Pen to generate the native peptidic linkages at the ligation sites. These results have demonstrated the flexibility of the chemical ligation approach to construct diverse peptide structural architectures. As ozone is a strong oxidant, the oxidizable amino acid acids (i.e. Trp, Met, and Cys) are not compatible with this method, which presents the limitation of this method. Nevertheless, we expect this strategy to provide an alternative strategic opportunity for synthetic peptide development.Foreseeing the vast potential of peptide drugs in modulating PPIs, this work presented here delineates a blueprint and serves as an inspiration for the structural design of PPI inhibitors with new modalities, which empowers future targeting of “undruggable” proteins targets. Based on the previous studies by other groups, having a rational structure-based design of therapeutic peptides is crucial for them to achieve higher specificity and biostability. Gratifyingly, our strategy has provided easy access to a diverse class of peptide structural motifs. More importantly, by ligating two functional peptides, the structural motifs synthesized would be a hybrid peptide with dual activities. In long term, the synthesis of more complex bioactive scaffolds will be studied while the developed modalities will be rationally optimized for potential therapeutic applications.
Author contributions
X. Li, D. Bierer and X. Huang conceived the presented prject. X. Li supervised the project. C. H. P. Cheung, J. Xu, C. L. Lee, Y. Zhang and R. Wei performed the chemical synthesis of building blocks and side chain-to-side chain cyclic peptides. C. H. P. Cheung performed the chemical synthesis of the peptide structural motifs.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the Hong Kong Research Grant Council of Hong Kong (HKUST C6009-15G, 17309616, AoE/M-09/12 and AoE/P-705/16).References
- D. R. Davies and G. H. Cohen, Proc. Natl. Acad. Sci. U. S. A., 1996, 93, 7–12 CrossRef CAS PubMed.
- S. Jones and J. M. Thornton, Proc. Natl. Acad. Sci. U. S. A., 1996, 93, 13–20 CrossRef CAS PubMed.
- D. M. Rosenbaum, S. G. Rasmussen and B. K. Kobilka, Nature, 2009, 459, 356–363 CrossRef CAS PubMed.
- J. A. Wells and C. L. McClendon, Nature, 2008, 450, 1001–1009 CrossRef PubMed.
- D. E. Scott, A. R. Bayly, C. Abell and J. Skidmore, Nat. Rev. Drug Discovery, 2016, 15, 533–550 CrossRef CAS PubMed.
- Z. Li, et al. , Nat. Commun., 2017, 8, 14356 CrossRef CAS PubMed.
- H. Lu, Q. Zhou, J. He, Z. Jiang, C. Peng, R. Tong and J. Shi, Signal Transduction Targeted Ther., 2020, 5, 213 CrossRef PubMed.
- Y. S. Tan, D. P. Lane and C. S. Verma, Drug Discovery Today, 2016, 21, 1642–1653 CrossRef CAS PubMed.
- G. Zinzalla and D. E. Thurston, Future Med. Chem., 2009, 1, 65–93 CrossRef CAS PubMed.
- O. Sillerud and R. S. Larson, Curr. Protein Pept. Sci., 2005, 6, 151–169 CrossRef PubMed.
- A. D. Cunningham, N. Qvit and D. Mochly-Rosen, Curr. Opin. Struct. Biol., 2017, 44, 59–66 CrossRef CAS PubMed.
- C. J. Brown, et al. , ACS Chem. Biol., 2012, 8, 506–512 CrossRef PubMed.
- A. C. Lee, J. L. Harris, K. K. Khanna and J. A. Hong, Int. J. Mol. Sci., 2019, 20, 2383 CrossRef PubMed.
- P. Vlieghe, V. Lisowski, J. Martinez and M. Khrestchatisky, Drug Discovery Today, 2010, 15, 40–56 CrossRef CAS PubMed.
- J. Zhu, et al. , Cell Rep., 2017, 21, 3781–3793 CrossRef CAS PubMed.
- G. L. Verdine and G. J. Hilinski, Methods Enzymol., 2012, 503, 3–33 CAS.
- T. A. Hill, N. E. Shepherd, F. Diness and D. P. Fairlie, Angew. Chem., Int. Ed., 2014, 53, 13020–13041 CrossRef CAS PubMed.
- A. M. Watkins, M. G. Wuo and P. S. Arora, J. Am. Chem. Soc., 2015, 137, 11622–11630 CrossRef CAS PubMed.
- L. A. Carvajal, et al. , Sci. Transl. Med., 2018, 10, eaao3003 CrossRef PubMed.
- Aileron, Pipeline ALRN-6924, 2019, retrieved from https://www.aileronrx.com/pipeline Search PubMed.
- S. K. Sia, et al. , Proc. Natl. Acad. Sci. U. S. A., 2002, 99, 14664–14669 CrossRef CAS PubMed.
- H. Zhang, et al. , J. Mol. Biol., 2008, 378, 565–580 CrossRef CAS PubMed.
- S. A. Kawamoto, A. Coleska, X. Ran, H. Yi, C. Y. Yang and S. Wang, J. Med. Chem., 2012, 55, 1137–1146 CrossRef CAS PubMed.
- D. Xin, L. M. Perez, T. R. Ioerge and K. Burgess, Angew. Chem., 2014, 126, 3668–3672 CrossRef.
- P. Timmerman, J. Beld, W. C. Puijk and R. H. Meloen, ChemBioChem, 2005, 6, 821–824 CrossRef CAS PubMed.
- J. W. Checco, et al. , Proc. Natl. Acad. Sci. U. S. A., 2015, 112, 4552–4557 CrossRef CAS PubMed.
- P. E. Dawson, T. W. Muir, I. Clark-Lewis and S. B. H. Kent, Science, 1994, 266, 776–779 CrossRef CAS PubMed.
- H. Chai, L. M. K. Hoang, M. D. Vu, K. Pasunooti, C. Liu and X. Liu, Angew. Chem., 2016, 128, 10519–10523 CrossRef.
- X. Li, T. Kawakami and S. Aimoto, Tetrahedron Lett., 1998, 39, 8669–8672 CrossRef CAS.
- (a) X. Li, H. Y. Lam, Y. Zhang and C. K. Chan, Org. Lett., 2010, 12, 1724–1727 CrossRef CAS PubMed; (b) Y. Zhang, C. Xu, H. Y. Lam, C. L. Lee and X. Li, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 6657–6662 CrossRef CAS PubMed; (c) C. L. Lee, H. Liu, C. T. T. Wong, H. Y. Chow and X. Li, J. Am. Chem. Soc., 2016, 138, 10477–10484 CrossRef CAS PubMed.
- (a) H. Liu and X. Li, Acc. Chem. Res., 2018, 51, 1643–1655 CrossRef CAS PubMed; (b) Y. Tan, T. Wei, H. Wu and X. Li, J. Am. Chem. Soc., 2020, 142, 20288–20298 CrossRef CAS PubMed.
- Y. Tan, J. Li, K. Jin, J. Liu, Z. Chen, J. Yang and X. Li, Angew. Chem., 2020, 132, 12841–12845 CrossRef.
- C. T. T. Wong, H. Y. Lam and X. Li, Org. Biomol. Chem., 2013, 11, 7616–7620 RSC.
- H. Y. Lam, Y. Zhang, H. Liu, J. Xu, C. T. T. Wong, C. Xu and X. Li, J. Am. Chem. Soc., 2013, 135, 6272–6279 CrossRef CAS PubMed.
- C. T. T. Wong, H. Y. Lam, T. Song, G. Chen and X. Li, Angew. Chem., 2013, 52, 10212–10215 CrossRef CAS PubMed.
- C. T. T. Wong, H. Y. Lam and X. Li, Tetrahedron Lett., 2014, 70, 7770–7773 CrossRef CAS.
- K. Jin, I. H. Sam, K. H. L. Po, D. Lin, E. H. Ghazvini Zadeh, S. Chen, Y. Yuan and X. Li, et al. , Nat. Commun., 2016, 7, 12394 CrossRef CAS PubMed.
- H. Y. Lam and X. Li, Synlett, 2014, 25, 1339–1344 CrossRef.
- C. L. Lee, H. Y. Lam and X. Li, Nat. Prod. Rep., 2015, 32, 1274–1279 RSC.
- B. C. J. Esseveldt, et al. , Adv. Synth. Catal., 2004, 346, 823–834 CrossRef.
- P. V. Podea, M. I. Tosa, C. Paizs and F. D. Irimie, Tetrahedron: Asymmetry, 2008, 19, 500–511 CrossRef CAS.
- V. K. Sharma and N. J. D. Graham, Ozone: Sci. Eng., 2010, 32, 81–90 CrossRef CAS.
- H. Y. Lam, Y. Zhang, H. Liu, J. Xu, C. T. T. Wong, C. Xu and X. Li, J. Am. Chem. Soc., 2013, 135, 6272–6279 CrossRef CAS.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1sc01174j |
| ‡ Equal contribution. |
| This journal is © The Royal Society of Chemistry 2021 |