Open Access Article
Open Access ArticleThe Ras dimer structure†
Till
Rudack‡
 *ab,
Christian
Teuber‡
ab,
Marvin
Scherlo
ab,
Jörn
Güldenhaupt
ab,
Jonas
Schartner
b,
Mathias
Lübben
ab,
Johann
Klare
*ab,
Christian
Teuber‡
ab,
Marvin
Scherlo
ab,
Jörn
Güldenhaupt
ab,
Jonas
Schartner
b,
Mathias
Lübben
ab,
Johann
Klare
 c,
Klaus
Gerwert
c,
Klaus
Gerwert
 *ab and
Carsten
Kötting
*ab and
Carsten
Kötting
 *ab
*ab
aBiospectroscopy, Center for Protein Diagnostics (PRODI), Ruhr University Bochum, 44801 Bochum, Germany. E-mail: Till.Rudack@ruhr-uni-bochum.de; Klaus.Gerwert@ruhr-uni-bochum.de; Carsten.Koetting@ruhr-uni-bochum.de
bDepartment of Biophysics, Ruhr University Bochum, 44801 Bochum, Germany
cDepartment of Physics, Osnabrück University, 49074 Osnabrück, Germany
First published on 4th May 2021
Abstract
Oncogenic mutated Ras is a key player in cancer, but despite intense and expensive approaches its catalytic center seems undruggable. The Ras dimer interface is a possible alternative drug target. Dimerization at the membrane affects cell growth signal transduction. In vivo studies indicate that preventing dimerization of oncogenic mutated Ras inhibits uncontrolled cell growth. Conventional computational drug-screening approaches require a precise atomic dimer model as input to successfully access drug candidates. However, the proposed dimer structural models are controversial. Here, we provide a clear-cut experimentally validated N-Ras dimer structural model. We incorporated unnatural amino acids into Ras to enable the binding of labels at multiple positions via click chemistry. This labeling allowed the determination of multiple distances of the membrane-bound Ras-dimer measured by fluorescence and electron paramagnetic resonance spectroscopy. In combination with protein–protein docking and biomolecular simulations, we identified key residues for dimerization. Site-directed mutations of these residues prevent dimer formation in our experiments, proving our dimer model to be correct. The presented dimer structure enables computational drug-screening studies exploiting the Ras dimer interface as an alternative drug target.
Introduction
Ras is a small G protein that acts as a molecular switch in the context of cell differentiation, proliferation, and growth. Ras is active in the guanosine triphosphate (GTP) bound state and transduces signals downstream to the cell nucleus. Furthermore, it catalyzes the hydrolysis from GTP to guanosine diphosphate (GDP) leading to an inactive state. This switch-off process is further accelerated by GTPase-activating proteins (GAPs).1 If the catalysis function of Ras is impaired by mutations, tumors can develop due its constitutively active state. The reaction mechanisms of the GTP hydrolysis have been well characterized using experimental techniques such as X-ray structure analysis,2,3 nuclear magnetic resonance spectroscopy,4 Fourier transform infrared (FTIR) spectroscopy,5 and theoretical methods.6 Both the allosteric regulation of Ras and its signaling7–10 as well as insights into catalysis of small GTPases have been previously reviewed.1,10–13Currently, there is no drug against constitutively active Ras approved and Ras is traditionally referred to as undruggable. Recently, with advancing methods and new targeting concepts, progress has been made in identifying the drug attacking points of Ras 14,15 and Ras targeting drug development.16 This progress has led to the first drug; however its effects are restricted to the G12C mutation17,18 that is e.g. found in lung cancer.19 In this case a covalent attachment to the C12 allows the interference with the otherwise hard to target catalytic center.
Because targeting the catalytic center is so difficult, many efforts are being made to study Ras in a larger context, including its processing, interaction partners, distribution, and organization at the plasma membrane.10 Ras dimerization and its impact on Raf-1 activation were described first in the year 2000.20 In 2013 the first in vitro structural model of a Ras dimer at a membrane was proposed based on attenuated total reflectance infrared spectroscopy measurements in combination with distances estimated by Förster resonance energy transfer (FRET) using fluorescently labeled nucleotides and biomolecular simulations.21 This study initiated several follow up studies that showed dimerization in vivo22–24 opening up a new strategy for cell growth signaling inhibition by disrupting the proposed dimerization interface. The enhanced dimerization was shown to activate downstream signaling23 while the inhibition of the dimer formation by a small protein22 led to signal interference. In vitro and in vivo the presence of K-Ras dimers was demonstrated by GFP based FRET measurements.25 The same study showed that a dimer preventing mutation of oncogenic K-Ras (K-RasG12D/D154Q) inhibits tumor growth in mice.25 Using quantitative photoactivated localization microscopy it was shown that K-Ras-GDP and K-Ras-GTP form dimers at a physiological expression level.23 Dynamic light scattering experiments showed the K-Ras4B dimer formation with the bound nucleotide analogue GTPγS.26 Very recently it was shown by a plethora of methods including small angle X-ray scattering (SAXS) and biomolecular simulations that K-Ras4B forms dimers in the presence of Raf-RBD that are crucial for cell signaling.27 Nevertheless, there have been conflicting reports about Ras dimerization.10,28
Kovrigina et al. analyzed the time-domain fluorescence anisotropy and NMR chemical shift perturbations of the Ras G-domain without a lipid anchor and they conclude that the Ras G-domain alone does not form dimers in solution.29 In addition, Groves and colleagues showed in vitro that full-length and fully processed K-Ras4B remains monomeric in supported lipid bilayer membranes under variable conditions.30 It was shown that photosensitized oxidation, due to experimental conditions such as high laser intensities, may lead to Ras dimerization.31 Furthermore, a high protein concentration might lead to artificial dimerization as well.
However, the majority of studies support the presence of membrane bound Ras dimers. Immune electron microscopy studies show that about 40% of the Ras proteins form nanoclusters with a radius of approximately 9 nm at the membrane.32,33 In one case even a trimeric organization of K-Ras has been observed,34 while the rest of the observed organizations showed dimerization independent of the isoform.21,26,35,36 The Ras clusters act as isoform specific signaling platforms that recruit and activate effectors.37–39 For targeted development of anti-cancer drugs that break dimers of malfunctioning Ras and thereby inhibit signaling, a detailed understanding of the exact dimer interaction network is desirable.40
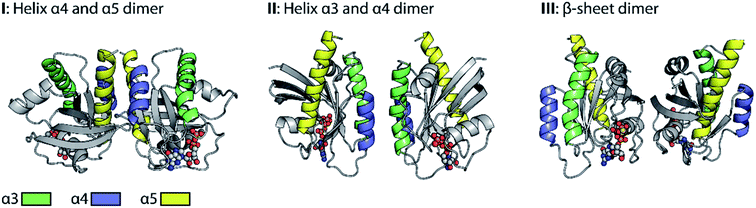
Promising drug targets can be proposed based on atomic level information regarding dimer interactions. However, the exact molecular structure of Ras-dimers remains elusive. Fig. 1 illustrates the diversity of the reported21,26,35,36 various different contradictory dimer structural models. We categorize these models into three main categories based on their dimer interaction sides. The models in category I undergo helix α4 and α5 dimerization, and the ones in category II undergo helix α3 and α4 dimerization, while the ones in category III undergo β-sheet dimerization. These categories are further subdivided with respect to their detailed dimer interactions in Fig. S1.† Structure I.1 was derived from the crystal contacts within most Ras X-ray structures,21I.2 (either N-Ras-GDP21 or K-Ras4B-GTP35) was obtained by biomolecular simulations, and I.3 (K-Ras4B-GDP) and I.4 (K-Ras4B-GTP) were both derived from NMR data.36 Structure II.1 (K-Ras4B-GTP) was obtained by biomolecular simulations,35 while II.2 (K-Ras4B-GppNHp) and III.1 (K-Ras4B-GTP-γ-S)26 were generated by the protein interactions by the structural matching (PRISM) algorithm.41 Details about the models are given in ESI Note 1.†
While various aforementioned experimental data indicate Ras dimerization, only a few studies provide insights into the structure of such dimers at the molecular level with atomic resolution. Two studies obtained NMR shifts for K-Ras4B dimers, one without the membrane26 and one using nanodiscs to mimic the membrane attachment.36 However, the structural models reported, namely models I.3, I.4, II.2, and III.1, vary over all three categories. In a previous study we measured a FRET distance of 46 ± 6 Å between the labeled (Mant/TNP) nucleotides.21 The aim of this study was to identify Ras dimerization and its orientation attachment to the membrane. However, the distance between labeled nucleotides measured with FRET is not sufficient to unambiguously determine the correct orientation of the monomers within a dimer. Overall, while there are various different dimer structural models reported none of them are clear-cut experimentally validated. Therefore, additional experimental data are required to allow an unambiguous validation of the proposed dimer structural models.
In order to obtain dimer structural models, we first used different established docking algorithms to predict dimer structural models with satisfactory statistical accuracy. Next, we employed FRET and electron paramagnetic resonance (EPR) spectroscopy to provide several experimental distances to enable an unambiguous validation for the predicted models. For these methods, fluorophores or spin labels need to be attached to various positions of the Ras surface to obtain sufficient distance variations for each structural model. To achieve membrane binding, a lipid anchor is attached to the Ras protein via maleimide chemistry at Cys181 limiting the chemical options to introduce labels. Therefore, to ensure flexible site specific labeling for the fluorophores and the EPR label we developed a strategy to incorporate unnatural amino acids into Ras. These amino acids were used as binding sites to which fluorophores or spin labels were attached via a second biorthogonal click reaction that does not cross-interfere with lipid coupling via the maleimide group. The identified dimer structural models that coincided with the available experimental FRET and EPR distances were further refined by biomolecular simulations to equilibrate the solvated membrane attached models. Based on these simulations we identified key dimer forming interaction residues. The effects of mutations of the computationally suggested interaction on dimerization were further checked experimentally by site-specific mutagenesis and FRET. A reliable characterization of the atomic details of Ras dimer interfaces is needed to guide the search for drug target sites for small molecules, which exert their anti-cancer effects by impairing Ras dimerization and constitutive signaling.
Results
Computational prediction of possible Ras dimer structural models
The literature shows that different workflows and docking algorithms provide numerous different Ras dimer models. We divide the seven published highly divers structural models into three main categories (I–III) based on their contact sides (Fig. 1). In order to obtain more potential dimer structures we used three different protein–protein docking web servers (PRISM,41 ZDOCK42 and Symmdock43,44). As input, we used six monomeric structural models of the Ras G-domain (residues 1 to 172) for H-Ras, K-Ras4B, and N-Ras with bound GTP and GDP. Table S1† summarizes the resulting 178 different dimer structural models reflecting nine different main interaction sides. While three of these interaction sides were already identified in the published models, six new possible interaction sides were detected by these algorithms (IV–IX). Details are given in ESI Note 2.†The experimental strategy to obtain different experimental distances
To reduce the number of possible dimer models and enable a clear-cut assignment of a unique structural dimer model, we developed a new strategy (Fig. 2) to obtain additional experimental distance information. The number of natural binding sites for labels to measure distances is limited and site specificity is difficult to achieve. Thus, the N-Ras lipid anchor containing a palmitoyl and a farnesyl group was attached to cysteine 181 of N-Ras via a maleimide group (see Fig. 2A and ESI Note 3†). The chemistry necessary to attach the anchor limits the chemical options to introduce labels for distance measurements. Individual labeling positions for FRET and EPR measurements were achieved by the incorporation of unnatural amino acids. The used strategy is described in Fig. S2,† and is based on previously reported protocols from Edward Lemke,45 which describe the site specific incorporation of the unnatural amino acids N-propargyl-L-lysine (PrK) and the cyclooctyne-lysine (SCO) into N-Ras using the amber codon suppression strategy. The unnatural amino acids bind the fluorophores or spin labels without interfering with lipid anchor binding. We identified S106 and T124 as the best incorporation sites yielding sufficient amounts of protein with the incorporated unnatural amino acid (Fig. S2B†).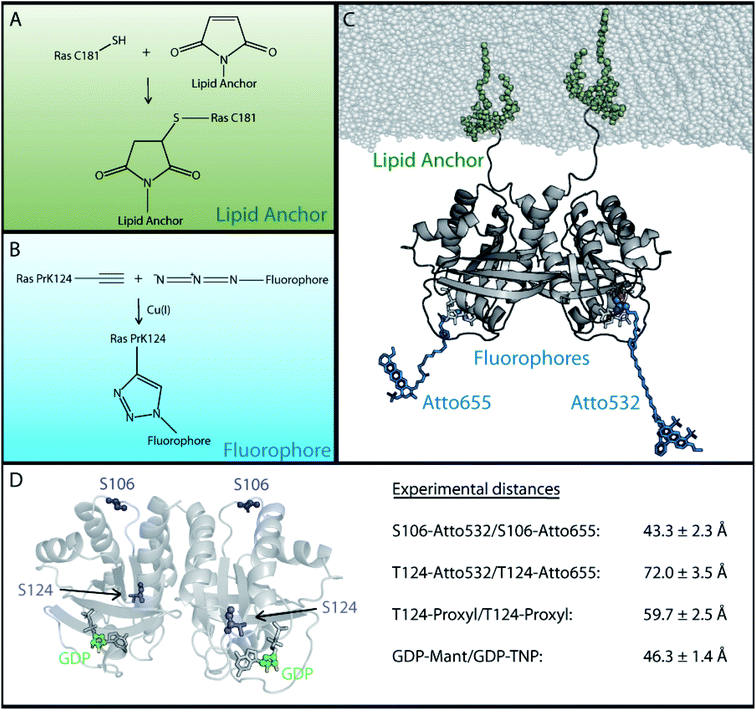 | ||
| Fig. 2 Experimental strategy for the attachment of lipid anchor and fluorophores to enable FRET or EPR measurements of membrane bound Ras proteins. In order to perform FRET or EPR measurements on membrane-bound Ras, two site specific protein modifications were necessary; one to attach the lipid anchor to the protein and another for the fluorophore or spin label. (A) The lipid anchor is attached via a maleimide group to ensure membrane binding. (B) The fluorophore is coupled site specifically via copper(I) catalyzed alkyne–azide cycloaddition to the previously incorporated unnatural amino acid (Fig. S2†). (C) A Ras dimer structural model with attached lipid anchors and the fluorophore pair T124-Atto 655/T124-Atto 532. (D) An overview of the labeling sites and the experimentally obtained FRET and EPR distances between two membrane bound full-length N-Ras-GDP proteins. The previously reported GDP distance was used.21 | ||
Next, the fluorophores and spin labels are bound to the unnatural amino acids via the copper(I) catalyzed alkyne–azide cycloaddition (CuAAC) as shown in Fig. 2B and ESI Note 3† or via the strain promoted alkyne–azide cycloaddition (SPAAC). One fraction of PrK labelled Ras was bound to Atto 532 azide (FRET donor) and another fraction was bound to Atto 655 azide (FRET acceptor). Similarly, the EPR spin label azido-proxyl was coupled to T124 of N-Ras via the unnatural amino acid SCO. Fig. 2C shows an exemplarily Ras dimer structural model with attached lipid anchor and the fluorophores Atto 655 and Atto 532 at position T124. Chemical drawings of the FRET labels are provided in Fig. S3.†
Then, we experimentally determined the FRET distances summarized in Fig. 2D. These distances are based on FRET efficiencies, which were derived from the fluorophore lifetimes measured with a time-correlated single photon counting (TCSPC) setup. The respective TCSPC histograms of all measured FRET distances and the experimental EPR data are presented in Fig. S4.† For these FRET measurements, we mixed the Ras protein to be investigated with 1-palmitoyl-2-oleoyl-sn-phosphatidylcholine (POPC) liposomes. The lifetime (τ) for membrane-bound N-Ras (GDP) S106-Atto 532 was analyzed in the presence (τDA) and absence (τD) of the acceptor N-Ras (GDP) S106-Atto 655 (Fig. S4A†). The analog experiment was performed for membrane-bound N-Ras (GDP) T124-Atto 532 and the acceptor N-Ras (GDP) T124-Atto 655 (Fig. S4B†). Both resulting histograms show a reduced τDA compared to τD due to FRET. The τ reduction is significantly stronger in samples with N-Ras (GDP) S106-Atto 532 (from 1.87 ns to 1.28 ns) than in samples with N-Ras (GDP) T124-Atto 532 (from 2.66 ns to 2.53 ns). Due to the large distance between the fluorophores at the T124 position, we verified the T124–T124 distance independently with an EPR spectroscopic double electron–electron resonance (DEER) measurement method that is more accurate in this distance regime (Fig. S4C†). This experiment resulted in a time-dependent echo amplitude that has been described by a Gaussian fit in order to obtain the distance information. The distance between the nucleotides was previously determined by FRET (18) (Fig. S4D†). The experimental details of the distance determination process are summarized in the Methods section, ESI Note 4, and in Table S2.†
The obtained distances between two membrane-bound lipidated N-Ras monomers (Fig. 2D) are 43.3 ± 2.3 Å between the FRET fluorophores coupled to S106–S106, 72 ± 3.5 Å between the fluorophores coupled to T124–T124, 59.7 ± 2.5 Å between the EPR proxyl labels at T124–T124, and 46.3 ± 1.4 Å between the fluorescent labeled nucleotides.21 The distance between the EPR labels at T124 is shorter than the one between the FRET labels at the same position, as the EPR label is shorter than the FRET label (Fig. S5†). The corresponding Cα distance is the same for both methods. The three different labeling positions are well distributed over the Ras surface as shown in Fig. 2D, which ensures a significantly more precise validation of possible Ras dimer structural models than the previously available single distance of the labeled nucleotides.
The obtained distances for two N-Ras monomers including the lipid anchor solely in solution without membrane are 49.7 ± 0.8 Å between the FRET fluorophores coupled to S106–S106 and 77 ± 0.5 Å between the ones coupled to T124–T124. All distances are summarized in Table S2.†
Comparison of experimental distances with calculated distances of the predicted dimer models
We utilized the four experimental distances for membrane-bound N-Ras-GDP to evaluate the 178 dimer conformations resulting from our protein–protein docking (Table S1†). The nine main conformations of the monomer orientation are further divided into 64 subcategories, representing slight variations in the dimer binding interface. Next, we calculated the respective distance distributions for each of the 64 conformations within the subcategories and compared them with the experimentally obtained values. Details regarding the generation of rotamer ensembles and calculation of the respective distance distributions for all experimentally used labels and positions are shown in Fig. S5 and ESI Note 5.† We used the PyMOL plugin MTSSL Wizard to calculate the rotamer ensembles. The distributions are fitted by a Gaussian function to obtain a mean value and standard deviation for each labeling position of each dimer conformation (Table S3†). As a further control of the label ensembles created by the MTSSL Wizard, we performed biomolecular simulations to analyze the conformational space sampled by SCO-proxyl at T124 (Fig. S6†).Fig. 3A–D represents the distance calculation for the seven published dimer models shown in Fig. S1.† Possible membrane interactions that potentially further restrict the conformational space of labels are excluded in the calculation. Also, protein label and label–label interactions are described by a simplified model. Employing more accurate models describing the label interactions will lead to a further restriction of the conformational space. Therefore, the bandwidths of the calculated curves reflect the outer limit of label flexibility for one single dimer. In contrast, the experimental bandwidth is the standard deviation of the average value of the protein ensembles (∼60 million photons per measurement) of eight measurements for two different samples. Therefore, the experimental bandwidth is substantially narrower than the calculated one. However, only for dimer models I.1 and I.2 the calculated distance distributions overlap with all four experimental distances. Model I.3 does not match the T124–T124 measurements, while model I.4 does not match the ones for T124–T124 and nucleotide–nucleotide. Model II.1 is in accordance with the nucleotide distance measurements and significantly out of the experimental region for the other three distances, while model II.2 is only in accordance with the S106–S106 distance. Model III.1 diverges significantly from the nucleotide–nucleotide distance. Besides models I.1 and I.2 only I.5 to I.9 are in accordance with all four values as revealed by the comparison of the 64 dimer conformations obtained through protein docking in Table S3,† while models I.10 to I.13 present only one value which deviates slightly from the four measurements. The rest of the obtained conformations displayed significantly different values compared to the experimental distances. Consistently, the category II and III models obtained for K-Ras4B do not match the distances for K-Ras4B-GTP (Table S4†) or K-Ras4B-GDP (Table S5†) that were derived from published NMR shifts.
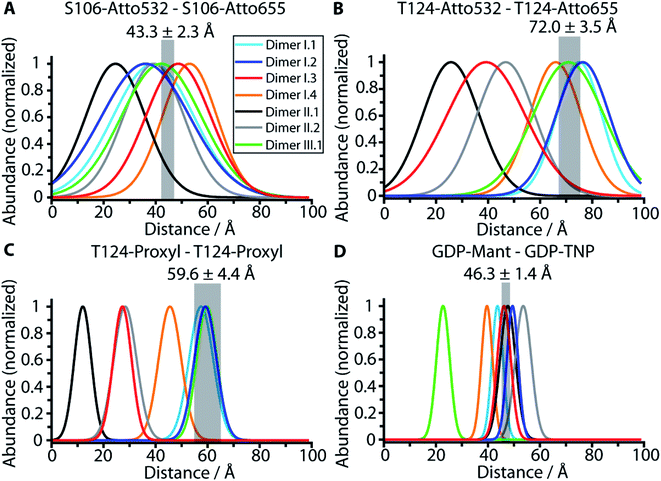 | ||
| Fig. 3 Comparison of the experimental distances with the calculated distances for predicted dimer structural models. The experimental values obtained by FRET and EPR measurements (Fig. S4†) are shown as transparent grey bars. Only dimer structural models I.1 and I.2 are in line with all four measured distances as revealed by the comparison of the measured distances S106–S106 (A), T124–T124 (B), T124–T124 (EPR) (C) and nucleotide–nucleotide (D) (grey bars) with the calculated distance distributions based on the dimer structural models I.1 (light blue), I.2 (dark blue), I.3 (cyan), II.1 (black), II.2 (grey) and III.1 (green) as detailed in ESI Note 5.† The width of the calculated distribution reflects the steric maximal possible conformational space of the labels that provide the outer limit. If the experimental value is not within the calculated distribution in one of the four plots the structure is ruled out. | ||
Based on their agreement with the experimental data, we probed the stability and further refined the detailed dimer model interaction interface of the conformations of category I through MD simulations, with the exception of I.2, that was already obtained by refinement through MD simulations,21 as well as I.3 and I.4 that were exclusively obtained for K-Ras4B whereas our experiments use N-Ras.
MD simulations used to obtain one equilibrated representative dimer model
In total nine MD simulations of N-Ras-GDP initiated by the dimer models of conformation I.1 and I.5 to I.12 were prepared, and were run for at least 550 ns. Simulations were setup, performed and analyzed as described in ESI Note 6.† Contact patterns were identified using the PyContact46 and the Maximoby contact matrix algorithm. Simulation times and system properties are summarized in Table S6.† Equilibration of the systems was checked based on the root mean square displacement (RMSD) from the respective starting structure of each conformation and the time evolution of the contact pattern (Fig. S7†). The distances between the ensembles of labels attached to equilibrated representative structures were calculated and compared to the experimental values (Table S7†) analogous to the procedure for the predicted docking structures. Simulations I.9 and I.10 converge to an almost identical representative equilibrium dimer structural model (Fig. 4), that is in total accordance with the four experimental distances mentioned above. All seven other simulations converge to equilibrium structures with almost identical monomer orientation among each other but slightly different compared to the equilibrium structures observed for I.9 and I.10 (Fig. S8†). The monomers are slightly tilted against each other, leading to divergence from the experimental nucleotide–nucleotide distance. The structural models of the representative structures of simulations I.9 and I.10 are provided within the ESI.†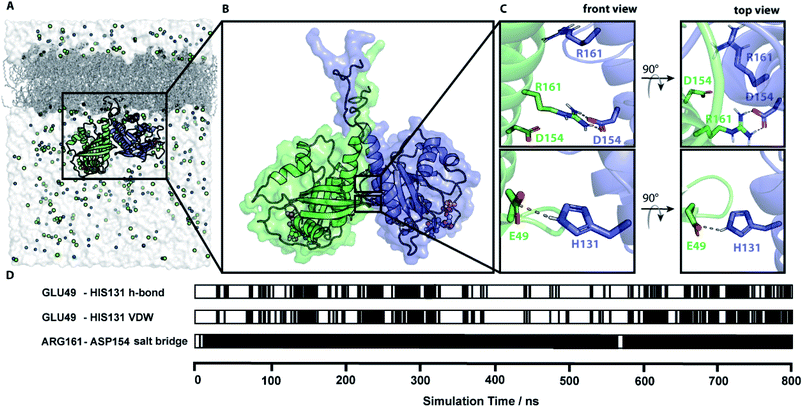 | ||
| Fig. 4 Simulation of a membrane-bound N-Ras dimer. (A) The simulation system of a lipidated N-Ras dimer structural model attached to a POPC membrane inside a solvation box with a physiological salt (NaCl) concentration. (B) Representative protein structure of an 800 ns long MD simulation started with conformation I.10 (representative structure in accordance with all four experimentally measured distances). (C) Key residues of the protein–protein interaction network identified by contact analysis of the simulation trajectory. (D) The time course of the key contacts within the simulation of conformation I.10. All contacts and the root mean square displacement (RMSD) are shown in Fig. S7.† | ||
The time course analysis of the dimer interface interaction pattern of simulation trajectories I.9 and I.10 identified a stable central salt bridge between D154 and R161. This contact was the key dimer stabilizing salt bridge in five out of the seven simulations leading to the second equilibrium structure. The contact between E49 and H131 is the second most prevalent contact involved in the dimer interface formation either through a hydrogen bond or van-der-Waals interactions. However, this contact is fluctuating over time due to its exposed position at the edge of the dimer contact interface resulting in competing solvent interactions. Therefore, this contact is less stable than the D154/R161 contact, which is deeply buried in the center of the dimer interface (Fig. S8†). The distance plots (Fig. S9†) between D154 and R161 as well as E49 and H131 within simulation I.9 further illustrates the fluctuating character of the E49/H131 interaction compared to the permanent D154/R161 contact. A detailed list of the relevant intra-protein contacts observed in the simulation trajectories is provided in Table S8.† Often protein bound water molecules or ions have a crucial structural or functional role.47 However, our contact analyses shown in Fig. S13† indicate that there are no bridging water molecules or ions involved in the formation of the dimer interface.
Validation of the predicted dimer interface by FRET measurements of the E49Q D154N N-Ras variant
The MD simulations revealed that the most prominent dimer contact is the salt bridge between D154 and R161 and the second most prominent interaction is between E49 and H131. Based on our predicted contacts, we expected the E49Q and D154N mutations in Ras to prevent dimerization (Fig. 5A). To validate our predicted contact interface, we performed FRET measurements of the lipidated membrane-bound N-Ras E49Q D154N variant. As the FRET signal with fluorophores at residue 106 was the most prominent in wild type, we chose this label position for our variant. No FRET signal was observed (Fig. 5B) for the N-Ras E49Q D154N variant, indicating that the E49Q D154N is the variant that prevents dimer formation. This dimer preventing variant provides a clear-cut experimental proof for our suggested dimer model.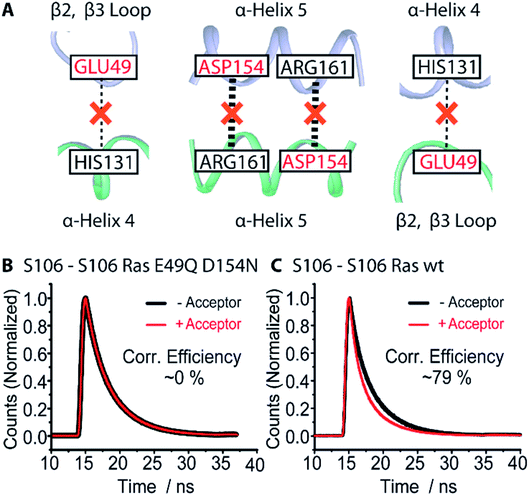 | ||
| Fig. 5 Dimerization preventing the E49Q D154N N-Ras variant. (A) Schematic summary of the key dimer interactions (Table S6†) obtained from MD simulations (Table S7†) suggesting E49 and D154 as promising candidates for dimer preventing mutations. (B) FRET measurements of the lipidated membrane-bound N-Ras E49Q D154N variant confirm that mutations of E49 and D154 prevent the dimer formation as no observable FRET signals were detected compared to N-Ras WT (C). The computationally predicted N-Ras dimer interface is experimentally validated. | ||
Discussion
The organization of Ras at the plasma membrane is decisive for its function. There is experimental evidence that Ras dimerization is crucial for nanoclustering at the plasma membrane and signal transmission.39 Our experiments with lipidated N-Ras WT attached to a POPC membrane (Fig. 2) provided further evidence for dimerization due to the evoked FRET-efficiencies or DEER signals (Table S2 and Fig. S4†). Our in vitro experimental setup using a POPC membrane does not perfectly match the in vivo conditions; however we are convinced that our found dimer interface is transferable to in vivo conditions. Dimerization of membrane-bound Ras was also shown for other completely different artificial membrane systems such as nanodiscs by NMR.36 Even more importantly, an in vivo study showed that an impaired dimerization of oncogenic K-Ras abolishes tumor growth.25 Dimerization at a membrane is indicated by the majority of experimental studies as previously described. The two most relevant studies for this matter indicate that K-Ras-GDP and K-Ras-GTP form dimers at a physiological expression level as revealed by quantitative photoactivated localization microscopy.22,23 These studies observed a connection between the Ras dimerization and the activation of the Raf–MAPK signal pathway. Furthermore, attenuated total reflectance (ATR) infrared spectroscopy measurements in combination with biomolecular simulations have shown that N-Ras dimerizes on POPC membranes.21 Some studies provide conflicting results that exclude dimerization, most of which were performed without a membrane in their experimental setup.29 The only biophysical study indicating monomeric Ras, while including a membrane, measured the diffusion of Ras in a solid supported bilayer in vitro.30 The authors of the study stated that Ras dimerization in vivo must depend on more factors than the presence of the protein alone. Furthermore, they noted that the glass support of the bilayer may damp large membrane undulations influencing the lateral organization of Ras leading to contradicting results compared to the aforementioned in vivo studies.There have been valid concerns that dimerization shown by FRET measurements is associated with high laser intensities30 or high protein concentrations. However, our measurements show that neither of these are the case for the used experimental setup. While high laser intensities are exerted by confocal microscopes, we used time correlated single photon counting (TCSPC) with low laser intensity or EPR which does not involve a laser. The concern regarding the protein concentration was eliminated since we obtained dimerization for one measurement (N-Ras WT S106–S106) and no dimerization for another (E49Q D154N N-Ras variant S106–S106) while using the same protein concentration and identical experimental setups. Thus, there is strong evidence that lipidated membrane-bound full-length Ras forms dimers.
The lack of a detectable FRET signal for the E49Q D154N N-Ras variant indicates that this interaction site is the only one present, as at least a somewhat visible FRET signal would then be expected. This implies that full-length N-Ras does not form oligomers at POPC membranes, as an additional interaction site, different from the dimer interaction site identified here, would be necessary to form oligomers in wt N-Ras.
Our results indicate a decisive role of the lipid anchor in dimerization with and without membrane. We obtained comparable FRET efficiencies of full-length lipidated N-Ras solely in solution (S106–S106: 60% and T124–T124: 10%) and membrane bound (S106–S106: 79% and T124–T124: 15%). However, the same FRET measurements in solution using N-Ras 1–181 without the lipid anchor showed no signal, indicating that the Ras G domain lacks the intrinsic propensity to form dimers in solution. The N-Ras anchor is very hydrophobic (Fig. S10†), indicating that the hydrophobic effects are driving forces for anchor dimerization. These findings are in line with previous studies on the self-association of the G domain in solution observed via analysis of the time-domain fluorescence anisotropy and NMR chemical shift perturbations.34 Overall, our data clearly indicate that the N-Ras lipid anchor is essential for the dimer formation in solution. We hypothesize that clustering of the lipid anchor is the first step towards dimerization. Clustering of the lipid anchor brings the G domains closer together and thus increases the local concentration of Ras proteins, which leads to the formation of one single otherwise comparably low affinity G domain interface. The lipid anchor substantially differs between the Ras isoforms leading to isoform specific lipid interactions.48,49 Thus isoform dependent clustering at specific membrane compartments depending on the membrane lipid composition is observed.
Gasper and Wittinghofer suggested as an argument against dimerization that different Ras crystals show different crystal packings.3 The C-terminal hypervariable region (HVR) including the lipid anchor is truncated in crystal structures due to their high flexibility. The remaining G domain has the potential to sample and form a huge variety of different dimer interfaces, due to the high protein concentrations during crystallization. This variety of dimer structural models is in line with the high variety of dimer conformations found by protein–protein docking algorithms (Table S1†). Both crystallography and docking studies miss the native conformational space restrictions as clustering of the lipid anchor and the space occupied by the membrane itself allows only limited possible G-domain dimer conformations for full length Ras. Therefore, crystal structures and protein–protein docking results lack the capacity to provide a single prominent structure, which we observe here for full length N-Ras. We note that our proposed dimer model is also a prominent motive within the crystal packing of Ras crystal structures. Thus, the lipid anchor steers the protein into an exclusive dimer structure at the membrane.
Knowledge of the precise amino acid interaction network of the dimer interface provides novel target sites for anti-cancer drugs that prevent Ras dimerization, which plays a crucial role in signal transduction in vivo.40 Our study shows that for N-Ras only those dimer models exhibiting an interface involving helices α4 and α5 (dimer category I) are in accordance with all four experimental distances. Dimer category II disagrees with both FRET and EPR distances obtained for position T124 and category III disagrees with the experimentally observed nucleotide–nucleotide distance. In addition, Fig. 6 shows that the binding sites for the three Ras interaction partners Raf, RasGAP and SOS are all accessible for dimer category I structural models. NMR distances obtained for K-Ras also show that helices α4 and α5 are involved in dimerization.36 The isoform independent motive of helices α4 and α5 dimerization interface is supported by a study showing the impact of disrupting this interface with a small synthetic protein affecting the signaling of both H-Ras and K-Ras.22 The importance of dimerization for in vivo signaling is emphasized by the study showing that the disruption of the oncogenic K-RasG12D dimer abolishes in vivo tumor growth.25 Taking together our results and the previously mentioned reports, we propose that the formation of the dimer interface by helices α4 and α5 is a universally valid motive for all Ras isoforms.
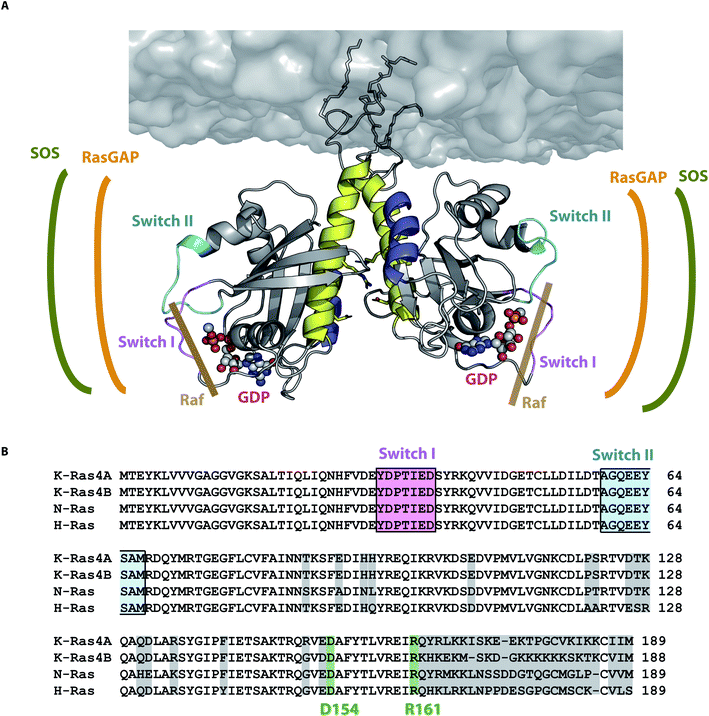 | ||
| Fig. 6 N-Ras dimer structural model and its interaction sites. (A) N-Ras dimer structural model in accordance with all four experimentally observed distances. The dimer interface is formed by helices α4 (blue) and α5 (yellow). The binding sites of SOS (green), Ras-GAP (orange) and Raf (brown) are accessible for the respective Ras interaction partners. The nucleotide binding site and the switch I (pink) and II (cyan) are not involved in the dimer interface as they are on the opposite sides of the dimer interface. (B) Alignment of all Ras isoforms with non-conserved amino acids highlighted in light gray. The key contact residues D154/R161 (green) stabilizing the Ras dimer (Fig. 4 and 5) are conserved among all Ras isoforms. | ||
Biomolecular simulations revealed one representative equilibrated lipidated membrane-bound full-length N-Ras-GDP dimer model (Fig. 6) that is in accordance with all experimental distances (Table S7†). The identified key residues (Fig. 4) forming the salt bridge D154/R161 are conserved among all Ras isoforms. This salt bridge was also identified as a key contact in our previously published simulations on N-Ras.21 The second most prominent contact residue E49 is also conserved and its interaction partner H131 is exchanged to Q131 in H- and K-Ras, still retaining the ability to form a hydrogen bond with E49. All these contact residues are positioned within a flat area of the Ras surface (Fig. S8†), which supports the proposed dimer interface. For N-Ras we confirmed the importance of D154 and E49 for dimerization as E49Q D154N mutations prevented dimerization (Fig. 5). Some but not all of the observed K-Ras4B NMR shifts36 are in agreement with our N-Ras models refined through biomolecular simulations (Tables S4 and S5†). These deviations indicate a difference between N-Ras and K-Ras4B. Furthermore, models I.3 and I.4 for K-Ras4B do not exhibit the D154/R161 key contact. Packer et al.27 showed the importance of the D154/R161 interaction within the formation of the K-Ras4B–Raf-RBD dimer. Further experimental proof regarding the presence of the D154/R161 interaction in K-Ras4B was provided by Ambrogio et al.25 They show the importance of the D154/R161 contact in K-Ras4B. Each of the two single mutants D154Q and R161E of oncogenic K-Ras4BG12D abolished in vivo tumor growth, implying that this is due to the impaired ability to form dimers. Thus, mutations in the dimer interface have the potential to eliminate the effect of oncogenic mutants at positions G12, G13, or Q61. The contact swapping mutant D154Q R161E showed tumor growth, though not as high as the wild type, possibly due to a weaker contact between Gln and Glu as opposed to the contact between Glu and Arg, though still allowing dimer formation. There might be slight difference in the detailed dimer interaction pattern between the Ras isoforms that still need further investigation. As shown by Spencer-Smith and colleagues, charge reversal mutation of D154 or R161 and R135 exhibited no effect on oncogenic H-RasG12V signaling, indicating the stabilization of its dimer interface through other residues for this isoform.22 We anticipate that N-Ras and K-Ras both dimerize through helices α4 and α5 with D154/R161 as the key contact; however, further research is required to unravel the detailed H-Ras dimer interaction pattern.
In summary, we developed a novel strategy to incorporate unnatural amino acids into Ras, allowing two orthogonal site-specific modifications of the same protein (Fig. 2); one to attach a lipid anchor and a second one for fluorophores or spin labels for distance measurements. Exploiting the gained flexibility of individual site-specific labeling we obtained three novel experimental distance information about N-Ras dimers. The S106–S106 distance of 43.3 ± 2.3 Å and T124–T124 distance of 72.0 ± 3.5 Å were obtained through FRET measurements and another T124–T124 distance of 59.6 ± 4.4 Å was obtained by EPR spectroscopy. All measurements indicate conclusively that lipidated full length N-Ras dimerizes at POPC membranes. We show that among 178 structural dimer models predicted by different protein–protein docking algorithms only those including helices α4 and α5 in the dimer interface are in accordance with the here obtained FRET and EPR data as well as previous FRET distances.21 Further refinement of the dimer interface through biomolecular simulations revealed that the salt bridge between D154/R161 together with the interactions of E49 with H131 are the key N-Ras dimerization contacts (Fig. 4). Our computationally predicted interface was experimentally validated as FRET measurements showed that the N-Ras E49Q D154N variant prevented dimerization (Fig. 5).
Taking together our results for N-Ras, the sequence similarities between the isoforms, and the described literature for H-Ras and K-Ras we propose that all Ras isoforms dimerize at the membrane through helices α4 and α5 (Fig. 6). Nevertheless, there might be isoform specific differences in the exact orientation of the monomers. These atomic resolution structural insights into the Ras dimer interface provide novel target sites to develop anti-cancer drugs inhibiting Ras signaling by preventing its dimerization.
Materials and methods
Experimental methods
Biochemical details of lipid anchor synthesis, sample preparation of lipidated N-Ras with incorporated unnatural amino acids, coupling of fluorophores, and sample composition for FRET and EPR measurements are described in ESI Note 3.† We have chosen hydrophilic dyes that have a very low membrane interaction factor50 to avoid artificial interactions.The TCSPC histograms were evaluated, and the FRET efficiencies were calculated as described in ESI Note 4.† The results are also displayed in Table S2.† In the presence of the fluorescence acceptor, the amplitude weighted and averaged fluorophore's lifetimes decreased from 1.87 ns to 1.28 ns in measurements with membrane-bound N-Ras (GDP) S106-Atto 532 and from 2.66 ns to 2.53 ns in measurements with membrane-bound N-Ras (GDP) T124-Atto 532. As detailed in ESI Note 4,† we have to consider that dimers are randomly formed between donor labeled Ras (D), acceptor labeled Ras (A) and unlabeled Ras. Presuming a dimer and considering the fraction of DA dimers these values lead to FRET efficiencies of ∼79% (S106–S106) and ∼15% (T124–T124).
We have performed the same experiment using proteins without the lipid anchor, but in the presence of POPC liposomes, and did not observe any FRET signals. This shows that the G-domain without the anchor does not form stable dimers. Measurements on the lipidated protein without POPC liposomes resulted in slightly lower FRET efficiencies as described above. Here, the amplitude weighted and averaged fluorophore's lifetime decreased from 1.86 ns to 1.41 ns in measurements with Ras S106-Atto 532 and from 3.56 ns to 3.44 ns in measurements with Ras T124-Atto 532 leading to FRET efficiencies of ∼60% for S106–S106 and ∼10% for T124–T124. This suggests that lipidated Ras proteins in solution interact with each other via the lipid anchor.
Experimental details of the EPR measurements are given in ESI Note 7.†
Computational methods
The protein–protein docking approach is described in ESI Note 2.† The protocol to calculate distance distributions for the predicted models employing the PyMOL plugin for MTSSL Wizard51 is described in ESI Note 5.†The dimer structures in accordance with the experimental data were prepared for the simulation by attachment of the membrane anchor, a side chain protonation based on a local pKa calculation and solvated using the Vedani algorithm using MAXIMOBY (CHEOPS, Germany). The membrane anchors were then embedded into a POPC membrane consisting of 512 lipids. The remaining simulation box was filled with water and a physiological salt concentration of 154 mmol l−1 and the simulations were run with Gromacs 2019.52 The structure preparation and the used protocol including all parameters for simulation runs are described in ESI Note 6.† Simulation results were visualized using VMD 1.9.4 (ref. 53) and PyMOL 2.2. Simulation trajectories were analyzed using the analysis tools of Gromacs 2019,52 VMD 1.9.4, and QwikMD.54 Contact patterns were identified using the PyContact46 and the MAXIMOBY (CHEOPS, Germany) contact matrix algorithm. Details about the input structures and the analysis of the biomolecular simulations are provided in ESI Note 6 and Table S6.†
Author contributions
TR, KG, and CK designed the research; TR, CT, MS, and JK performed research; CT performed FRET measurements and labeling experiments supervised by CK. TR and MS performed MD simulations. MS performed protein docking, and MTSSL Wizard calculations supervised by TR. JG assisted with the MTSSL Wizard analysis. JS synthesized the lipid anchor. JK performed EPR experiments and related MD simulations. ML assisted with protein expression. TR, CT, MS, JG, JK, and CK analyzed data; TR, CT, MS, KG, and CK wrote the paper with edits from all co-authors. KG and CK obtained funding.Funding
This work was supported by Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) Individual Research Grant GE 599/20-1 to KG and KO 3813/1-1 to CK. Further support was provided by the Ministry for Culture and Science (MKW) of North Rhine-Westphalia (Germany) through PRODI to KG and the Protein Research Unit Ruhr within Europe (PURE) funded by the Ministry of Innovation, Science and Research (MIWF) of North-Rhine Westphalia (Germany) to KG.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We thank Herbert Waldmann and Sascha Gentz for their support with synthesizing the lipid anchor. We thank Iris Bourdos for her support in the creation of N-Ras mutants and expression tests. We thank Léon Beyer and Konstantin Gavriljuk for MALDI measurements and Gregor Hagelueken for technical support with the MTSSL Wizard. We thank Udo Höweler for fruitful discussion of the biomolecular simulation and docking data as well as support with the contact analysis. The plasmids encoding tRNAPyl/PylRSWT/AF were obtained through an MTA with Prof. Dr Edward Lemke (Johannes Gutenberg University, Biocentre & IMB, Mainz).References
- A. K. Mishra and D. G. Lambright, Invited review: small GTPases and their GAPs, Biopolymers, 2016, 105(8), 431–448 CrossRef CAS PubMed.
- K. Scheffzek, M. R. Ahmadian, W. Kabsch, L. Wiesmüller, A. Lautwein and F. Schmitz, et al., The Ras-RasGAP complex: structural basis for GTPase activation and its loss in oncogenic Ras mutants, Science, 1997, 277(5324), 333–338 CrossRef CAS PubMed.
- R. Gasper and F. Wittinghofer, The Ras switch in structural and historical perspective, Biol. Chem., 2019, 401(1), 143–163 CrossRef PubMed.
- R. Thapar, J. G. Williams and S. L. Campbell, NMR characterization of full-length farnesylated and non-farnesylated H-Ras and its implications for Raf activation, J. Mol. Biol., 2004, 343(5), 1391–1408 CrossRef CAS PubMed.
- C. Kötting, J. Güldenhaupt and K. Gerwert, Time-resolved FTIR spectroscopy for monitoring protein dynamics exemplified by functional studies of Ras protein bound to a lipid bilayer, Chem. Phys., 2012, 396, 72–83 CrossRef , available from: http://www.sciencedirect.com/science/article/pii/S0301010411003636.
- A. R. Calixto, C. Moreira, A. Pabis, C. Kötting, K. Gerwert and T. Rudack, et al., GTP Hydrolysis Without an Active Site Base: A Unifying Mechanism for Ras and Related GTPases, J. Am. Chem. Soc., 2019, 141(27), 10684–10701 CrossRef CAS PubMed.
- A. D. Cox and C. J. Der, Ras history: the saga continues, Small GTPases, 2010, 1(1), 2–27 CrossRef PubMed.
- S. Lu, H. Jang, S. Muratcioglu, A. Gursoy, O. Keskin and R. Nussinov, et al., Ras Conformational Ensembles, Allostery, and Signaling, Chem. Rev., 2016, 116(11), 6607–6665 CrossRef CAS PubMed.
- I. A. Prior and J. F. Hancock, Ras trafficking, localization and compartmentalized signalling, Semin. Cell Dev. Biol., 2012, 23(2), 145–153 CrossRef CAS PubMed.
- D. K. Simanshu, D. V. Nissley and F. McCormick, RAS Proteins and Their Regulators in Human Disease, Cell, 2017, 170(1), 17–33 CrossRef CAS PubMed , available from: https://pubmed.ncbi.nlm.nih.gov/28666118/.
- K. A. Maegley, S. J. Admiraal and D. Herschlag, Ras-catalyzed hydrolysis of GTP: a new perspective from model studies, Proc. Natl. Acad. Sci. U. S. A., 1996, 93(16), 8160–8166 CrossRef CAS PubMed.
- M. R. Ahmadian, P. Stege, K. Scheffzek and A. Wittinghofer, Confirmation of the arginine-finger hypothesis for the GAP-stimulated GTP-hydrolysis reaction of Ras, Nat. Struct. Mol. Biol., 1997, 4(9), 686–689 CrossRef CAS PubMed.
- K. Gerwert, D. Mann and C. Kötting, Common mechanisms of catalysis in small and heterotrimeric GTPases and their respective GAPs, Biol. Chem., 2017, 398(5–6), 523–533 CAS.
- S. Lu, D. Ni, C. Wang, X. He, H. Lin and Z. Wang, et al., Deactivation Pathway of Ras GTPase Underlies Conformational Substates as Targets for Drug Design, ACS Catal., 2019, 9(8), 7188–7196 CrossRef CAS.
- B. Mörchen, O. Shkura, R. Stoll and I. Helfrich, Targeting the “undruggable” RAS - new strategies - new hope?, Cancer Drug Resist., 2019, 2, 813–826 Search PubMed.
- A. R. Moore, S. C. Rosenberg, F. McCormick and S. Malek, RAS-targeted therapies: is the undruggable drugged?, Nat. Rev. Drug Discovery, 2020, 19(8), 533–552 CrossRef CAS PubMed , available from: https://pubmed.ncbi.nlm.nih.gov/32528145/.
- J. Canon, K. Rex, A. Y. Saiki, C. Mohr, K. Cooke and D. Bagal, et al., The clinical KRAS(G12C) inhibitor AMG 510 drives anti-tumour immunity, Nature, 2019, 575(7781), 217–223 CrossRef CAS PubMed.
- L. Goebel, M. P. Müller, R. S. Goody and D. Rauh, KRasG12C inhibitors in clinical trials: a short historical perspective, RSC Med. Chem., 2020, 11(7), 760–770 RSC.
- I. A. Prior, P. D. Lewis and C. Mattos, A comprehensive survey of Ras mutations in cancer, Cancer Res., 2012, 72(10), 2457–2467 CrossRef CAS PubMed.
- K. Inouye, S. Mizutani, H. Koide and Y. Kaziro, Formation of the Ras dimer is essential for Raf-1 activation, J. Biol. Chem., 2000, 275(6), 3737–3740 CrossRef CAS PubMed.
- J. Güldenhaupt, T. Rudack, P. Bachler, D. Mann, G. Triola and H. Waldmann, et al., N-Ras forms dimers at POPC membranes, Biophys. J., 2012, 103(7), 1585–1593 CrossRef PubMed.
- R. Spencer-Smith, A. Koide, Y. Zhou, R. R. Eguchi, F. Sha and P. Gajwani, et al., Inhibition of RAS function through targeting an allosteric regulatory site, Nat. Chem. Biol., 2017, 13(1), 62–68 CrossRef CAS PubMed.
- X. Nan, T. M. Tamgüney, E. A. Collisson, L.-J. Lin, C. Pitt and J. Galeas, et al., Ras-GTP dimers activate the Mitogen-Activated Protein Kinase (MAPK) pathway, Proc. Natl. Acad. Sci. U. S. A., 2015, 112(26), 7996–8001 CrossRef CAS PubMed.
- M. Chen, A. Peters, T. Huang and X. Nan, Ras Dimer Formation as a New Signaling Mechanism and Potential Cancer Therapeutic Target, Mini-Rev. Med. Chem., 2016, 16(5), 391–403 CrossRef CAS PubMed.
- C. Ambrogio, J. Köhler, Z.-W. Zhou, H. Wang, R. Paranal and J. Li, et al., KRAS Dimerization Impacts MEK Inhibitor Sensitivity and Oncogenic Activity of Mutant KRAS, Cell, 2018, 172(4), 857–868 CrossRef CAS PubMed.
- S. Muratcioglu, T. S. Chavan, B. C. Freed, H. Jang, L. Khavrutskii and R. N. Freed, et al., GTP-Dependent K-Ras Dimerization, Structure, 2015, 23(7), 1325–1335 CrossRef CAS PubMed.
- M. R. Packer, J. A. Parker, J. K. Chung, Z. Li, Y. K. Lee and T. Cookis, et al., Raf promotes dimerization of the Ras G-domain with increased allosteric connections, Proc. Natl. Acad. Sci. U. S. A., 2021, 118(10), e2015648118 CrossRef PubMed.
- J. M. Rhett, I. Khan and J. P. O'Bryan, Biology, pathology, and therapeutic targeting of RAS, Adv. Cancer Res., 2020, 148, 69–146 CrossRef PubMed.
- E. A. Kovrigina, A. R. Galiakhmetov and E. L. Kovrigin, The Ras G Domain Lacks the Intrinsic Propensity to Form Dimers, Biophys. J., 2015, 109(5), 1000–1008 CrossRef CAS PubMed , available from: https://pubmed.ncbi.nlm.nih.gov/26331257/.
- J. K. Chung, Y. K. Lee, J.-P. Denson, W. K. Gillette, S. Alvarez and A. G. Stephen, et al., K-Ras4B Remains Monomeric on Membranes over a Wide Range of Surface Densities and Lipid Compositions, Biophys. J., 2018, 114(1), 137–145 CrossRef CAS PubMed.
- J. K. Chung, Y. K. Lee, H. Y. M. Lam and J. T. Groves, Covalent Ras Dimerization on Membrane Surfaces through Photosensitized Oxidation, J. Am. Chem. Soc., 2016, 138(6), 1800–1803 CrossRef CAS PubMed.
- S. J. Plowman, C. Muncke, R. G. Parton and J. F. Hancock, H-ras, K-ras, and inner plasma membrane raft proteins operate in nanoclusters with differential dependence on the actin cytoskeleton, Proc. Natl. Acad. Sci. U. S. A., 2005, 102(43), 15500–15505 CrossRef CAS PubMed.
- I. A. Prior, C. Muncke, R. G. Parton and J. F. Hancock, Direct visualization of Ras proteins in spatially distinct cell surface microdomains, J. Cell Biol., 2003, 160(2), 165–170 CrossRef CAS PubMed.
- E. Barklis, A. G. Stephen, A. O. Staubus, R. L. Barklis and A. Alfadhli, Organization of Farnesylated, Carboxymethylated KRAS4B on Membranes, J. Mol. Biol., 2019, 431(19), 3706–3717 CrossRef CAS PubMed.
- P. Prakash, A. Sayyed-Ahmad, K.-J. Cho, D. M. Dolino, W. Chen and H. Li, et al., Computational and biochemical characterization of two partially overlapping interfaces and multiple weak-affinity K-Ras dimers, Sci. Rep., 2017, 7(1), 40109 CrossRef CAS PubMed , available from: https://www.nature.com/articles/srep40109.
- K.-Y. Lee, Z. Fang, M. Enomoto, G. Gasmi-Seabrook, L. Zheng and S. Koide, et al., Two Distinct Structures of Membrane-Associated Homodimers of GTP- and GDP-Bound KRAS4B Revealed by Paramagnetic Relaxation Enhancement, Angew. Chem., Int. Ed., 2020, 59(27), 11037–11045 CrossRef CAS PubMed.
- T. Tian, A. Harding, K. Inder, S. Plowman, R. G. Parton and J. F. Hancock, Plasma membrane nanoswitches generate high-fidelity Ras signal transduction, Nat. Cell Biol., 2007, 9(8), 905–914 CrossRef CAS PubMed , available from: https://pubmed.ncbi.nlm.nih.gov/17618274/.
- T. Tian, S. J. Plowman, R. G. Parton, Y. Kloog and J. F. Hancock, Mathematical modeling of K-Ras nanocluster formation on the plasma membrane, Biophys. J., 2010, 99(2), 534–543 CrossRef CAS PubMed.
- Y. Zhou and J. F. Hancock, Ras nanoclusters: versatile lipid-based signaling platforms, Biochim. Biophys. Acta, 2015, 1853(4), 841–849 CrossRef CAS PubMed.
- M. Zuberi, I. Khan and J. P. O’Bryan, Inhibition of RAS: proven and potential vulnerabilities, Biochem. Soc. Trans., 2020, 48(5), 1831–1841 CrossRef CAS PubMed.
- A. Baspinar, E. Cukuroglu, R. Nussinov, O. Keskin and A. Gursoy, PRISM: a web server and repository for prediction of protein-protein interactions and modeling their 3D complexes, Nucleic Acids Res., 2014, 42(web server issue), W285–W289 CrossRef CAS PubMed.
- B. G. Pierce, K. Wiehe, H. Hwang, B.-H. Kim, T. Vreven and Z. Weng, ZDOCK server: interactive docking prediction of protein-protein complexes and symmetric multimers, Bioinformatics, 2014, 30(12), 1771–1773 CrossRef CAS PubMed.
- D. Schneidman-Duhovny, Y. Inbar, R. Nussinov and H. J. Wolfson, PatchDock and SymmDock: servers for rigid and symmetric docking, Nucleic Acids Res., 2005, 33(web server issue), W363–W367 CrossRef CAS PubMed.
- D. Schneidman-Duhovny, Y. Inbar, R. Nussinov and H. J. Wolfson, Geometry-based flexible and symmetric protein docking, Proteins, 2005, 60(2), 224–231 CrossRef CAS PubMed.
- T. Plass, S. Milles, C. Koehler, C. Schultz and E. A. Lemke, Genetically encoded copper-free click chemistry, Angew. Chem., Int. Ed., 2011, 50(17), 3878–3881 CrossRef CAS PubMed.
- M. Scheurer, P. Rodenkirch, M. Siggel, R. C. Bernardi, K. Schulten and E. Tajkhorshid, et al., PyContact: Rapid, Customizable, and Visual Analysis of Noncovalent Interactions in MD Simulations, Biophys. J., 2018, 114(3), 577–583 CrossRef CAS PubMed.
- B. M. Kearney, C. W. Johnson, D. M. Roberts, P. Swartz and C. Mattos, DRoP: a water analysis program identifies Ras-GTP-specific pathway of communication between membrane-interacting regions and the active site, J. Mol. Biol., 2014, 426(3), 611–629 CrossRef CAS PubMed.
- L. Brunsveld, H. Waldmann and D. Huster, Membrane binding of lipidated Ras peptides and proteins--the structural point of view, Biochim. Biophys. Acta, 2009, 1788(1), 273–288 CrossRef CAS PubMed.
- Z.-L. Li and M. Buck, Computational Modeling Reveals that Signaling Lipids Modulate the Orientation of K-Ras4A at the Membrane Reflecting Protein Topology, Structure, 2017, 25(4), 679–689 CrossRef CAS PubMed.
- L. D. Hughes, R. J. Rawle and S. G. Boxer, Choose your label wisely: water-soluble fluorophores often interact with lipid bilayers, PLoS One, 2014, 9(2), e87649 CrossRef PubMed.
- G. Hagelueken, R. Ward, J. H. Naismith and O. Schiemann, MtsslWizard: In Silico Spin-Labeling and Generation of Distance Distributions in PyMOL, Appl. Magn. Reson., 2012, 42(3), 377–391 CrossRef CAS PubMed.
- M. J. Abraham, T. Murtola, R. Schulz, S. Páll, J. C. Smith and B. Hess, et al., GROMACS: high performance molecular simulations through multi-level parallelism from laptops to supercomputers, SoftwareX, 2015, 1–2, 19–25 CrossRef , available from: http://www.sciencedirect.com/science/article/pii/S2352711015000059.
- W. Humphrey, A. Dalke and K. Schulten, VMD: visual molecular dynamics, J. Mol. Graphics, 1996, 14(1), 33–38 CrossRef CAS PubMed.
- J. V. Ribeiro, R. C. Bernardi, T. Rudack, J. E. Stone, J. C. Phillips and P. L. Freddolino, et al., QwikMD - Integrative Molecular Dynamics Toolkit for Novices and Experts, Sci. Rep., 2016, 6(1), 26536 CrossRef CAS PubMed , available from: https://www.nature.com/articles/srep26536.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1sc00957e |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2021 |