Open Access Article
Open Access ArticleMultiplexed droplet loop-mediated isothermal amplification with scorpion-shaped probes and fluorescence microscopic counting for digital quantification of virus RNAs†
Ya-Ling
Tan
,
A-Qian
Huang
,
Li-Juan
Tang
* and
Jian-Hui
Jiang
 *
*
State Key Laboratory of Chemo/Biosensing and Chemometrics, College of Chemistry and Chemical Engineering, Hunan University, Changsha 410082, P. R. China. E-mail: jianhuijiang@hnu.edu.cn; tanglijuan@hnu.edu.cn; Fax: +86-731-88822577; Tel: +86-731-88822872
First published on 14th May 2021
Abstract
Highly sensitive digital nucleic acid techniques are of great significance for the prevention and control of epidemic diseases. Here we report the development of multiplexed droplet loop-mediated isothermal amplification (multiplexed dLAMP) with scorpion-shaped probes (SPs) and fluorescence microscopic counting for simultaneous quantification of multiple targets. A set of target-specific fluorescence-activable SPs are designed, which allows establishment of a novel multiplexed LAMP strategy for simultaneous detection of multiple cDNA targets. The digital multiplexed LAMP assay is thus developed by implementing the LAMP reaction using a droplet microfluidic chip coupled to a droplet counting microwell chip. The droplet counting system allows rapid and accurate counting of the numbers of total droplets and the positive droplets by collecting multi-color fluorescence images of the droplets in a microwell. The multiplexed dLAMP assay was successfully demonstrated for the quantification of HCV and HIV cDNA with high precision and detection limits as low as 4 copies per reaction. We also verified its potential for simultaneous digital assay of HCV and HIV RNA in clinical plasma samples. This multiplexed dLAMP technique can afford a useful platform for highly sensitive and specific detection of nucleic acids of viruses and other pathogens, enabling rapid diagnosis and prevention of infectious diseases.
Introduction
Infectious diseases are among the most critical global health problems.1,2 Current and rapidly evolving public health threats from HIV to emerging pathogens, including the ongoing pandemic of COVID-19, have impelled urgent needs of sensitive, fast and cost-effective techniques for nucleic acid detection (NAD).3–6 Digital NAD (dNAD) techniques hold promise for high-precision, high-sensitivity and absolute quantification of pathogens.7–12 Originating from the digital polymerase chain reaction (dPCR) using precise thermal-control instruments,13–15 dNAD techniques have evolved to combine isothermal amplification reactions, such as rolling circle amplification (RCA),16 recombinase polymerase amplification (RPA)17 and loop-mediated isothermal amplification (LAMP).18 These isothermal dNAD techniques afford advantages of simpler instruments and faster reactions, enabling a useful point-of-care (POC) analytical platform for rapid diagnosis and prevention of infectious diseases.LAMP is a robust field-amenable and user-friendly technique for highly efficient nucleic acid amplification.19–22 Some of the NAD kits based on LAMP have been commercialized for pathogen identification.23 Combining the design of microwell arrays and droplet generators, digital LAMP has also been developed for the detection of various pathogens.24–31 However, the specific amplification chemistry of LAMP creates difficulties in designing multi-colored fluorescent primers for multiplexed detection. Slight modifications of primers could significantly alter its amplification efficiency due to the complex but exquisite design of LAMP. Attempts have been made for a few real-time multiplex/multicolor LAMP techniques, such as MethyLight that uses a fluorophore-labelled loop primer quenched by neighboring guanine nucleotides,32 assimilating or DARQ probes that use duplex-structured inner primers with terminally labeled fluorophores and quenchers,33 and one-step strand displacement or mediator displacement probes that use duplex-structured inner or loop primers with terminally labelled fluorophores and quenchers.34–36 Nevertheless, these multiplexed LAMP strategies have, to our knowledge, not been demonstrated for dNAD. The adaptation of multiplexed LAMP to digital format is quite challenging. In order to achieve accurate quantification, digital droplet assays typically require highly specific probes to improve stability against emulsification reagents and consistently efficient reactions in numerous parallel reaction units. Multiplexed LAMP probes that can be generated according a simple and universal design and have high stability are advantageous in specificity and efficiency consistency. However, the above methods either suffer from limitations of careful redesign of probes according to different target genes, or have increased susceptibility to non-specific displacement reactions and substantial reduction of amplification efficiency due to coexisting components in the droplet systems. Moreover, multiplexed detection in single digitalized units can not only allow simultaneous assays for multiple nucleic acid targets, but also offer internal control for contaminants or experimental conditions. Therefore, development of new multiplexed dLAMP techniques represents a significant effort to address the challenges of rapid diagnosis and prevention of infectious diseases.
Here we develop a novel multiplexed droplet LAMP (dLAMP) technique for the quantification of virus cDNAs using fluorescence-activable scorpion-shaped probes (SPs) with fluorescence microscopic counting. A real-time multiplexed LAMP strategy is first proposed by using a set of target-specific fluorescence-activable SPs as the loop primers. The LAMP reaction can open the hairpin structure of SP, activating a fluorescence signal of a specified color to indicate the presence of target DNA. Then, this new multiplexed LAMP is for the first time integrated with droplets to develop a digital LAMP platform, by implementing the LAMP reaction using a droplet microfluidic chip coupled to a droplet counting microwell chip. The droplet counting system allows rapid and accurate counting of the numbers of total droplets and the positive droplets by collecting multi-color fluorescence images of the droplets in a microwell. The multiplexed dLAMP assay was successfully demonstrated for the quantification of HCV and HIV cDNAs. To our knowledge, it is the first time that the multiplexed dLAMP technique has been developed using fluorescence-activable SPs for nucleic acid quantification. This multiplexed dLAMP technique can afford a useful platform for highly sensitive and specific detection of nucleic acids of viruses and other pathogens, enabling rapid diagnosis and prevention of infectious diseases.
Results and discussion
Design of the multiplexed dLAMP strategy
The multiplexed dLAMP technique relies on a novel design that combines a multiplexed LAMP reaction using fluorescence-activable SPs with a droplet-generating and counting microfluidic platform, as illustrated in Scheme 1. As a proof-of-principle, the multiplexed dLAMP assay was demonstrated for the quantification of HCV and HIV cDNAs. To develop the multiplexed LAMP strategy, we designed a set of target-specific loop primers using fluorescence-activable SPs (Scheme 1A). Each SP has a target-specific sequence in the downstream region and a common hairpin structure in the upstream region with a BHQ2 quencher plus a fluorescent tag of a prescribed color. Based on this design, the SPs are all fluorescence quenched in the intact state, delivering a low fluorescence background. During the LAMP reaction, each SP acts as the loop primer for amplification of the corresponding target. The amplification reaction allows opening of the hairpin structure of SPs and thus activating a fluorescence signal of a specified color to indicate the presence of the target cDNA. Using the real-time fluorescence curve recorded for the corresponding colors, quantification of multiple target cDNA can be achieved. Beyond the purpose of activatable and multiplexed fluorescence detection, the scorpion-shaped design has a unique advantage of increasing the stability of the probes due to intramolecular folding. And it is shown that it does not reduce the amplification efficiency. Although the synthesis of double labels in primers would slightly increase the cost for each assay, it affords a universal reporter, as it is simply obtained by extending the loop primer at the 3′ terminal with a fixed hairpin structure with no need of optimization for any target gene or primer sets. Therefore, our strategy allows simpler assay design with easy implementation for various targets, more consistent amplification efficiency and better stability, which allows its adaptation to digital assays.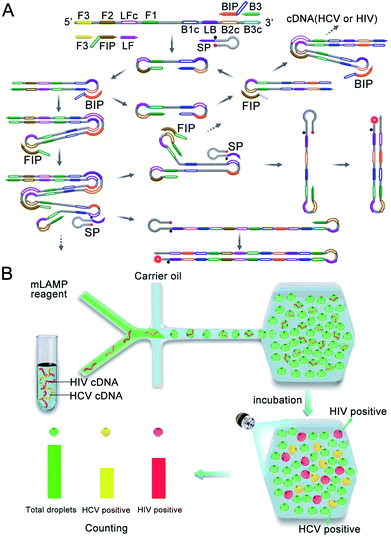 | ||
| Scheme 1 (A) Design and reaction of multiplexed LAMP using fluorescence-activable SPs. (B) Design of the multiplexed dLAMP platform with a microscopic counting system. | ||
Based on the developed multiplexed LAMP technique, we then establish a digital droplet platform using a droplet microfluidic chip coupled to a droplet counting microwell chip (Scheme 1B). The multiplexed LAMP reagents are digitalized into picoliter-sized droplets using a Y-shape generator and delivered into a droplet counting microwell (Fig. S1†). After incubation to allow completion of the LAMP reaction, the droplets are counted using a fluorescence microscopic system in the specified fluorescence channels. A green fluorescent dye, fluorescein, is introduced into the LAMP reagents, which facilitates accurate counting of the total droplets from the green fluorescence image. Rhodamine and Cy5 are used as the fluorescent tags in the SPs for HCV and HIV cDNA, respectively. Hence, the HCV and HIV cDNA positive droplets are counted from the orange and near infrared (NIR) fluorescence images, respectively. Compared to traditional flow counting platforms which have large variability and more working time, and microscopic counting platforms which require separate differential interference contrast imaging of the total droplet number, our fluorescent counting design allows facile and reproducible counting of the total droplet number and the positive droplets. In addition, the presented dLAMP has other advantages such as improved detection precision, improved resistance to polymerase inhibitors and the capability of absolute quantification of cDNA targets without the need of standard samples, making the dLAMP technique a useful platform for ultrasensitive nucleic acid quantification.
Multiplexed SP-based LAMP assay for HCV and HIV cDNAs
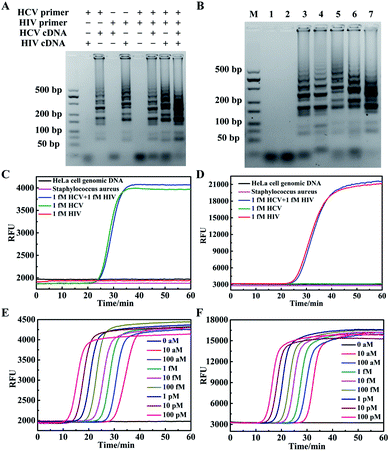
We firstly developed a multiplexed LAMP strategy for simultaneous detection of HCV and HIV cDNAs. Two sets of LAMP primers with SP-based loop primers were designed separately (Table S1†). The optimized reaction temperature of the multiplexed LAMP assay was found to be 64 °C (Fig. S2†). To test the specificity of these two sets of primers, we performed gel electrophoresis analysis of the amplification products for samples in the presence of a single cDNA component or both cDNA components (Fig. 1A). As anticipated, with the HCV primer and SP set we only obtained ladder-like bands with large molecular weights for HCV cDNA containing samples, with no amplification products obtained for samples without cDNA or only containing HIV cDNA. Likewise, with the HIV primer and SP set amplification products appeared for samples containing HIV cDNA, and no amplification products were obtained for samples without cDNA or only containing HCV cDNA. This result validated the specificity of the primer and SP sets for HCV and HIV cDNAs. To further examine its ability for detecting complex samples, multiplexed LAMP was used for the detection of HeLa cell genomic DNA, and cDNAs reversely transcribed from extracted RNAs in HCV or HIV-infected plasma samples (Fig. 1B). The gel electrophoresis image displayed clear amplification products for HCV-infected and HIV-infected plasma, with no amplification products for HeLa cell genomic DNA. This observation confirmed the specificity of multiplexed LAMP for HCV or HIV cDNA detection.Then, the specificity of multiplexed LAMP was investigated using real-time fluorescence curves obtained for TAMRA and Cy5, respectively, throughout the reactions. The real-time fluorescence curve in the TAMRA channel showed a typical sigmoidal curve with a point of inflection (POI) of 25.7 min for a sample containing only 1 fM HCV cDNA (Fig. 1C). By contrast, no apparent inflection of fluorescence response appeared in the TAMRA channel for a sample containing only 1 fM HIV cDNA. We also did not obtain remarkable fluorescence signal inflection in the reaction with genomic DNA of HeLa cells and Staphylococcus aureus. This result verified that the fluorescence curve in the TAMRA channel was a specific indicator for the presence of HCV cDNA. Moreover, in the assay for a sample containing 1 fM HCV cDNA and 1 fM HIV cDNA, we obtained a POI of 26.3 min, which slightly deviated from that for 1 fM HCV cDNA. A further statistical test of three repetitive assays for 1 fM HCV cDNA and 1 fM HCV cDNA plus 1 fM HIV cDNA showed insignificant deviations of POI between two sets of assays. This finding implied that the fluorescence curve in the TAMRA channel was highly specific for detecting HCV cDNA, and the presence of HIV cDNA had no interference with the detection of HCV cDNA. The fluorescence curves obtained in the Cy5 channel also displayed high specificity for detecting HIV cDNA, and the presence of HCV cDNA had no interference with its detection (Fig. 1D). Together, these results suggested the specific amplification and detection of HCV and HIV cDNAs using the multiplexed LAMP method.
The multiplexed LAMP assay was further applied to the detection of cDNA reversely transcribed from extracted RNA of three plasma samples from an HCV-infected but HIV-noninfected patient, an HIV-infected but HCV-noninfected patient, and a volunteer noninfected with HCV or HIV (Fig. S3†). We found that the HCV-infected sample gave a POI of 23.7 min in the fluorescence curve in the TAMRA channel with no infection in the fluorescence curve in the Cy5 channel. The HIV-infected sample gave a POI of 34.6 min only in the fluorescence curve in the Cy5 channel, and no infection appeared in the fluorescence curve in the two channels for the non-infected specimen. These results implied the potential of our multiplexed LAMP assay for specific detection of HCV or HIV infections for clinical diagnosis. The multiplexed LAMP assay was further investigated for its ability to quantitatively detect HCV and HIV cDNAs. We found that the real-time fluorescence curves in the two channels gave POIs dynamically correlated with HCV and HIV cDNA concentrations in the range from 1.0 × 10−17 to 1.0 × 10−10 M, respectively (Fig. 1E, F and S4†). The detection limits were estimated to be 5 × 10−18 M and 7 × 10−18 M for HCV and HIV cDNAs, respectively, which were also the lowest target concentration that can be 100% detected in four repetitive assays. A closer interrogation revealed that use of the SPs to replace the corresponding loop primers had little effects on the amplification (Fig. S5†). This finding suggested that the SP provided a useful design of the multiplexed LAMP assay.
Multiplexed dLAMP assay for HCV and HIV cDNAs
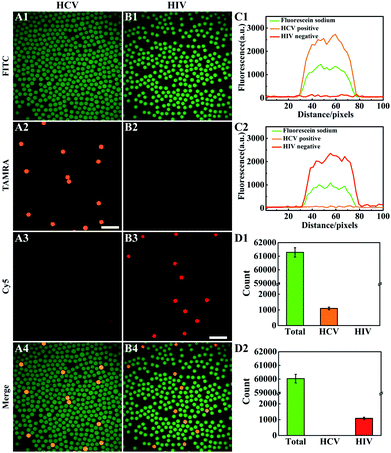
We then explored the possibility of using the multiplexed LAMP assay for the development of the multiplexed dLAMP platform for simultaneous digital detection of HCV and HIV cDNA. According to the multiplexed LAMP design, the droplets with orange and red fluorescence were positive for HCV and HIV cDNA, respectively. Moreover, we introduced fluorescein into the LAMP reaction reagent such that all the droplets could be detected using green fluorescence through fluorescence imaging in the droplet counting microwell. The fluorescein staining of droplets facilitated our counting of the entire droplets. To collect the fluorescence image of the whole microwell, 300 small images were scanned and combined into the large image. From the green fluorescence image in the fluorescein channel, we observed monodisperse droplets with an average diameter of ∼50 μm (Fig. S6 and S7†). After thorough optimization of the reaction time, we found that the counting of positive droplets became stabilized after 60 min (Fig. S8†). With the optimized reaction time, we obtained typical small fluorescence images in fluorescein, TAMRA and Cy5 channels for two samples containing 600 copies per μL of HCV cDNA and HIV cDNA, respectively (Fig. 2A and B). The droplets were readily detected in the green fluorescence images, with high mono-dispersity and uniform size. In the orange and NIR fluorescence images, most of the droplets exhibited very weak fluorescence, and only a small number of fluorescent droplets were detected. A closer examination of fluorescence intensity profiles was performed for one of the bright fluorescent droplets (Fig. 2C). We observed intense fluorescence profiles in the fluorescein channel for both droplets, but intense fluorescence profiles in TAMRA and Cy5 channels were merely obtained in the HCV cDNA positive and HIV cDNA positive samples, respectively. This result verified that bright fluorescent droplets in the orange image implied the presence of HCV cDNA, while bright fluorescent droplets in the NIR image implied the presence of HIV cDNA. We then conducted a further quantitative analysis of the large fluorescence images of the two HCV and HIV cDNA samples (Fig. 2D). For the HCV cDNA sample, 61![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 282 droplets were counted in the FITC channel, no droplets were found to display appreciable Cy5 fluorescence, implying high specificity of the multiplexed LAMP reaction. Droplets with bright TAMRA fluorescence were counted as 1095, indicating 1.79% of the percentage of HCV-positive droplets, p. According to the Poisson distribution, the copy number of HCV cDNA could be calculated by the equation m = n × ln[1/(1 − p)],37 where n is the total number of droplets. Hence, we expected that there was 1105 copy number of HCV cDNA in the sample input to the droplet generator. The total volume of the sample was estimated using V = n/2 × Vd where Vd was the droplet size. Then, the concentration of HCV cDNA was calculated to be 551 copies per μL in the sample, indicating a detection efficiency of 91.8% for the multiplexed LAMP reaction. Similarly, for the HIV cDNA sample the number of green fluorescent droplets was 60
282 droplets were counted in the FITC channel, no droplets were found to display appreciable Cy5 fluorescence, implying high specificity of the multiplexed LAMP reaction. Droplets with bright TAMRA fluorescence were counted as 1095, indicating 1.79% of the percentage of HCV-positive droplets, p. According to the Poisson distribution, the copy number of HCV cDNA could be calculated by the equation m = n × ln[1/(1 − p)],37 where n is the total number of droplets. Hence, we expected that there was 1105 copy number of HCV cDNA in the sample input to the droplet generator. The total volume of the sample was estimated using V = n/2 × Vd where Vd was the droplet size. Then, the concentration of HCV cDNA was calculated to be 551 copies per μL in the sample, indicating a detection efficiency of 91.8% for the multiplexed LAMP reaction. Similarly, for the HIV cDNA sample the number of green fluorescent droplets was 60![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 032 and NIR fluorescent droplets were 1079, suggesting 1.80% of HIV-positive droplets in all the droplets. Therefore, the expected copy number of HIV cDNA was in the input sample, and the concentration of HIV cDNA was 555 copies per μL with a detection efficiency of 92.5%. These imply the ability of the multiplexed dLAMP for simultaneous absolute quantification of multiple nucleic acid analytes.
032 and NIR fluorescent droplets were 1079, suggesting 1.80% of HIV-positive droplets in all the droplets. Therefore, the expected copy number of HIV cDNA was in the input sample, and the concentration of HIV cDNA was 555 copies per μL with a detection efficiency of 92.5%. These imply the ability of the multiplexed dLAMP for simultaneous absolute quantification of multiple nucleic acid analytes.
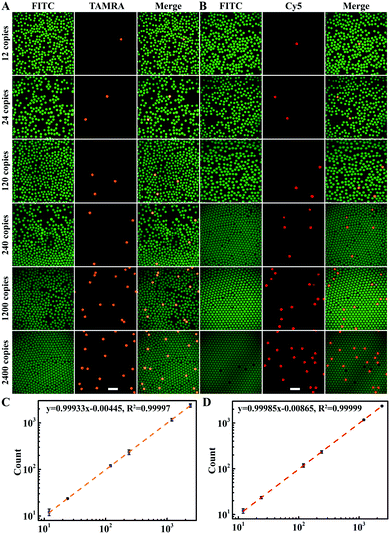
Then, the ability of multiplexed dLAMP for quantitative digital detection HCV and HIV cDNA was evaluated (Fig. 3A, B and S9, Table S2†). With samples containing increasing concentrations of HCV and HIV cDNA, we obtained increased counts of positive droplets in the fluorescence images. The estimated copy numbers for HCV and HIV cDNA were in good consistency with the actual copy numbers of cDNA targets in the samples from 12 copies to 1200 copies (Fig. 3C and D). A detection limit of 4 copies was estimated for both HCV and HIV cDNA according to triple standard deviation over the average count in four repetitive assays for the blank samples (Table S3†). Moreover, multiplexed dLAMP was found to have comparable reliability and precision with digital PCR.14,38 In four repetitive assays, the relative error was −2.51% and −2.03% for 1200 copies of HCV and HIV, respectively, and coefficients of variation (CV) of range from 12 to 1200 copies were below 16.57%. These results demonstrated high sensitivity and superb precision of multiplexed dLAMP for the quantification of nucleic acids.
Multiplexed dLAMP for simultaneous quantitation of samples containing two cDNA targets
Next, we investigated the performance of multiplexed dLAMP for simultaneous quantitation of samples containing both cDNA targets. For a sample containing 120 copies per μL HCV cDNA and 120 copies per μL HIV cDNA, the positive droplets were counted to be 227 for HCV cDNA and 224 for HIV cDNA among the total 60![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 738 droplets, with estimated copy numbers being 228 copies and 225 copies for HCV cDNA and HIV cDNA, respectively (Fig. 4A). The concentrations of HCV and HIV cDNA were calculated to be 115 copies per μL and 113 copies per μL in the sample, indicating a detection efficiency of 95.8% and 94.2%, respectively. For a sample containing 120 copies per μL HCV cDNA and 1200 copies per μL HIV cDNA, there were 226 HCV positive droplets and 2197 HIV positive droplets in the total 61
738 droplets, with estimated copy numbers being 228 copies and 225 copies for HCV cDNA and HIV cDNA, respectively (Fig. 4A). The concentrations of HCV and HIV cDNA were calculated to be 115 copies per μL and 113 copies per μL in the sample, indicating a detection efficiency of 95.8% and 94.2%, respectively. For a sample containing 120 copies per μL HCV cDNA and 1200 copies per μL HIV cDNA, there were 226 HCV positive droplets and 2197 HIV positive droplets in the total 61![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 746 droplets (Fig. 4B). These found cDNA copy numbers exhibited little discrepancy from the actual values. A similar quantification result was obtained for the sample containing 1200 copies per μL HCV and 1200 copies per μL HIV cDNA with 2162 HCV positive droplets and 2239 HIV positive droplets in the total 60
746 droplets (Fig. 4B). These found cDNA copy numbers exhibited little discrepancy from the actual values. A similar quantification result was obtained for the sample containing 1200 copies per μL HCV and 1200 copies per μL HIV cDNA with 2162 HCV positive droplets and 2239 HIV positive droplets in the total 60![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 237 droplets (Fig. 4C). Interestingly, we found one droplet containing both HCV cDNA and HIV cDNA, for which the fluorescence images displayed intense profiles in both TAMRA and Cy5 channels (Fig. S10†). The results demonstrated the ability of the multiplexed dLAMP strategy for simultaneous and precise quantification of HCV and HIV cDNAs.
237 droplets (Fig. 4C). Interestingly, we found one droplet containing both HCV cDNA and HIV cDNA, for which the fluorescence images displayed intense profiles in both TAMRA and Cy5 channels (Fig. S10†). The results demonstrated the ability of the multiplexed dLAMP strategy for simultaneous and precise quantification of HCV and HIV cDNAs.
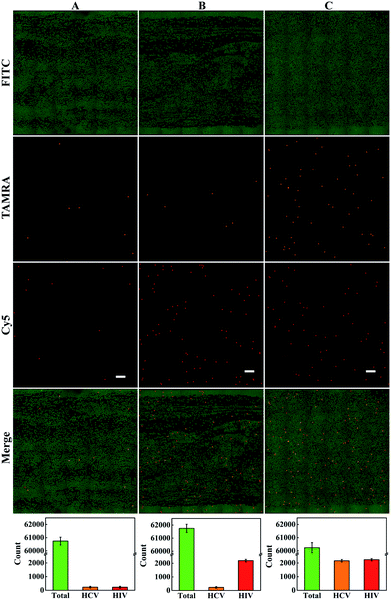
Furthermore, we explored the potential of the multiplexed dLAMP strategy for the quantification of HCV and HIV viruses in clinical plasma samples. In the assays, 16 plasma samples were obtained from The Third Xiangya Hospital of Central South University, with RNA extracted and cDNA obtained using a commercialized reverse transcription kit. The assays were performed in a double-blind experiment with no qPCR result labelled to the samples, and the samples were tested independently using the dLAMP assay. The results of qPCR and dLAMP were directly compared to evaluate the performance of the dLAMP method. The copy numbers of HCV and HIV viruses found using multiplexed dLAMP assays showed deviations ranging from −20% to 2.5% from those obtained using qPCR (Table S4 and Fig. S11–S14†). These results implied the promise of the multiplexed dLAMP strategy for highly sensitive quantification of HCV and HIV viruses in complicated clinical samples.
Conclusions
We have developed a multiplexed droplet LAMP assay (dLAMP) strategy for simultaneous quantification of multiple nucleic acids. The dLAMP strategy relied on a set of scorpion-shaped loop primers for isothermal amplification, a droplet microfluidic chip for droplet generation, and a droplet counting microwell chip for enumerating the positive droplets. The multiplexed dLAMP strategy was successfully demonstrated for the quantification of HCV and HIV cDNA with high precision and detection limits as low as 4 copies. This strategy also demonstrated the potential for the simultaneous digital assay of HCV and HIV RNA in clinical plasma samples. Combination of the small fluorescence imaging module and the presented dLAMP will make it possible to develop a multiplexed dLAMP platform for POC applications. This multiplexed dLAMP technique can afford a useful platform for highly sensitive and specific detection of nucleic acids of viruses and other pathogens, implying its potential for rapid diagnosis and prevention of infectious diseases.Author contributions
Y. L. T., L. J. T., and J. H. J. designed the project, discussed the results and contributed to writing the manuscript. Y. L. T. and A. Q. H. conducted experiments and performed the graphic design. All authors participated in the reviewed and have approved the final version of the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the National Key Research Program (Grant 2019YFA0905800) and NSFC Programs (Grants 21874040, 22074037 and 21527810).Notes and references
- D. E. Bloom, S. Black and R. Rappuoli, Proc. Natl. Acad. Sci. U. S. A., 2017, 114, 4055–4059 CrossRef CAS PubMed.
- K. Shah, E. Bentley, A. Tyler, K. S. R. Richards, E. Wright, L. Easterbrook, D. Lee, C. Cleaver, L. Usher, J. E. Burton, J. K. Pitman, C. B. Bruce, D. Edge, M. Lee, N. Nazareth, D. A. Norwood and S. A. Moschos, Chem. Sci., 2017, 8, 7780–7797 RSC.
- E. Petersen, M. Koopmans, U. Go, D. H. Hamer, N. Petrosillo, F. Castelli, M. Storgaard, S. Al Khalili and L. Simonsen, Lancet Infect. Dis., 2020, 20, e238–e244 CrossRef CAS PubMed.
- I. Pereiro, A. Bendali, S. Tabnaoui, L. Alexandre, J. Srbova, Z. Bilkova, S. Deegan, L. Joshi, J.-L. Viovy, L. Malaquin, B. Dupuy and S. Descroix, Chem. Sci., 2017, 8, 1329–1336 RSC.
- D. D. Nguyen, K. Gao, J. Chen, R. Wang and G.-W. Wei, Chem. Sci., 2020, 11, 12036–12046 RSC.
- R. Liu, L. He, Y. Hu, Z. Luo and J. Zhang, Chem. Sci., 2020, 11, 12157–12164 RSC.
- F. Shen, B. Sun, J. E. Kreutz, E. K. Davydova, W. Du, P. L. Reddy, L. J. Joseph and R. F. Ismagilov, J. Am. Chem. Soc., 2011, 133, 17705–17712 CrossRef CAS PubMed.
- Y.-T. Yeh, K. Gulino, Y. Zhang, A. Sabestien, T.-W. Chou, B. Zhou, Z. Lin, I. Albert, H. Lu, V. Swaminathan, E. Ghedin and M. Terrones, Proc. Natl. Acad. Sci. U. S. A., 2020, 117, 895–901 CrossRef CAS PubMed.
- B. Sun, F. Shen, S. E. McCalla, J. E. Kreutz, M. A. Karymov and R. F. Ismagilov, Anal. Chem., 2013, 85, 1540–1546 CrossRef CAS PubMed.
- Y. Wang, Q. Ruan, Z.-C. Lei, S.-C. Ling, Z. Zhu, L. Zhou and C. Yang, Anal. Chem., 2018, 90, 5224–5231 CrossRef CAS PubMed.
- G. Gines, R. Menezes, K. Nara, A.-S. Kirstetter, V. Taly and Y. Rondelez, Sci. Adv., 2020, 6, eaay5952 CrossRef CAS PubMed.
- L. Shang, Y. Cheng and Y. Zhao, Chem. Rev., 2017, 117, 7964–8040 CrossRef CAS PubMed.
- M. Li, W. D. Chen, N. Papadopoulos, S. N. Goodman, N. C. Bjerregaard, S. Laurberg, B. Levin, H. Juhl, N. Arber, H. Moinova, K. Durkee, K. Schmidt, Y. He, F. Diehl, V. E. Velculescu, S. Zhou, L. A. Diaz Jr, K. W. Kinzler, S. D. Markowitz and B. Vogelstein, Nat. Biotechnol., 2009, 27, 858–863 CrossRef CAS PubMed.
- K. Yang, J. Li, J. Zhao, P. Ren, Z. Wang, B. Wei, B. Dong, R. Sun, X. Wang, H. J. M. Groen, J. Ma and Y. Guo, Anal. Chem., 2018, 90, 11203–11209 CrossRef CAS PubMed.
- N. G. Schoepp, E. M. Khorosheva, T. S. Schlappi, M. S. Curtis, R. M. Humphries, J. A. Hindler and R. F. Ismagilov, Angew. Chem., Int. Ed., 2016, 55, 9557–9561 CrossRef CAS PubMed.
- J. Jarvius, J. Melin, J. Goransson, J. Stenberg, S. Fredriksson, C. Gonzalez-Rey, S. Bertilsson and M. Nilsson, Nat. Methods, 2006, 3, 725–727 CrossRef CAS PubMed.
- F. Schuler, F. Schwemmer, M. Trotter, S. Wadle, R. Zengerle, F. von Stetten and N. Paust, Lab Chip, 2015, 15, 2759–2766 RSC.
- Y. Zhang, L. Zhang, J. Sun, Y. Liu, X. Ma, S. Cui, L. Ma, J. J. Xi and X. Jiang, Anal. Chem., 2014, 86, 7057–7062 CrossRef CAS PubMed.
- T. Notomi, H. Okayama, H. Masubuchi, T. Yonekawa, K. Watanabe, N. Amino and T. Hase, Nucleic Acids Res., 2000, 28, e63 CrossRef CAS PubMed.
- J. Rodriguez-Manzano, M. A. Karymov, S. Begolo, D. A. Selck, D. V. Zhukov, E. Jue and R. F. Ismagilov, ACS Nano, 2016, 10, 3102–3113 CrossRef CAS PubMed.
- Y. Zhao, F. Chen, J. Qin, J. Wei, W. Wu and Y. Zhao, Chem. Sci., 2018, 9, 392–397 RSC.
- Y. Zhao, F. Chen, Q. Li, L. Wang and C. Fan, Chem. Rev., 2015, 115, 12491–12545 CrossRef CAS PubMed.
- X. Fang, H. Chen, S. Yu, X. Jiang and J. Kong, Anal. Chem., 2011, 83, 690–695 CrossRef CAS PubMed.
- H. Yuan, Y. Chao, S. Li, M. Y. H. Tang, Y. Huang, Y. Che, A. S. T. Wong, T. Zhang and H. C. Shum, Anal. Chem., 2018, 90, 13173–13177 CrossRef CAS PubMed.
- J. Chen, X. Xu, Z. Huang, Y. Luo, L. Tang and J. H. Jiang, Chem. Commun., 2018, 54, 291–294 RSC.
- M. Yu, X. Chen, H. Qu, L. Ma, L. Xu, W. Lv, H. Wang, R. F. Ismagilov, M. Li and F. Shen, Anal. Chem., 2019, 91, 8751–8755 CrossRef CAS PubMed.
- Y. Hu, P. Xu, J. Luo, H. He and W. Du, Anal. Chem., 2016, 89, 745–750 CrossRef PubMed.
- R. Wang, R. Zhao, Y. Li, W. Kong, X. Guo, Y. Yang, F. Wu, W. Liu, H. Song and R. Hao, Lab Chip, 2018, 18, 3507–3515 RSC.
- H. Yuan, Y. Chao and H. C. Shum, Small, 2020, 16, 1904469 CrossRef CAS PubMed.
- N. G. Schoepp, T. S. Schlappi, M. S. Curtis, S. S. Butkovich, S. Miller, R. M. Humphries and R. F. Ismagilov, Sci. Transl. Med., 2017, 9, eaal3693 CrossRef PubMed.
- Y.-D. Ma, W.-H. Chang, K. Luo, C.-H. Wang, S.-Y. Liu, W.-H. Yen and G.-B. Lee, Biosens. Bioelectron., 2018, 99, 547–554 CrossRef CAS PubMed.
- F. Zerilli, C. Bonanno, E. Shehi, G. Amicarelli, D. Adlerstein and G. M. Makrigiorgos, Clin. Chem., 2010, 56, 1287–1296 CrossRef CAS PubMed.
- N. A. Tanner, Y. Zhang and T. C. Evans Jr, BioTechniques, 2012, 53, 81–89 CrossRef CAS PubMed.
- Y. S. Jiang, S. Bhadra, B. Li, Y. R. Wu, J. N. Milligan and A. D. Ellington, Anal. Chem., 2015, 87, 3314–3320 CrossRef CAS PubMed.
- L. Becherer, M. Bakheit, S. Frischmann, S. Stinco, N. Borst, R. Zengerle and F. von Stetten, Anal. Chem., 2018, 90, 4741–4748 CrossRef CAS PubMed.
- C. S. Ball, Y. K. Light, C.-Y. Koh, S. S. Wheeler, L. L. Coffey and R. J. Meagher, Anal. Chem., 2016, 88, 3562–3568 CrossRef CAS PubMed.
- B. J. Hindson, K. D. Ness, D. A. Masquelier, P. Belgrader, N. J. Heredia, A. J. Makarewicz, I. J. Bright, M. Y. Lucero, A. L. Hiddessen, T. C. Legler, T. K. Kitano, M. R. Hodel, J. F. Petersen, P. W. Wyatt, E. R. Steenblock, P. H. Shah, L. J. Bousse, C. B. Troup, J. C. Mellen, D. K. Wittmann, N. G. Erndt, T. H. Cauley, R. T. Koehler, A. P. So, S. Dube, K. A. Rose, L. Montesclaros, S. Wang, D. P. Stumbo, S. P. Hodges, S. Romine, F. P. Milanovich, H. E. White, J. F. Regan, G. A. Karlin-Neumann, C. M. Hindson, S. Saxonov and B. W. Colston, Anal. Chem., 2011, 83, 8604–8610 CrossRef CAS PubMed.
- F. Wang, L. Zhu, B. Liu, X. Zhu, N. Wang, T. Deng, D. Kang, J. Pan, W. Yang, H. Gao and Y. Guo, Anal. Chem., 2018, 90, 8919–8926 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1sc00616a |
| This journal is © The Royal Society of Chemistry 2021 |