Open Access Article
Open Access ArticleOne-step hexaplex immunoassays by on-line paper substrate-based electrospray ionization mass spectrometry for combined cancer biomarker screening†
Shuting
Xu
ac,
Mingxia
Liu
a,
Jie
Feng
b,
Guangtao
Yan
b,
Yu
Bai
 *a and
Huwei
Liu
*a and
Huwei
Liu
 a
a
aBeijing National Laboratory for Molecular Sciences, Key Laboratory of Bioorganic Chemistry and Molecular Engineering of Ministry of Education, College of Chemistry and Molecular Engineering, Peking University, Beijing 100871, P. R. China. E-mail: yu.bai@pku.edu.cn; Tel: +86 10 6275 8198
bDepartment of Clinical Laboratory Center, The First Medical Center of the Chinese People's Liberation Army General Hospital, Beijing 100853, P. R. China
cInstitute of Analytical Food Safety, School of Food Science and Technology, Jiangnan University, Wuxi 214122, P. R. China
First published on 17th February 2021
Abstract
Mass spectrometry (MS) is attractive as a multiplexed immunoassay readout benefiting from its high sensitivity, speed and mass resolution. Here, a simple paper-based hexaplex immunoassay with an on-line MS readout was proposed, using functionalized paper as the immune substrates, along with rhodamine-based mass tags assembled on gold nanoparticles prepared as the mass probes (MPs). Simultaneous immune capture and labeling were conducted in one step on paper substrates in 96-well plates with a high throughput within 30 minutes, and the on-line efficient dissociation of the mass tags highly facilitated the hexaplex readout of the immune signals by a newly established on-line paper substrate-based electrospray ionization-MS setup. Six MPs were synthesized for the simultaneous quantification of six important cancer protein markers (cancer antigen 15-3, cancer antigen 19-9, carcinoma embryonic antigen, cancer antigen 125, human epididymis protein 4, and alpha fetoprotein) using only 10 μL serum, presenting satisfactory sensitivity, accuracy and specificity. This platform was further tested in screening for the six biomarkers in serum samples of patients with breast, liver and gastric cancers, showing its high potential for sensitive and specific early cancer diagnosis.
Introduction
Cancer survival is greatly dependent on effective detection at an early stage, so early cancer screening has become more and more popularized in regular physical exams.1,2 Diagnosis techniques with higher throughput, broader spectra and user-friendliness are in high demand. Serologic protein markers are common indicators in blood examination for early cancer diagnosis,3,4 but they are always harder to quantify because of their low abundance and interference from the complicated matrix.Immunoassays have been employed as gold-standards for protein quantification, which increase the protein signal by labeling tags and reduce the matrix interference based on the specific recognition between antigens and antibodies.5–7 Various commercial immunoassay kits and high throughput readout instruments have been developed for clinical use, represented by enzyme-linked immunosorbent assays (ELISAs)5 and electrochemiluminescence immunoassays (ECLIAs).7 For signal amplification, horse radish peroxidase (HRP) and ruthenium tris(bipyridine) [Ru(bpy)32+] are the top-two labeling tags, which possess the advantages of high sensitivity, high stability, and universality for optical readout.8,9 However, measurements based on the optical readout are often restricted in multi-target simultaneous detection (limited to five targets), an important aspect of throughput, because of scant optical tags and overlap between their emission spectra.10 An obvious trend for early cancer diagnosis is to monitor more and more biomarkers for broad disease screening or accurate diagnosis,11,12 which means repeated measurements of one sample have to be performed using these advanced methods with larger sample consumption, longer detection time or a complex multi-channel design.13,14
Mass spectrometry (MS) allows for unfettered resolution of targets and is an attractive one-spot high-throughput readout technique for immunoassays using mass tags for protein labeling.15–19 However, MS-based methods are encumbered by complex instruments compared with universal optical-based techniques. Especially, inductively coupled plasma (ICP) or laser ablation sources are usually employed for tag dissociation and ionization.20,21 The emergence of ambient ionization makes a simple, flexible and sample-oriented setup possible for MS detection.22,23 Among these techniques, paper spray ionization (PSI) is recognized as one of the simplest ionization forms24–27 and has been demonstrated to be suitable for multiplexed immunoassays carried out on paper,28–30 but challenges remain for clinical detection. One big challenge is the paper-to-paper reproducibility because the paper tip shapes and the reactive areas are critical for accurate quantitative detection. Another challenge is the lack of efficient mass tag series compatible with the PSI system, which is directly related to the simultaneous targets in a single test. High energy is hard to provide in the soft PSI process for the direct cleavage of mass-tag bonds. As an alternative, bond cleavage by rapid and universal chemical reactions has become the major way to realize it, but the reactions may limit the design of the tag and introduce additional pretreatment. Our previous work indicated that Au–S bonds could be cleaved by chemical or electrochemical processes under electrospray ionization, which has been recognized as a soft and efficient mode.31,32
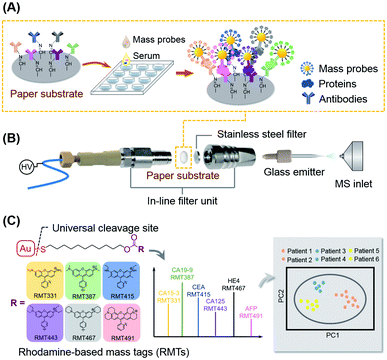
To meet the challenges above in PSI MS-based immunoassays, paper substrates were designed in round pieces and were functionalized for multiplexed sandwich-type immunoassays in this work. The immune reactions on the paper substrates proceeded in 96-well plates in a one-step, high-throughput and routine method based on ELISA with good reproducibility (Fig. 1A). In contrast to the typical PSI setups that employ the special triangle paper tip for ionization, paper substrate-based electrospray ionization (PS-ESI) was established (Fig. 1B). The PS-ESI setup was specially designed for the paper substrates, combining the in-line filter unit for target dissociation and glass ESI emitters for ionization, which greatly improved the stability during spraying and the reproducibility. For the multiplexed immunoassays, mass probes (MPs) were synthesized by assembling a series of rhodamine-based mass tags (RMTs) on gold nanoparticles (GNPs) by Au–S bonds. The highly efficient online cleavage of Au–S bonds facilitated the mass tag detection, which became the basis for the immune signal MS readout. Based on the feature of Au–S bond dissociation, RMT homologous on the synthesized MPs were dissociated simultaneously for the detection of multiple cancer protein markers (Fig. 1C). As a demonstration, six MPs were synthesized and applied for the quantification of six important cancer protein markers in serum, including cancer antigen 15-3 (CA15-3), cancer antigen 19-9 (CA19-9), carcinoma embryonic antigen (CEA), cancer antigen 125 (CA125), human epididymis protein 4 (HE4), and alpha fetoprotein (AFP). Serum samples of cancer patients were screened for further evaluation of the proposed hexaplex immunoassay, and combined biomarkers were preliminarily examined for cancer diagnosis, which is significant and valuable for the application of the MS-based multiplexed immunoassays in highly efficient clinical serological examination.
Experimental section
Chemicals and materials
Six rhodamine-based mass tags (RMTs), including RMT331, RMT387, RMT415, RMT443, RMT467, and RMT491, were synthesized according to our previous work31,32 and details of the chemicals and synthetic routes are described in the ESI (Fig. S1†). Six target proteins, including CA15-3, CA19-9, CEA, CA125, HE4, and AFP, and their capture antibodies (cAbs) and detection antibodies (dAbs) were purchased from Novus Biologicals, Inc. (Colorado, USA) and Fitzgerald, Inc. (Acton, MA, USA) (Table S1†). Other chemicals and materials for immune preparation and detection are summarized in the ESI† and all chemicals were used without further purification. Serum samples were collected from healthy donors and breast (malignant: B1, B2, and B3; benign: B4, B5, and B6), liver (L1, L2, and L3) and gastric (G1, G2, and G3) cancer patients from the Chinese People's Liberation Army General Hospital.Apparatus
A paper substrate-based ESI setup (PS-ESI) was established in front of the MS inlet for on-line dissociation of the mass tags and the detection of immune paper substrates. The setup consisted of a refitted in-line filter unit (part no. 205000343, Waters Corporation, USA), a home-made glass ESI emitter, a cross joint for applied voltage and solvent supplied by a syringe pump (Fig. 1B and S2A†). The in-line filter unit was able to accommodate the round piece of paper substrate (4 mm diameter) inside along with the original 0.2 μm stainless-steel filter with good sealing and pressure resisting (Fig. S2B†). The stainless-steel filter protected the emitter from clogging. The glass emitter was pulled from borosilicate glass capillaries (1.0 mm o.d., 0.2 mm i.d., Sutter Instrument, USA) by the P-2000 (Sutter Instrument, USA). The parameters of P-2000 were as follows: HEAT = 400, FIL = 5, VEL = 28, DEL = 125, and PUL = 80 to make an emitter with a tip of about 30 μm o.d. (Fig. S2C†). The spray voltage was applied by an electrode inserted through the cross joint. The voltage was supplied from the mass spectrometer and could be controlled by the MS control software. The solvent from the injection syringe flowed through the cross joint (place of high voltage), the paper substrate (platform of the sandwich-type immunoassay), and the stainless-steel filter to finally be sprayed by the glass ESI emitter.LTQ XL MS (Thermo Scientific, USA) was employed for an immune readout. During detection, the on-line dissociation and ionization setup was installed on the commercial Nanospray Flex source (Thermo Scientific, USA) which was equipped with a 3D moving platform allowing the horizontal and vertical position adjustment and digital microscopes to monitor the emitters and spray. All mass spectra were recorded in the positive full scan mode with in-source collision induced dissociation (in-source CID). Argon gas (99.995%) was used as the collision gas. The MS parameters of LTQ were set as follows: scan range: m/z = 100–1000, capillary temperature: 275 °C, capillary voltage: 39 V, tube lens voltage: 130 V, maximum injection time: 10 ms, microscans: 1, and scan rate: enhanced. After optimization, the spray voltage was set at 5 kV, and the fragmentation voltage of the in-source CID was set at 100 V. Xcalibur software (version 4.0) was used for the control of the MS system and data analysis.
Chromogenic results for the ELISAs with HRP-conjugated antibodies were recorded by a microplate reader (Bio-Rad model 550, USA) with a 450 nm interference filter.
Transmission electron microscopy (TEM) images were collected using a field-emission high-resolution transmission electron microscope (JEM-2100F, Japan).
Preparation of capture antibody modified paper substrates
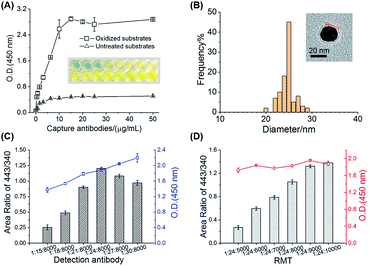
As sandwich-type immunoassay substrates, the 4 mm diameter round paper pieces were prepared using Whatman #1 chromatography paper (GE Healthcare, USA). The size was appropriately adapted to the downstream 96-well plates and the in-line filter unit. The paper substrates were washed with acetone, methanol, and water for 10 min in sequence. The preparation route of the capture antibody modified paper substrates is shown in Fig. S3A.†The cellulose on paper substrates was oxidized to aldehyde groups for antibody-functionalization. The paper substrates were soaked in 0.03 mol L−1 KIO4 solution at 65 °C for 3 hours and then washed with water three times. The functionalization of aldehyde groups was tested with a 2,4-dinitrophenylhydrazine (2,4-DNP) solution. 10 μL 2,4-DNP solution was added to each paper substrate and incubated for 1 min. After washing with water five times, the color of each substrate was observed (Fig. S4†).
For the antibody-functionalization, an oxidized paper substrate was placed in each well of the 96-well plates (part no. 3599-1, Corning, USA), and subsequent modification and immune recognition were carried out in the wells. 20 μL of the capture antibodies in PBS buffer (+10% v/v glycerol) was added into the wells. The concentrations of the capture antibodies were optimized from 2.5 μg mL−1 to 50 μg mL−1, and a final concentration of 15 μg mL−1 was used for modification. For the simultaneous detection of the six targets, a mixture of the six capture antibodies each with a concentration of 2.5 μg mL−1 was used. The paper substrates immersed in the capture antibody solution were incubated at 4 °C with gentle shaking for 12 h. The capture antibody modified paper substrates (PS@cAb) were washed with PBS four times and then immersed in 100 μL tris–HCl buffer containing 0.1% bovine serum albumin (BSA) for 2 h to block the unreacted active sites and reduce nonspecific adsorption. After blocking, the substrates were washed with PBS buffer three times and used immediately or kept in PBS buffer at 4 °C for a week.
Preparation of the mass probes
Spherical gold nanoparticles (GNPs) with a diameter of about 25 nm were synthesized using citrate as a reducing agent as the core of the MPs and they were sequentially modified by detection antibodies (dAbs) and RMTs. The GNP functionalization was as follows (Fig. S3B†): 120 μg mL−1 antibodies was reacted with 4,7,10,13,16,19,22,25,32,35,38,41,44,47,50,53-hexadecaoxa-28,29-dithiahexapentacontanedioic acid di-N-succinimidyl ester (NHS-PEG-S-S-PEG-NHS) in a 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 molar ratio in 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) buffer for 12 h. Then, a certain volume of reacted antibody-PEG-SH (with a concentration of 120 μg mL−1) was added into 1 mL 1 nmol L−1 GNPs, followed by the addition of K2CO3 with a final concentration of 1.8 mmol L−1. The mixture was incubated for 12 h with gentle shaking in the dark at room temperature, followed by the addition of a certain volume of 100 μmol L−1 RMTs for continued incubation for another 12 h. The added volume of reacted antibodies and RMTs was optimized from 18.75–37.5 μL and 50–100 μL to obtain the highest sensitivity and specificity, and 30 μL reacted antibodies and 90 μL RMTs were finally selected for GNP probe preparation. Afterwards, the dAbs/RMTs modified GNPs (GNPs@dAbs/RMTs) were centrifuged (6000 rpm, 15 min) and washed with water 3 times, then the resulting GNPs@dAbs/RMTs were re-suspended in 1 mL PBS buffer. Six MPs (GNPs@anti-CA15-3-2/RMT331, GNPs@CA19-9-2/RMT387, GNPs@anti-CEA-2/RMT415, GNPs@anti-CA125-2/RMT443, GNPs@anti-HE4-2/RMT467, and GNPs@anti-AFP-2/RMT491) were prepared by the same process, and six MPs were mixed with equal volumes for simultaneous detection. The GNPs@dAbs/RMTs were used immediately or kept at 4 °C for a week.
1 molar ratio in 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) buffer for 12 h. Then, a certain volume of reacted antibody-PEG-SH (with a concentration of 120 μg mL−1) was added into 1 mL 1 nmol L−1 GNPs, followed by the addition of K2CO3 with a final concentration of 1.8 mmol L−1. The mixture was incubated for 12 h with gentle shaking in the dark at room temperature, followed by the addition of a certain volume of 100 μmol L−1 RMTs for continued incubation for another 12 h. The added volume of reacted antibodies and RMTs was optimized from 18.75–37.5 μL and 50–100 μL to obtain the highest sensitivity and specificity, and 30 μL reacted antibodies and 90 μL RMTs were finally selected for GNP probe preparation. Afterwards, the dAbs/RMTs modified GNPs (GNPs@dAbs/RMTs) were centrifuged (6000 rpm, 15 min) and washed with water 3 times, then the resulting GNPs@dAbs/RMTs were re-suspended in 1 mL PBS buffer. Six MPs (GNPs@anti-CA15-3-2/RMT331, GNPs@CA19-9-2/RMT387, GNPs@anti-CEA-2/RMT415, GNPs@anti-CA125-2/RMT443, GNPs@anti-HE4-2/RMT467, and GNPs@anti-AFP-2/RMT491) were prepared by the same process, and six MPs were mixed with equal volumes for simultaneous detection. The GNPs@dAbs/RMTs were used immediately or kept at 4 °C for a week.
Sandwich-type immunoassays and detection
The recognition and labeling of the target proteins was conducted on PS@cAb in each well of the 96-well plate. 10 μL sample and 40 μL MPs with an optimized concentration of 0.5 nmol L−1 were both added to the well for simultaneous protein recognition and labeling. The 96-well plate was then incubated in the dark at 37 °C for 30 min. Then, the substrates were washed with PBS five times and washed with water quickly one time with controlling the total washing within 10 min. After washing, the substrates were dried in air (within 10 min), and 1 μL crystal violet (CV) methanol solution with a concentration of 1 μg mL−1 was dropped onto the substrates as an internal standard (IS) for quantification.Dried substrates were placed in the in-line filter, and methanol was flowed through the substrates with an optimized flow rate of 10 μL min−1. The dissociated RMTs and IS were ionized using the glass emitter and detected by MS, and each test lasted 2 min.
Along with MS detection, chromogenic detection by a microplate reader similar to the ELISA was utilized during condition optimization and protein detection. The HRP-conjugated antibodies participated in the sandwich-type immunoassays. Other immune preparations on papers were conducted as those for MS detection. After immune preparation, the chromogenic reaction details were as follows: the 3,3′,5,5′-tetramethylbenzidine (TMB) liquid and buffers were mixed in accordance with the kit instructions, and 100 μL mixed solution was added to each well with paper substrate. The plate was incubated with gentle shaking in the dark for 20 min, and the paper substrates were removed after incubation. Then 100 μL 1 mol L−1 HCl was added to quench the chromogenic reaction in the wells, and the plate was detected by a microplate reader, providing cross-validation of the MS results with the chromogenic results.
Data analysis
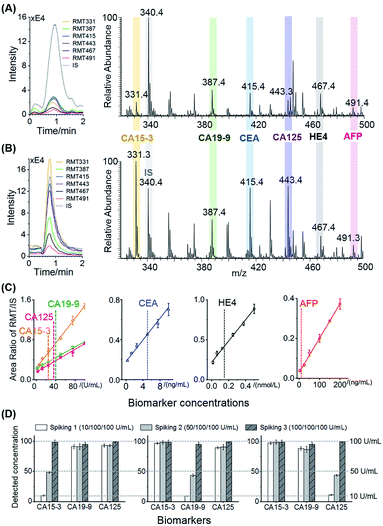
The quantification of the target proteins after MS detection was based on the peak areas of the dissociated RMT signals and IS signals. The extracted ion chromatograms (EICs) of the dissociated RMTs (m/z = 331.1 ± 0.3, m/z = 387.2 ± 0.3, m/z = 415.2 ± 0.3, m/z = 443.2 ± 0.3, m/z = 467.2 ± 0.3, and m/z = 491.2 ± 0.3) and IS (m/z = 340.2 ± 0.3) were obtained from the total ion chromatograms (TICs) for each test. The integral peak areas of the six dissociated RMTs were calibrated with the integral peak areas of the IS, and the ratios of the peak areas were used for further quantitative analysis. In order to estimate the nonspecific binding in the sample, negative controls (buffer/serum) were prepared and detected in parallel.The peak area ratios of the six RMTs and IS were further used for cancer screening. Using the combined six area ratios from the different patient serum samples as the dataset, the differentiation of the different patients was visualized by principal component analysis (PCA) (SIMCA 14.1), and the clustering of different cancer sites was achieved by the hierarchical clustering analysis (Ward's method, square Euclidean distance; SPSS 23).
Results and discussion
Characterization of versatile sandwich-type immune substrates and probes
Filter paper is one of the cheapest and readily available substrates for immunoassays and is easy to functionalize based on the cellulose.28,33 The 2,4-DNP color reaction clearly showed the formation of orange phenylhydrazones on the oxidized papers compared with the untreated papers, demonstrating the successful aldehyde-functionalization (Fig. S4†). The aldehyde groups in the oxidized papers were allowed to react with the amino groups of cAbs to form imine bonds. The modification of antibodies could be visualized after reacting with the HRP-conjugated anti-IgG and examining with the TMB substrate chromogenic kit. The optical density (O.D. 450 nm) values of the paper substrates obviously increased after the modification of HRP-conjugated anti-IgG and was employed for the optimization of antibody amounts on the paper substrates (Fig. 2A). The antibodies reached saturation with an incubation concentration of 15 μg mL−1. Compared to the untreated substrates with minor absorption of antibodies on the papers, the antibodies on the oxidized substrates were modified through stable covalent bonds. The protein activity (>95%) on the modified substrates can be kept for a week in PBS buffer, which provides convenience for batch sample screening (Fig. S5†). Following this general route for substrate modification, six capture antibodies were simultaneously modified for hexaplex capture of six targets on one substrate.Another important component of the sandwich-type immune system was the MPs (GNPs@dAbs/RMTs), which were directly related to the specificity and sensitivity of the MS immunoassays. Monoclonal antibodies with highly specific recognition of targets were utilized as dAbs, while a series of RMTs were used as mass tags, one of which, namely RMT443, was proven to provide a 107 increased sensitivity in our previous work.31 TEM images suggested spherical and monodispersed MPs with a particle diameter of about 25 nm and a modified corona with a 2 nm thickness (Fig. 2B and S7†). The dAbs and RMTs on GNPs had a clear division of responsibilities for protein recognition and signal conversion/amplification, respectively. Thus, an adjustment in their amounts on the GNPs was key to the concurrent high specificity and sensitivity. The O.D. (450 nm) signal of the HRP and MS signal of RMT443 demonstrated their successful modification on GNPs, and these signals were used to evaluate MPs with different amounts of HRP-conjugated anti-IgG and RMT443 for IgG detection. The amount of dAb with a GNP/dAbs ratio of 1/24 was employed because of inefficient recognition of the target protein with less dAbs and reduced signal amplification with more dAbs (Fig. 2C). For the RMTs, the higher amounts the better, but too many RMTs would compete for the sites of dAbs on the GNP surfaces and affect the recognition of the probes. Finally, the molar ratio for GNPs, dAbs, and RMTs was set as 1/24/9000, balancing the high specific recognition to target proteins and high sensitivity of tags (Fig. 2D). Besides the signal amplifiers, RMTs on the GNP probes possessed the function of reducing nonspecific adsorption for the paper-based immunoassays, showing negligible signals for the blank control, which were attributed to the hydrophobicity of the MP surfaces. On the contrary, the addition of the hydrophilic blocking agent (11-mercaptoundecyl)hexa(ethylene glycol) (MUDOL)34,35 on the GNPs instead increased the nonspecific adsorption (Fig. S6†), which was probably because of the strong hydrophilicity of the paper cellulose.36 Without extra blocking, the MPs could be prepared with a simple two-step assembly of dAbs and RMTs on GNPs and were versatile for different dAbs and RMTs.
On-line dissociation and sensitive hexaplex signal detection based on PS-ESI MS
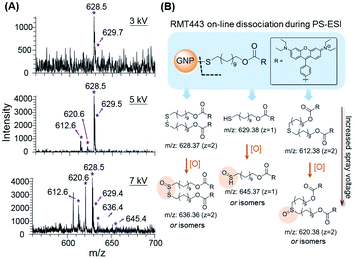
MS with high mass resolution is an ideal multiplexed readout system for immunoassays, but the dissociation of mass tags has always made MS-based techniques more complicated, which employ laser,16,19 plasma,20,37 or specific chemical reactions28 for the bond cleavage. In this system, a simple, rapid and universal dissociation method was developed based on PS-ESI, an on-line dissociation and ionization setup. The cleavage of the Au–S bonds between RMTs and GNPs was discovered completely through the chemical or electrochemical process during ESI without the help of extra laser irradiation, plasma or other reagents. To test the dissociation performance of RMTs on paper substrates, RMTs modified GNPs were loaded on substrates and investigated by PS-ESI MS. Using RMT443 as an example, the signals of the typical dimer derivatives [M–S–S–M]2+ (m/z = 628.5, z = 2, M = C28H30N2O3(CH2)11) were found in the mass spectra when the spray voltages were up to 3 kV (Fig. 3A), demonstrating the successful cleavage of the Au–S bonds on the paper substrates when the charged ESI solvent flowed through the substrates. The on-line dissociation ensured the rapid readout of immune signals, avoiding the extra chromogenic time in conventional ELISA.The mechanism of on-line dissociation was further discussed. Similar Au–S cleavage phenomena have been discovered on substrates in the open air with a charged solvent spray31 or in a flowing solution during ESI32 in our previous work, and one possible mechanism suggested the cleavage of the Au–S bond undergoing chemically or electrochemically controlled processes with the catalysis of GNPs. According to this mechanism, increased spray voltages, an important parameter for electrochemical processes, were applied for the PS-ESI system. More RMT443 dissociated derivatives were generated with increased spray voltages (Fig. 3A) and further identified to be RMT443 derivatives using tandem MS (Table S2†). More dissociated derivatives appearing with higher voltages were highly probable to be oxidation products of dissociated RMT443, the proposed structures are shown in Fig. 3B. The appearance of oxidized derivatives was consistent with the chemical or electrochemical mechanism mentioned above. However, different from the abundant deeply oxidized products reported in the ambient environment,31 fewer deep oxidized derivatives were produced in this system, attributed to the closed solution environment.
The Au–S cleavage could be employed as a universal site for thiol-based mass tags self-assembling on GNPs. Besides RMT443, more RMTs, including RMT331, RMT387, RMT415, RMT467, and RMT491, were designed and synthesized. These mass tag series had the same thiol group and undecyl chain but were alterable in the rhodamine head groups which provided distinguishable signals in the MS readout for the simultaneous detection of different target proteins. The six RMTs were proven to be in situ dissociated simultaneously by the PS-ESI with high efficiency (Fig. S8A†).
Higher sensitivity was achieved when in-source CID was applied after dissociation and ionization. With the help of in-source CID, the dissociated RMTs generated their specific fragments of rhodamine head groups, and the rhodamine fragments accumulated the dissociated signals. Six RMTs underwent similar fragmentation processes but RMTs with different head groups presented differences in the fragmentation degree under increased fragmentation energy (Fig. S8B†). Higher energy was required for the fragmentation of RMTs with larger conjugated head structures. For example, a large proportion of RMT331 derivatives cleaved beside the ester bonds under a fragmentation energy of 80 V but for RMT491, its derivatives were not completely cleaved even under the highest fragmentation energy of 100 V. Following the overall trends for the six RMTs, the fragmentation energy was set as 100 V for the simultaneous detection, providing six specific rhodamine fragments (m/z = 331.1, 387.2, 415.2, 443.2, 467.2, and 491.2 in Fig. S8C†) as the final mass reporters. After fragmentation, the signal-to-noise ratios for representative ions of six RMTs were about 2- to 5-fold larger than before (Fig. S8D†). These mass reporters were easily distinguished by LTQ mass spectrometry with a relatively lower resolution (full width at half maximum = 2000) and the complex immune condition caused a negligible interference with the mass reporter readout (Fig. S9†), which reduced instrumental requirements for protein detection, showing its high potential to be combined with portable mass spectrometry for point-of-care testing (POCT).
Parameters that importantly influenced the intensities of mass reporters were optimized, including the spray voltage and the flow rate of the solvent. The spray voltage was related to the efficiency of RMT dissociation and ionization, and a higher spray voltage (5 kV, Fig. S10A†) was needed for this system when compared with the conventional ESI. Flow rates of solvent should match the emitter size, the spray voltage and the signal duration. After optimization, a flow rate of 10 μL min−1 was utilized (Fig. S10B†), which ensured the highest sensitivity and stability for mass tag detection and a shortened duration of mass reporters within 1 min (Fig. S10C†).
Along with six RMTs, CV was selected as IS for quantification. Six RMT modified GNPs were detected simultaneously at different concentrations together with IS. The signal area ratios of RMTs to IS showed a good linear relationship (Fig. S11†), which was essential for further multiplexed immune quantification.
One-step and high throughput immunoassays for six cancer biomarkers
The immune reactions on the paper substrates were designed with reference to the universal ELISA or ECLIA. The size of the paper substrates was well-suited to the 96-well plates, enabling immune capture and labeling in a high-throughput and reproducible way. Furthermore, since the cAb–dAb pairs have been demonstrated to recognize different domains of target proteins with high specificity,38,39 the capture and labeling of target proteins were conducted together as one step in the wells, which shortened the immune preparation time compared to many multi-step sandwich immunoassays.40–42 Samples, such as 10 μL serum, were added into the wells, followed by the immediate addition of 40 μL GNP probe dispersions with an optimized concentration of 0.5 nmol L−1 (Fig. S12†). The whole immune preparation could be finished within 50 minutes with high throughput processing of array samples for up to 96 tests.To validate the immunoassays, IgG in PBS was first detected as the target. When HRP-conjugated anti-IgG was used as the detection antibody, the sandwich-type immune complex could be detected both by the chromogenic and MS readout. Good linearity between the MS signal area ratios and the logarithm of concentrations was obtained for IgG spiked into PBS, ranging from 0.2 ng mL−1 to 200 ng mL−1 (Fig. S13†). The limit of detection (LOD) based on the PS-ESI MS was 0.111 ng mL−1. Compared with the chromogenic results (linearity from 0.8 ng mL−1 to 50 ng mL−1 with an LOD of 0.603 ng mL−1), the MS results obtained 16-fold broader linearity ranges and 6-fold lower LODs. The robustness of the PS-ESI MS immunoassay system was further validated using paper substrates and MPs kept for some days, and the paper substrates and MPs presented almost unchanged activity for a week (R2 < 9.5%, n = 3; Fig. S14†). Good reproducibility was obtained using different batches of substrates and MPs, which further ensured their robustness and was significant for the application of this immunoassay for clinical use (R2 < 8.3%, n = 6; Fig. S14†). Since the six RMT series had predictably similar performances, the PS-ESI MS was further applied to the simultaneous quantification of six protein biomarkers for clinical diagnosis.
Six biomarkers, including four specific biomarkers (CA19-9, CA15-3, CA125, and HE4) and two relative universal biomarkers (CEA and AFP), were simultaneous quantified using this PS-ESI MS immune system. Among the six combined biomarkers, CA19-9 is recognized as a specific indicator for pancreatic cancer;43 CA15-3 is often specific for breast cancer;44 CA125 and HE4 are two specific combined markers for ovarian cancer;45 and CEA and AFP are relative universal markers for colorectal cancer, lung cancer, liver cancer, pancreatic cancer, and gastric cancer.46,47 Efficient cancer screening would be achieved when these six proteins are detected simultaneously. Six mass reporters, each representing one protein marker, were easily identified in the mass spectra under the complex sandwich-type immune system (Fig. 4A and B). After the calibration of the signals with IS, six proteins in PBS buffer were all quantified with good linear relationships (R2 > 0.98, Fig. 4C and Table S3†). Considering the clinical applications, unlike the larger linear ranges for the IgG detection, accurate quantification relationships around the clinical cut-off values for the six protein targets were focused on, which is practical for cancer diagnosis at early or later stages. As a result, all of the linear ranges corresponded well with the clinical cut-off values, including 25 U mL−1 for CA15-3, 40 U mL−1 for CA19-9, 35 U mL−1 for CA125, 5 ng mL−1 for CEA, 140 pmol L−1 for HE4, and 10 ng mL−1 for AFP.43–47 The LODs were calculated as three times the signal-to-noise ratios, which were much lower than the cut-off values (Table S3†). These biomarker cut-off values have been demonstrated to be valuable references for early disease diagnosis and evaluation. For example, concentrations larger than the cut-off value (10 ng mL−1) of AFP or continuously increasing around this value indicate a high possibility of suffering from primary liver cell cancer.47 The accurate quantification methods around these values could be applied well in clinical use.
The accuracy and specificity of the immunoassays were verified by the multi-target standard addition measurements in the complex serum matrix. The recoveries were obtained as 87.7% to 114% (RSD < 13%, Tables S4–S9†) for six target proteins, which were acceptable for multiplexed quantitative assays in biological samples. For each target, it could be accurately quantified (recoveries from 88.2% to 114%, RSD < 13%) at concentrations lower than the clinical cut-off values with other proteins 10 times greater than it. Especially for CA15-3, CA19-9, and CA125, although they are all cancer antigen (CA) species with high structural similarity, such as up to 70% glycans in their structures, these three CAs could be quantified in relatively low concentrations with interference from other protein markers and the matrix (Fig. 4D). These results have thereby illustrated the high accuracy and specificity of the paper-based sandwich-type immune processes and PS-ESI MS readout for the multiplexed detection of six protein biomarkers.
Combined screening for six biomarkers in patient serum
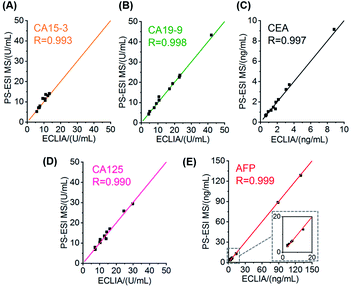
The high throughput immune preparation and simultaneous detection of six cancer protein biomarkers made the cancer screening more efficient and patient-friendly. As a further demonstration of clinical application, serum samples from 12 patients who have been diagnosed with cancers at several sites, including breast tumors (benign and malignant), gastric cancer, and liver cancer, were tested with these hexaplex immunoassays. Following the procedure of PS-ESI MS-based immunoassays, only 10 μL serum was consumed for each test, and the simultaneous immune recognition and labeling of six biomarkers in the 12 samples (each sample taking six parallel tests) was carried out in one 96-well plate and finished within 50 min, coupled with a 2 min MS read-out for each test. Compared with representative optical-/electrochemistry-based and MS-based multiplexed immunoassays for clinical cancer biomarkers (listed in Table S10†), the PS-ESI MS-based immunoassays achieved hexaplex immunoassays in one step, which represents progress for immunoassays. This method also has the advantages of high throughput preparation, low sample consumption, rapid detection and satisfactory linear ranges and sensitivity for clinical use. Six clinical concerned biomarkers in serum were successfully quantified simultaneously by the PS-ESI MS-based immunoassays, and the quantification results of the six biomarkers were compared with the clinical detection results by ECLIA (Table S11†). Good correlations with all high correlation coefficients (R > 0.990) were obtained between the PS-ESI MS-based methods and ECLIA (Fig. 5A–E), and the most clinically significant results around the cut-off values were covered by the PS-ESI MS-based methods, demonstrating the reliability of the proposed immunoassays in clinical diagnosis.As newly popular indicators for cancer diagnosis, protein biomarkers are sometimes controversial for cancer diagnosis, especially when only using single biomarkers. It is factually confusing to distinguish cancer sites with the detected concentrations of single CA15-3 (known to be specific for breast cancer), CA19-9 (known to be specific for pancreatic cancer) or CEA (known to indicate colorectal cancer, lung cancer, pancreatic cancer, gastric cancer and so on) (Fig. S15A–C†). As an improvement, combined biomarker testing has attracted more attention as it could achieve higher sensitivity and specificity for cancer diagnosis.3,4 The PS-ESI MS-based immunoassays ensured the simultaneous quantification of the combined biomarkers efficiently with a micro-volume serum sample and with no need to obtain the exact biomarker concentration. The calibrated RMT peak areas could be directly employed for analysis, which provides a simple cancer screening approach. With the 12 samples as a preliminary test, tumors at different sites could be distinguished by hierarchical clustering and visualized by principal component analysis, showing the potential of the combined evaluation of six biomarkers for cancer diagnosis (Fig. S15D†). Although a larger sample dataset is needed to validate the combined biomarker validation and establish the automatic machine learning program, the multiplexed MS-based immunoassay has shown its potential and usefulness for efficient cancer diagnosis.
Conclusions
In summary, one-step hexaplex sandwich-type immunoassays with high throughput, accuracy, sensitivity and specificity were fabricated based on array-prepared paper substrates and an on-line dissociation/ionization MS system. The paper substrates underwent sandwich-type immune reactions conducted in 96-well plates with high efficiency and high throughput. With the stable prepared paper substrates and mass probes, a batch of immune preparation could be finished within 50 minutes with a 10 μL sample, which compares favourably with commercial immune kits or automatic immune analyzers. Six mass probes constructed by thiol compounds and antibodies assembled on GNPs enabled the one-spot simultaneous readout of six cancer protein markers. The Au–S bond was discovered to be efficiently on-line dissociated in the charged solvent flow during ESI, which was expanded as a universal site for multiple mass tag dissociation and immune signal amplification. Using a series of distinguishable mass reporters, six important cancer protein markers were quantified simultaneously in serum, which corresponded well with the cut-off values for cancer diagnosis. Furthermore, the hexaplex immunoassays were applied for the screening of patient serum samples. It is highly probable that high sensitivity and specificity for diagnosis could be achieved with combined biomarker signals with small microliter-level sample consumption by this MS-based immune system. With further extension of mass tags, simplification of the MS-based immune readout instruments and automated data analysis, these types of immunoassays show great potential for the early disease screening and accurate diagnosis with simultaneous detection of dozens of biomarkers.Ethical statement
The study was approved by the Ethics Committee of Chinese People's Liberation Army General Hospital and was conducted in accordance with the Helsinki Declaration of 1964 with later revisions.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was financially supported by the National Natural Science Foundation of China (21874003, 21775008, and 22074003) and the Beijing Natural Science Foundation Essential Research Project (Z170002).References
- R. L. Siegel, K. D. Miller and A. Jemal, Ca-Cancer J. Clin., 2019, 69, 7–34 CrossRef.
- F. Cardoso, S. Kyriakides, S. Ohno, F. Penault-Llorca, P. Poortmans, I. T. Rubio, S. Zackrisson, E. Senkus and E. G. Comm, Ann. Oncol., 2019, 30, 1194–1220 CrossRef CAS.
- C. Nunez, Clin. Chim. Acta, 2019, 490, 113–127 CrossRef CAS.
- M. Bhardwaj, K. Weigl, K. Tikk, A. Benner, P. Schrotz-King and H. Brenner, Mol. Oncol., 2020, 14, 8–21 CrossRef CAS.
- K. Iha, M. Inada, N. Kawada, K. Nakaishi, S. Watabe, Y. H. Tan, C. Shen, L.-Y. Ke, T. Yoshimura and E. Ito, Diagnostics, 2019, 9, 78 CrossRef CAS.
- P. Rubisz, M. Ciebiera, L. Hirnle, M. Zgliczynska, T. Lozinski, P. Dziegiel and C. Kobierzycki, Int. J. Mol. Sci., 2019, 20, 1136 CrossRef CAS.
- D. Sadighbayan, K. Sadighbayan, M. R. Tohid-Kia, A. Y. Khosroushahi and M. Hasanzadeh, TrAC, Trends Anal. Chem., 2019, 118, 73–88 CrossRef CAS.
- S. Ye, Y. Mao, Y. Guo and S. Zhang, TrAC, Trends Anal. Chem., 2014, 55, 43–54 CrossRef CAS.
- H. Ju, J. Anal. Test., 2017, 1, 7 CrossRef.
- D. Geissler, S. Stufler, H.-G. Loehmannsroeben and N. Hildebrandt, J. Am. Chem. Soc., 2013, 135, 1102–1109 CrossRef CAS.
- K. R. Khondakar, S. Dey, A. Wuethrich, A. A. Ibn Sina and M. Trau, Acc. Chem. Res., 2019, 52, 2113–2123 CrossRef CAS.
- W. Wang, J. Chang, B. Jia and J. Liu, Cancer Manage. Res., 2020, 12, 5431–5438 CrossRef CAS.
- A. Jones, L. Dhanapala, R. N. T. Kankanamage, C. V. Kumar and J. F. Rusling, Anal. Chem., 2020, 92, 345–362 CrossRef CAS.
- W. Lv, H. Ye, Z. Yuan, X. Liu, X. Chen and W. Yang, TrAC, Trends Anal. Chem., 2020, 123, 115767 CrossRef CAS.
- C. Benoist and N. Hacohen, Science, 2011, 332, 677–678 CrossRef CAS.
- X. Zhong, L. Qiao, N. Gasilova, B. Liu and H. H. Girault, Anal. Chem., 2016, 88, 6184–6189 CrossRef CAS.
- W. Ma, S. Xu, H. Nie, B. Hu, Y. Bai and H. Liu, Chem. Sci., 2019, 10, 2320–2325 RSC.
- Y. Wang, R. Du, L. Qiao and B. Liu, Chem. Commun., 2018, 54, 9659–9662 RSC.
- J. Sun, H. Liu, L. Zhan, C. Xiong, X. Huang, J. Xue and Z. Nie, Anal. Chem., 2018, 90, 6397–6402 CrossRef CAS.
- R. Liu, S. Zhang, C. Wei, Z. Xing, S. Zhang and X. Zhang, Acc. Chem. Res., 2016, 49, 775–783 CrossRef CAS.
- B. Unnikrishnan, C.-Y. Chang, H.-W. Chu, A. Anand and C.-C. Huang, Anal. Methods, 2016, 8, 8123–8133 RSC.
- F. Pu, S. Chiang, W. Zhang and Z. Ouyang, Analyst, 2019, 144, 1034–1051 RSC.
- C. R. Ferreira, K. E. Yanne, A. K. Jarmusch, V. Pirro, Z. Ouyang and R. G. Cooks, Clin. Chem., 2016, 62, 99–110 CrossRef CAS.
- H. Wang, J. Liu, R. G. Cooks and Z. Ouyang, Angew. Chem., Int. Ed., 2010, 49, 877–880 CrossRef CAS.
- Y.-N. Yao, D. Di, Z.-C. Yuan, L. Wu and B. Hu, Anal. Chem., 2020, 92, 6207–6212 CrossRef CAS.
- Y.-C. Huang, H.-H. Chung, E. P. Dutkiewicz, C.-L. Chen, H.-Y. Hsieh, B.-R. Chen, M.-Y. Wang and C.-C. Hsu, Anal. Chem., 2020, 92, 1653–1657 CrossRef CAS.
- Y. Yang, H. Liu, Z. Chen, T. Wu, Z. Jiang, L. Tong and B. Tang, Anal. Chem., 2019, 91, 12874–12881 CrossRef CAS.
- S. Chen, Q. Wan and A. K. Badu-Tawiah, J. Am. Chem. Soc., 2016, 138, 6356–6359 CrossRef CAS.
- C. Zhang, T. Glaros and N. E. Manicke, J. Am. Chem. Soc., 2017, 139, 10996–10999 CrossRef CAS.
- R. Chen, Y. Xiao, H. Liu, L. Fang, J. Liu, X. Ruan, B. Chen and T. Luan, Anal. Chem., 2020, 92, 13298–13304 CrossRef CAS.
- S. Xu, W. Ma, Y. Bai and H. Liu, J. Am. Chem. Soc., 2019, 141, 72–75 CrossRef CAS.
- S. Xu, M. Liu, Y. Bai and H. Liu, Angew. Chem., Int. Ed., 2020, 60, 1806–1812 CrossRef.
- Y. Kitkulnumchai, A. Ajavakom and M. Sukwattanasinitt, Cellulose, 2008, 15, 599–608 CrossRef CAS.
- C. Roberts, C. S. Chen, M. Mrksich, V. Martichonok, D. E. Ingber and G. M. Whitesides, J. Am. Chem. Soc., 1998, 120, 6548–6555 CrossRef CAS.
- Z. Han, S. Sarkar and A. M. Smith, ACS Nano, 2020, 14, 3227–3241 CrossRef CAS.
- K. Kaneko, M. Hara, T. Nishino and T. Maruyama, Anal. Chem., 2020, 92, 1978–1987 CrossRef CAS.
- R. K. Manova, S. Joshi, A. Debrassi, N. S. Bhairamadgi, E. Roeven, J. Gagnon, M. N. Tahir, F. W. Claassen, L. M. W. Scheres, T. Wennekes, K. Schroen, T. A. van Beek, H. Zuilhof and M. W. F. Nielen, Anal. Chem., 2014, 86, 2403–2411 CrossRef CAS.
- D. V. Gold, G. Newsome, D. Liu and D. M. Goldenberg, Mol. Cancer, 2013, 12, 143 CrossRef.
- O. Hosu, A. Ravalli, G. M. Lo Piccolo, C. Cristea, R. Sandulescu and G. Marrazza, Talanta, 2017, 166, 234–240 CrossRef CAS.
- C. Li, Y. Yang, D. Wu, T. Li, Y. Yin and G. Li, Chem. Sci., 2016, 7, 3011–3016 RSC.
- W. Guo, H. Ding, C. Gu, Y. Liu, X. Jiang, B. Su and Y. Shao, J. Am. Chem. Soc., 2018, 140, 15904–15915 CrossRef CAS.
- Z. Hu, G. Sun, W. Jiang, F. Xu, Y. Zhang, M. Xia, X. Pan, Z. Xing, S. Zhang and X. Zhang, Anal. Chem., 2019, 91, 5980–5986 CrossRef CAS.
- M. J. Duffy, Clin. Chem., 2006, 52, 345–351 CrossRef CAS.
- K. E. Poruk, D. Z. Gay, K. Brown, J. D. Mulvihill, K. M. Boucher, C. L. Scaife, M. A. Firpo and S. J. Mulvihill, Curr. Mol. Med., 2013, 13, 340–351 CAS.
- R. G. Moore, D. S. McMeekin, A. K. Brown, P. DiSilvestro, M. C. Miller, W. J. Allard, W. Gajewski, R. Kurman, R. C. Bast Jr and S. J. Skates, Gynecol. Oncol., 2009, 112, 40–46 CrossRef CAS.
- Y. Wang, X. Jiang, S. Dong, J. Shen, H. Yu, J. Zhou, J. Li, H. Ma, E. He and S. Skog, Cell Biochem. Biophys., 2016, 16, 529–536 Search PubMed.
- F. Trevisani, P. E. D'Intino, A. M. Morselli-Labate, G. Mazzella, E. Accogli, P. Caraceni, M. Domenicali, S. De Notariis, E. Roda and M. Bernardi, J. Hepatol., 2001, 34, 570–575 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0sc06784a |
| This journal is © The Royal Society of Chemistry 2021 |