Open Access Article
Open Access ArticleDesign and enhanced gene silencing activity of spherical 2′-fluoroarabinose nucleic acids (FANA-SNAs)†
Hassan H.
Fakih
 ,
Adam
Katolik
,
Elise
Malek-Adamian
,
Johans J.
Fakhoury
,
Sepideh
Kaviani
,
Masad J.
Damha
* and
Hanadi F.
Sleiman
,
Adam
Katolik
,
Elise
Malek-Adamian
,
Johans J.
Fakhoury
,
Sepideh
Kaviani
,
Masad J.
Damha
* and
Hanadi F.
Sleiman
 *
*
Department of Chemistry, McGill University, Montreal, Quebec H3A 0B8, Canada. E-mail: Hanadi.sleiman@mcgill.ca; Masad.Damha@mcgill.ca
First published on 15th January 2021
Abstract
Drug delivery vectors for nucleic acid therapeutics (NATs) face significant barriers for translation into the clinic. Spherical nucleic acids (SNAs) – nanoparticles with an exterior shell made up of DNA strands and a hydrophobic interior – have recently shown great potential as vehicles to improve the biodistribution and efficacy of NATs. To date, SNA design has not taken advantage of the powerful chemical modifications available to NATs. Here, we modify SNAs with 2′-deoxy-2′-fluoro-D-arabinonucleic acid (FANA-SNA), and show increased stability, enhanced gene silencing potency and unaided uptake (gymnosis) as compared to free FANA. By varying the spacer region between the nucleic acid strand and the attached hydrophobic polymer, we show that a cleavable DNA based spacer is essential for maximum activity. This design feature will be important when implementing functionalized nucleic acids into nanostructures for gene silencing. The modularity of the FANA-SNA was demonstrated by silencing two different targets. Transfection-free delivery was superior for the modified SNA compared to the free FANA oligonucleotide.
Introduction
Nucleic acid therapeutics (NATs), such as small interfering RNA (siRNA) and antisense oligonucleotides (ASOs) offer unique potential as therapeutics, because they can selectively and efficiently silence genes involved in disease.1–3 An essential strategy for translating NATs to the clinic has been the chemical modification of their natural DNA or RNA backbone. This improves their stability in biological media and augments their therapeutic efficacy.4,5 One salient sugar-modified DNA analogue is 2′-deoxy-2′-fluoro-β-D-arabinonucleic acid (FANA), which is compatible with both ASO and siRNA modalities.4–9 The FANA modification increases hydrolytic and nuclease stability,10 and decreases immune side-effects and toxicity.5,9–11 Importantly, it increases both binding affinity to the target mRNA and recruitment of RNase-H for its destruction, thereby improving the silencing mechanism.4–6,8–11 In addition, carrier-free cellular internalization (gymnosis) of FANA oligonucleotides permits the sequence-specific silencing of multiple targets in many cell types, both at the protein and mRNA level, at concentrations in the low micromolar range.11–15While chemical modifications of nucleic acid therapeutics have radically improved their stability and activity, there are still critical issues that need to be addressed, including biodistribution, pharmacokinetics and extra-hepatic delivery.1–3,16,17 NATs are rapidly cleared by the kidneys and those left in circulation accumulate in the liver.4,18 While this is useful for hepatic diseases, other diseases require biodistribution to critical organs such as brain, muscle, heart, tumour, etc. all of which still present significant challenges for nucleic acid therapies.
Nano-sized drug delivery systems have emerged as promising technologies to guide NATs to targets beyond the liver.4,18–21 Nanoparticles such as liposomes, gold nanoparticles, viral delivery systems and polymers16,22–24 can encapsulate various drugs including combination therapies, and incorporate targeting ligands that allow them to specifically bind to disease cells.16,22,23 Spherical nucleic acids (SNAs) are promising nanocarriers for nucleic acid therapeutics.21,25 They consist of a nanoparticle core, which can be liposomal, metallic, or polymeric, and a dense nucleic acid corona. This 3D structure has unique biological and physiological properties. SNAs have higher affinity to their nucleic acid target, are less susceptible to nucleases, can be taken up by many cell lines, and are less toxic and immunogenic.21,25 In some cases, they have favourable biodistribution beyond the liver in vivo (brain, skin, etc.).21,26,27
One of our laboratories has developed a new class of spherical nucleic acids assembled from monodisperse, sequence-controlled polymers. They are created by adapting DNA solid-phase automated synthesis whereby monomer units are attached in a sequence-controlled fashion to a growing oligonucleotide chain28via phosphoramidite chemistry. This modular control allows the synthesis of amphiphilic oligonucleotide polymers that self-assemble into a variety of nanostructures such as fibres and sheets,29 hydrophobic DNA cubes,30–32 and SNAs.33–36 In particular, conjugating twelve units of dodecane (punctuated with phosphates) to DNA or RNA gives amphiphiles that spontaneously form monodisperse SNAs in aqueous media, with a hydrophobic core and a nucleic acid corona.34–36 These SNAs can encapsulate small molecule drugs and are stable in biological media. They are readily taken up into cells and have a favourable biodistribution profile in mouse models.34–36 They also show higher activity (2–3-fold), requiring less transfection agents compared to linear nucleic acids,33 and they can be designed to release cargo in the presence of specific biomolecules.36 Furthermore, these SNAs are assembled from a single component in comparison to other nucleic acid based structures such as origami,37 making them DNA-minimal and favourable in terms of large-scale production and translation into clinical applications.18,38 In comparison to other SNAs, our sequence-controlled polymeric SNAs are made from biocompatible, biodegradable materials, can encapsulate small molecule drugs and allow for near infinite variation in polymer chemistry and sequence in an automated fashion.39 This gives them distinct advantages in biocompatibility and ease of synthesis, as well as control of size, shape, and function.28
Despite these benefits, our SNAs and most other SNAs reported in literature lack chemical modifications to the nucleic acids beyond a simple phosphorothioate backbone.26,27,40–47 We reasoned that combining SNAs with the powerful nucleic acid chemical modifications that have enabled clinical applications will provide NATs with improved biodistribution and gene silencing activity. However, the design rules that ensure optimal activity while maintaining the SNA morphology are not clear.
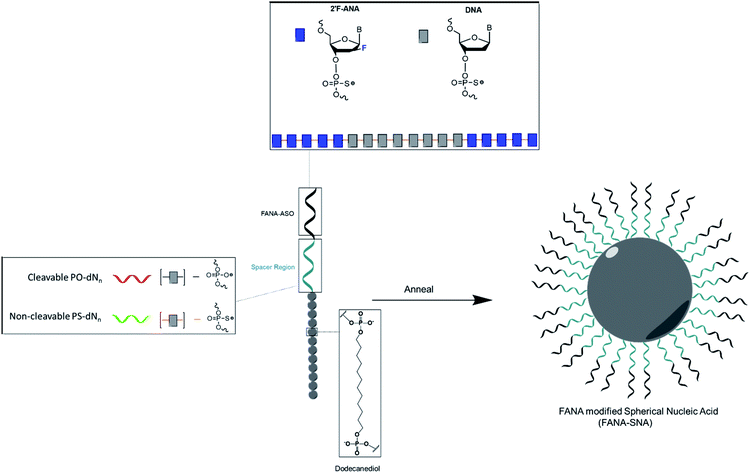
In this work, we prepared FANA spherical nucleic acids (FANA-SNA) by covalently attaching a FANA-ASO to twelve [dodecanediol phosphate] units (Fig. 1). The FANA-ASO consists of 5 nucleotide (nt) FANA “wings” flanking a 8 nt DNA “gap” in the middle of the sequence. This architecture was shown to have low nanomolar silencing activity.9 In aqueous media this construct spontaneously assembles into a SNA micelle. We found that a cleavable spacer between the FANA strand and the hydrophobic polymer modification is essential for reaching full activity of the SNA. Furthermore, carrier-free cellular internalization of FANA-SNAs resulted in superior gene silencing activity relative to the free (unconjugated) FANA-ASO. The modularity of our FANA-SNAs was demonstrated by successfully silencing three different cellular targets. Thus, the combination of ASO modification and a cleavable linker are required for optimal activity. Combined with SNA's stability, low polydispersity, ability to deliver small molecule drugs, improved biodistribution and design simplicity, we anticipate that FANA-SNAs will be powerful gene silencing tools with enhanced therapeutic potential.
Results and discussion
Antisense oligonucleotides (ASOs)
The table below shows all the strands and variations of ASO used throughout the manuscript (Table 1). More information on the sequences used can be found in the Methods section (ESI Table 1†).Synthesis and characterization of FANA-modified SNAs (FANA-SNAs)
We first verified that introducing a FANA-modified oligonucleotide in the SNA does not affect the self-assembly. We synthesized FANA-SNA with different spacer regions (no spacer or PO-dN4 spacer) between the FANA portion and the hydrophobic polymer, and we assembled the nanoparticles in a magnesium containing buffer (1× TA).28 Characterization was carried out using dynamic light scattering (DLS) and atomic force microscopy (AFM) (Fig. 2). As seen in Fig. 2, the ASO-conjugates form well-defined spherical structures with a comparable size to previously characterized SNAs.48,49 The addition of spacer led to a slight increase in size for the FANA-(PO-dN4)-SNA. In both cases, the larger features observed by AFM are attributed to dimers or small aggregates resulting from the AFM conditions (dried sample on mica surface) and the palindromic nature of part of the sequence. We verified by agarose gel electrophoresis the monodisperse nature of the assembly (ESI Fig. 7†).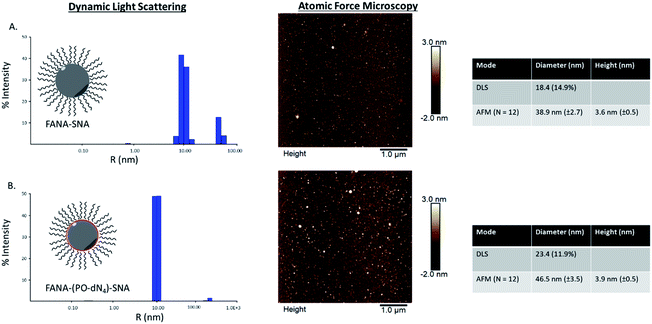 | ||
| Fig. 2 Dynamic light scattering and atomic force microscopy characterization of (A) FANA-SNA (no spacer) and (B) FANA-(PO-dN4)-SNA. | ||
Gene silencing activity of the SNAs
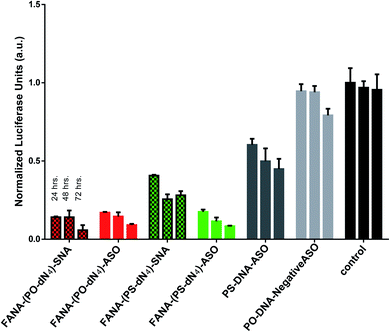
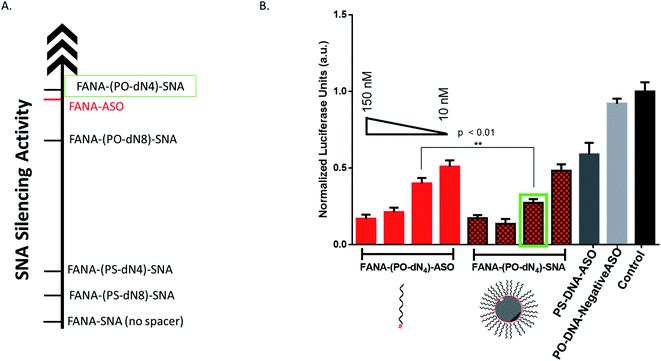
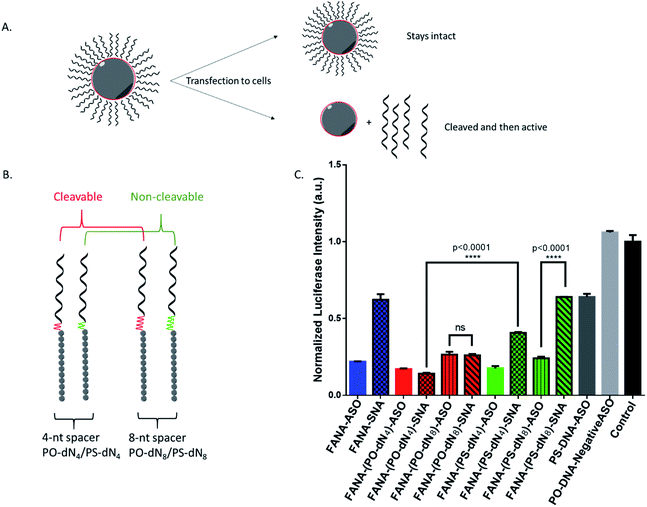
After confirming the formation of the nanoparticles, we tested and compared the activity of FANA-SNA and FANA-ASO targeting luciferase mRNA. The ASOs and SNAs were transfected into luciferase expressing HeLa cells for 24 hours and assessed for their gene-silencing activity (luciferase activity, Fig. 3A) and cell-death/cytotoxicity (CellTiter-Blue assay, Fig. 3B). Results show that even though the FANA-SNA is active, its activity is significantly lower than its free FANA-ASO counterpart. However, when a 4 nt spacer with a phosphate backbone was introduced (FANA-(PO-dN4)-SNA), the activity fully recovered; in fact it was superior to the activity seen for free FANA-ASO. None of the ASOs or the SNAs showed any significant toxicity to the cells, as the metabolic activity was equivalent across all samples.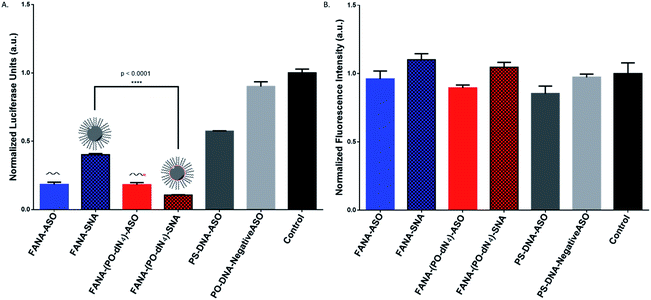 | ||
| Fig. 3 Firefly luciferase knockdown activity comparison between various free FANA-ASO and FANA-SNA (see Table 1 for description of the different strands). (A) Luciferase activity was measured after 24 h incubation and normalized to CellTiter-Blue and control (no ASO). (B) Fluorescence measurement of CellTiter-Blue cytotoxicity results normalized to control (no ASO). Error bars represent SD of triplicate experiments for each sample. ASO final concentration is 150 nM for all samples. | ||
Steric vs. stability of spacer
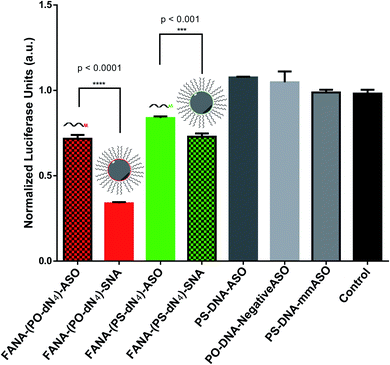
We reasoned that the superior activity FANA-SNAs with a (PO-dN4) spacer was possibly due to (a) the separation of the ASO portion from the hydrophobic core, thus increasing flexibility and reducing steric hindrance of the ASO strands, and/or (b) the metabolically cleavable nature of the PO-DNA spacer, which would release off the free ASO strand from the SNA (Fig. 4A). To assess these scenarios, we synthesized four new conjugates: (i) FANA-(PS-dN4)-SNA, where the phosphate linkages that connect the dodecanediol units to the ASO are replaced by phosphorothioate (PS) linkages. Because of their superior resistance to nucleases, PS linkages are expected to increase spacer stability and significantly slow down the release of the FANA from the SNA. (ii) FANA-(PO-dN8)-SNA and FANA-(PS-dN8)-SNA, where the spacer length is increased, allowing us to test the effect of increased separation and decreased steric hindrance (Fig. 4B). The gene silencing experiment showed that the PO-dN4/8 spacers are superior (more activity seen) than the more stable PS-dN4/8 spacers (Fig. 4C). The longer spacers (PO or PS) were less active than shorter ones, possibly due to increased off–target interactions. (iii) Next, we tested a FANA-SNA containing a non-cleavable hexaethylene glycol (HEG) spacer, which also showed lower activity compared to FANA-(PO-dN4)-SNA (ESI Fig. 2†). These results support the notion that the more metabolically cleavable PO-DNA spacer is required for the increased activity of FANA-SNA.Assessing the cellular uptake of SNAs with varying spacer structure
To examine if the superior activity of the FANA-(PO-dN4)-SNAs was due to enhanced cellular uptake relative to the FANA-(PS-dN4)-SNAs, cellular uptake was assessed via flow cytometry in 3 different cell lines. The SNAs were modified with a Cy3 dye on the interface between the ASO and conjugate to avoid any influence on uptake from the dye itself.The data shown in Fig. 5 shows that both SNAs (cleavable and non-cleavable spacer) have similar uptake in all 3 cell lines, suggesting that the higher activity imparted by the (PO-dN4) spacer likely results from the release of the ASO from the SNA and endosomes, and not from increase in uptake. It is important to note that the higher uptake signal seen for the free ASOs is due to having an exposed Cy3 dye, which is known to cleave over time and increase intracellular fluorescence as studied in our previous work.50,51 Additional data analysis and plots are included in ESI (Fig. 3†).
Duration of effect
Next, we compared the activities of the various FANA-SNAs over an extended period of time (Fig. 5). The results of the silencing experiment show that FANA-(PO-dN4)-SNA remains the most active, and that its activity increased over the 72 h period; in contrast, the activity of FANA-(PS-dN4)-SNA plateaued at 48 h, with no significant change after 72 h (Fig. 6).FANA-SNA exhibits greater activity compared to free FANA-ASO
We measured the activity of FANA-(PO-dN4)-SNA and FANA-(PO-dN4)-ASO at 150, 100, 50 and 10 nM after 24 hours of incubation (Fig. 7). The free ASO showed the expected gradual loss of activity as its concentration was decreased from 150 to 10 nM. The SNA shows superior silencing activity over FANA-(PO-dN4)-ASO at most of the concentrations tested (e.g., 50–100 nM; Fig. 7B).Serum stability
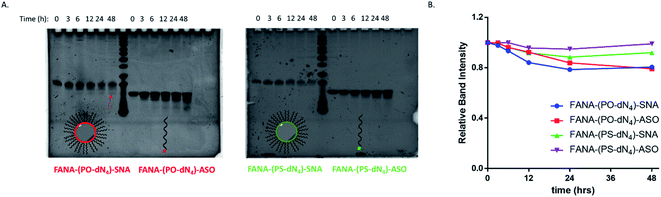
FANA-SNAs were incubated in cell media containing fetal bovine serum (10%) and collected aliquots at different timepoints were assessed on a denaturing polyacrylamide gel (PAGE). As seen in Fig. 8, little degradation is observed for SNAs with the PS-dN4 spacer over 48 hours. However, the SNA with a PO-dN4 spacer undergoes more extensive cleavage (around 20%) relative to the SNA with a PS-dN4 spacer, consistent with the notion that the PO linker releasing the ASO more readily. This cleavage is expected to be more extensive intracellularly, considering the acidity of the endosomal-lysosomal compartments and the presence of additional nucleases in this environment. No significant differences were observed between the FANA-SNAs and the corresponding free FANA demonstrating the excellent nuclease stability of the FANA modification under these conditions.Testing against different mRNA targets
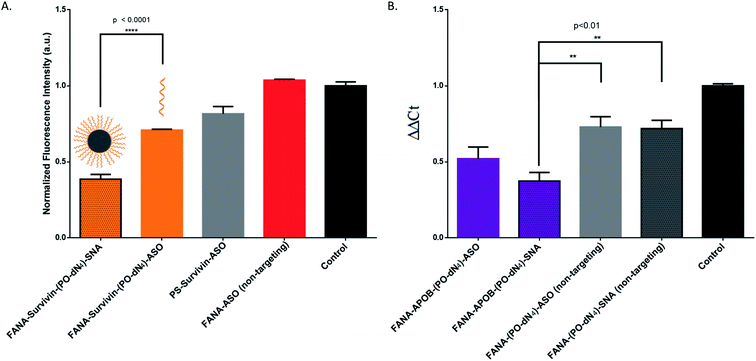
The SNAs' design is versatile and modular, as they can theoretically silence any target of interest by changing their oligonucleotide sequence. To show this, we changed the sequence to target survivin, an overexpressed oncogene in numerous tumour cells responsible for cell proliferation and inhibition of apoptosis, allowing them to build resistance to therapy.41,52,53 If silenced, the cancer cells will stop proliferating, which can be measured via their metabolic activity using the CellTiter-Blue assay. We opted for the design with the PO-dN4 cleavable spacer that shows the highest activity. The results show that the SNAs are indeed effective against an endogenous target (Fig. 9A). In fact, the SNA has significantly higher activity compared to the free ASO counterpart. Titration of various concentrations was also performed, showing that the SNA retains higher activity at decreasing concentrations (ESI Fig. 4†).In order to provide evidence of gene knockdown at the RNA level, we targeted the APOB gene in HepG2 cells.35 We quantified the relative silencing by measuring mRNA degradation with RT-qPCR. The data (Fig. 9B) shows the successful decrease of RNA level using FANA-APOB-(PO-dN4)-ASO/SNA to a greater extent than the non-targeting controls (we used the mismatched FANA-(PO-dN4)-ASO/SNA of the luciferase target).
Hence the SNAs display modularity by showing effective silencing against two other targets, both at the protein and RNA level.
SNA improves transfection-free delivery (gymnosis)
FANA ASOs are known to penetrate cells without transfection (gymnosis).11–15 Furthermore, we have previously shown that DNA-SNAs are taken up by cells to a greater extent than free DNA in absence of transfection agents, and that they are able to efficiently deliver small molecule drugs in cells. However, our previous ASO-SNAs did not show silencing activity without transfection agents.49 We tested the ability of FANA-SNAs for gymnotic silencing of luciferase mRNA in HeLa cells. In this experiment (Fig. 10), the ASO and SNAs were incubated at 1 uM (strand concentration) for 3 days, following a previously reported protocol.12Fig. 10 shows that gymnotic delivery of FANA-(PO-dN4)-SNA and FANA-ASO downregulated luciferase expression compared to control untreated cells; however, silencing was more significant for the SNA. In contrast, a targeting PS-DNA with no FANA modifications and a mismatch control (PS-DNA) did not produce silencing under similar conditions.Discussion
To our knowledge, only one reported example has previously shown incorporation of FANA modifications into the corona rather than the interior of a nanoparticle delivery system, which was a poly (D,L-lactide-co-glycolide) particle.54 The inhibitory effect of FANA ASOs is attributed to RNase H1 activation or steric hindrance depending on the mRNA target site and ASO design.9,11 The current work investigates for the first time the behaviour and silencing activity of a FANA modified ASO as part of a spherical nucleic acid (SNA) previously developed in our group.28,49 This system arises from sequentially synthesizing an ASO conjugated to twelve dodecanediol artificial “nucleotide” units in sequence-defined and automated manner. This allows it to assemble into an exceptionally stable monodisperse particle of tuneable properties (size, shape, functionality, biodistribution) for delivery.48,49,55 FANA has the benefit of increasing gene silencing efficacy, nuclease resistance and reduced immune-stimulatory side-effects.4–9,11We first verified that such a modification did not impact the assembly of the SNA (Fig. 2). We then showed that, while still capable of gene silencing, the activity of the FANA-SNA particle was significantly reduced compared to free ASO (Fig. 3). However, when a PO-DNA tetranucleotide spacer was introduced between the ASO and the SNA (i.e., FANA-(PO-dN4)-SNA), the activity fully recovered and, in fact, was superior to the activity of the free FANA-ASO. The spacer design between the oligonucleotide and the core is therefore crucial in many aspects to the SNA's overall activity performance, as it can modulate the crowding and loading capacity on its surface.21,45,56,57 This work suggests that release of the ASO from our SNA is another important factor. In support of this, we observed greater silencing activity with FANA-SNAs containing a PO-DNA spacer relative to a more stable PS-DNA (Fig. 3). By changing the length (4- vs. 8-nucleotide) and the stability (PO vs. PS) of the spacer, we showed that a less stable and shorter phosphate linker is better than a longer spacer for activity (Fig. 4). A hexaethylene glycol spacer was also detrimental for our SNAs, similar to what is reported in literature.42,44,45,58
The increased activity for the SNA with PO spacer is not due to increased uptake into cells, as both PO-dN4 and PS-dN4 have similar uptake levels across three different cell lines (Fig. 5). This indicates that the cleavability of the spacer plays a major role after the uptake of SNAs into cells. The activity of the FANA-(PO-dN4)-SNA was consistent and improved up to 72 hours of incubation with cells (Fig. 6).
The cleavability of siRNA conjugates has recently been touched upon briefly by Prakash et al.59 and Biscans et al.,60 in an investigation of siRNA–hydrophobic conjugates extra-hepatic delivery. They also noticed that a cleavable portion between the ASO and the conjugate gave the most active version, with no impact on biodistribution. Wada et al.61 have also recently investigated the effect of linker stability in a cholesterol–ASO conjugate, and found that the release of the conjugate from the ASO was essential for enhanced activity. A recent study by Wang et al.62 discusses that lipid conjugated ASOs have higher uptake into cells than their unconjugated counterpart, but they traffic through the same pathway of late endosomes. However, they found that ASO-conjugates, in comparison to free ASOs, tend to remain more associated with plasma and endosomal membranes rather than getting digested by the endosomes themselves. This causes their increased activity and release from endosomes to find their target RNA. This faster release from endosomes is critical to reach full activity, and in the case of our SNA, we hypothesize that having a metabolically cleavable spacer allows for the ASO to be cleaved and released, while the hydrophobic dodecanediol-based core may remain bound to the endosomal membrane and to proteins.62 It is important to note that the SNA did not significantly shed its ASO corona up to the measured 48 hours of incubation in serum containing cell culture (Fig. 8), suggesting that the ASO on the SNA circulates as a nanoparticle and acts as an ASO once cleaved and released in the cytosol.
Furthermore, the FANA-(PO-dN4)-SNA shows increased activity at lower concentration compared to its free ASO counterpart (Fig. 7). This is another advantage for using a nanoparticle, where the dose of the oligonucleotide drug can be decreased without compromising its effect.62 The SNA is modular in its design, and it can be adapted to target other endogenous targets inside cells, such as survivin and APOB (Fig. 9).
Oligonucleotides typically require targeting ligands or transfection agents to aid their uptake into cells.12,21,38,63 Here, we demonstrate that our FANA-SNAs exhibit more potent gymnotic gene silencing relatively to the free FANA-ASO (Fig. 10). In comparison to the previously reported gymnotic silencing with FANA-ASOs, we obtained slightly better activity (70% silencing) compared to 60% at 1 μM.12 We anticipate that changing the chemistry of the sequence-controlled polymer in the SNA core has the potential to further increase this activity.
In conclusion, we have demonstrated that carefully designed FANA modified SNAs are highly efficient particles for gene silencing. They combine the efficacy of modified oligonucleotides with the enhanced stability and uptake of spherical nucleic acids, and they show higher activity at lower concentrations, modularity in targeting different mRNAs, as well as unaided uptake into cells. Additionally, taking advantage of the hydrophobic core and FANA shell, surface modifications such as targeting ligands, and small encapsulated molecules that enhance endosomal release could readily be implemented. We envisage this system to see applications in delivery and therapy of oligonucleotide therapeutics.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
We would like to acknowledge the help of Dr Violeta Toader on the synthesis of the dodecanediol phosphoramidite conjugate. We are grateful to Henrietta Lacks, now deceased, and to her surviving family members for their contributions to biomedical research. We thank the Natural Sciences and Engineering Research Council of Canada (Discovery and NSERC CREATE Training grant to H. H. F., H. F. S. and M. J. D.), FRQNT, and the Canada Research Chairs program.References
- J. B. Opalinska and A. M. Gewirtz, Nucleic-acid therapeutics: basic principles and recent applications, Nat Rev Drug Discov, 2002, 1, 503–514 CrossRef CAS.
- E. Blanco, H. Shen and M. Ferrari, Principles of nanoparticle design for overcoming biological barriers to drug delivery, Nat Biotech, 2015, 33, 941–951 CrossRef CAS.
- M. Riley and W. Vermerris, Recent Advances in Nanomaterials for Gene Delivery—A Review, Nanomaterials, 2017, 7, 94 CrossRef.
- G. F. Deleavey and M. J. Damha, Designing Chemically Modified Oligonucleotides for Targeted Gene Silencing, Chem. Biol., 2012, 19, 937–954 CrossRef CAS.
- A. Kalota, L. Karabon, C. R. Swider, E. Viazovkina, M. Elzagheid, M. J. Damha and A. M. Gewirtz, 2′-Deoxy-2′-fluoro-β-d -arabinonucleic acid (2′F-ANA) modified oligonucleotides (ON) effect highly efficient, and persistent, gene silencing, Nucleic Acids Res., 2006, 34, 451–461 CrossRef CAS.
- T. Dowler, D. Bergeron, A.-L. Tedeschi, L. Paquet, N. Ferrari and M. J. Damha, Improvements in siRNA properties mediated by 2′-deoxy-2′-fluoro-β-d-arabinonucleic acid (FANA), Nucleic Acids Res., 2006, 34, 1669–1675 CrossRef CAS.
- G. F. Deleavey, J. K. Watts, T. Alain, F. Robert, A. Kalota, V. Aishwarya, J. Pelletier, A. M. Gewirtz, N. Sonenberg and M. J. Damha, Synergistic effects between analogs of DNA and RNA improve the potency of siRNA-mediated gene silencing, Nucleic Acids Res., 2010, 38, 4547–4557 CrossRef CAS.
- K.-L. Min, E. Viazovkina, A. Galarneau, M. A. Parniak and M. J. Damha, Oligonucleotides comprised of alternating 2′-Deoxy-2′-fluoro-β-d-arabinonucleosides and d-2′-deoxyribonucleosides (2′F-ANA/DNA ‘Altimers’) induce efficient RNA cleavage mediated by RNase H, Bioorg. Med. Chem. Lett., 2002, 12, 2651–2654 CrossRef CAS.
- C.-N. Lok, E. Viazovkina, K.-L. Min, E. Nagy, C. J. Wilds, M. J. Damha and M. A. Parniak, Potent Gene-Specific Inhibitory Properties of Mixed-Backbone Antisense Oligonucleotides Comprised of 2‘-Deoxy-2‘-fluoro-d-arabinose and 2‘-Deoxyribose Nucleotides, Biochemistry, 2002, 41, 3457–3467 CrossRef CAS.
- J. K. Watts, A. Katolik, J. Viladoms and M. J. Damha, Studies on the hydrolytic stability of 2′-fluoroarabinonucleic acid (2′F-ANA), Org. Biomol. Chem., 2009, 7, 1904–1910 RSC.
- M. Takahashi, H. Li, J. Zhou, P. Chomchan, V. Aishwarya, M. J. Damha and J. J. Rossi, Dual Mechanisms of Action of Self-Delivering, Anti-HIV-1 FANA Oligonucleotides as a Potential New Approach to HIV Therapy, Mol. Ther. Nucleic Acids, 2019, 17, 615–625 CrossRef CAS.
- N. Souleimanian, G. F. Deleavey, H. Soifer, S. Wang, K. Tiemann, M. J. Damha and C. A. Stein, Antisense 2'-Deoxy, 2'-Fluroarabino Nucleic Acids (2'F-ANAs) Oligonucleotides: In Vitro Gymnotic Silencers of Gene Expression Whose Potency Is Enhanced by Fatty Acids, Mol Ther Nucleic Acids, 2012, 1, e43 CrossRef.
- K. Schmidt, C. A. Weidmann, T. A. Hilimire, E. Yee, B. M. Hatfield, J. S. Schneekloth Jr, K. M. Weeks and C. D. Novina, Targeting the Oncogenic Long Non-coding RNA SLNCR1 by Blocking Its Sequence-Specific Binding to the Androgen Receptor, Cell Rep., 2020, 30, 541–554 CrossRef CAS.
- F. Della Valle, M. P. Thimma, M. Caiazzo, S. Pulcrano, M. Celii, S. A. Adroub, P. Liu, G. Alanis-Lobato, V. Broccoli and V. Orlando, Transdifferentiation of Mouse Embryonic Fibroblasts into Dopaminergic Neurons Reactivates LINE-1 Repetitive Elements, Stem Cell Rep., 2020, 14, 60–74 CrossRef CAS.
- G. Smaldone, G. Beneduce, M. Incoronato, K. Pane, M. Franzese, L. Coppola, A. Cordella, R. Parasole, M. Ripaldi and G. Nassa, et al., KCTD15 is overexpressed in human childhood B-cell acute lymphoid leukemia, Sci. Rep., 2019, 9, 20108 CrossRef CAS.
- S. Ganta, H. Devalapally, A. Shahiwala and M. Amiji, A review of stimuli-responsive nanocarriers for drug and gene delivery, J. Controlled Release, 2008, 126, 187–204 CrossRef CAS.
- T. Tokatlian and T. Segura, siRNA applications in nanomedicine, Wiley Interdiscip. Rev.: Nanomed. Nanobiotechnol., 2010, 2, 305–315 CAS.
- E. Blanco, H. Shen and M. Ferrari, Principles of nanoparticle design for overcoming biological barriers to drug delivery, Nat. Biotechnol., 2015, 33, 941–951 CrossRef CAS.
- Y. Weng, Q. Huang, C. Li, Y. Yang, X. Wang, J. Yu, Y. Huang and X.-J. Liang, Improved Nucleic Acid Therapy with Advanced Nanoscale Biotechnology, Mol Ther Nucleic Acids, 2020, 19, 581–601 CrossRef CAS.
- A. D. Springer and S. F. Dowdy, GalNAc–siRNA Conjugates: Leading the Way for Delivery of RNAi Therapeutics, Nucleic Acid Ther., 2018, 28, 109–118 CrossRef CAS.
- C. H. Kapadia, J. R. Melamed and E. S. Day, Spherical Nucleic Acid Nanoparticles: Therapeutic Potential, BioDrugs, 2018, 32, 297–309 CrossRef CAS.
- M. W. Tibbitt, J. E. Dahlman and R. Langer, Emerging Frontiers in Drug Delivery, J. Am. Chem. Soc., 2016, 138, 704–717 CrossRef CAS.
- D. Peer, J. M. Karp, S. Hong, O. C. Farokhzad, R. Margalit and R. Langer, Nanocarriers as an emerging platform for cancer therapy, Nat. Nanotechnol., 2007, 2, 751–760 CrossRef CAS.
- R. Juliano, M. R. Alam, V. Dixit and H. Kang, Mechanisms and strategies for effective delivery of antisense and siRNA oligonucleotides, Nucleic Acids Res., 2008, 36, 4158–4171 CrossRef CAS.
- S. Gudipati, K. Zhang and J. L. Rouge, Towards Self-Transfecting Nucleic Acid Nanostructures for Gene Regulation, Trends Biotechnol., 2019, 37, 983–994 CrossRef CAS.
- S. A. Jensen, E. S. Day, C. H. Ko, L. A. Hurley, J. P. Luciano, F. M. Kouri, T. J. Merkel, A. J. Luthi, P. C. Patel and J. I. Cutler, et al., Spherical nucleic acid nanoparticle conjugates as an RNAi-based therapy for glioblastoma, Sci Transl Med, 2013, 5, 209ra152 Search PubMed.
- K. T. Lewandowski, R. Thiede, N. Guido, W. L. Daniel, R. Kang, M. I. Guerrero-Zayas, M. A. Seeger, X. Q. Wang, D. A. Giljohann and A. S. Paller, Topically Delivered Tumor Necrosis Factor-α-Targeted Gene Regulation for Psoriasis, J. Invest. Dermatol., 2017, 137, 2027–2030 CrossRef CAS.
- T. G. Edwardson, K. M. Carneiro, C. J. Serpell and H. F. Sleiman, An efficient and modular route to sequence-defined polymers appended to DNA, Angew. Chem. Int. Ed., 2014, 53, 4567–4571 CrossRef CAS.
- D. Bousmail, P. Chidchob and H. F. Sleiman, Cyanine-Mediated DNA Nanofiber Growth with Controlled Dimensionality, J. Am. Chem. Soc., 2018, 140, 9518–9530 CrossRef CAS.
- T. G. Edwardson, K. M. Carneiro, C. K. McLaughlin, C. J. Serpell and H. F. Sleiman, Site-specific positioning of dendritic alkyl chains on DNA cages enables their geometry-dependent self-assembly, Nat. Chem., 2013, 5, 868–875 CrossRef CAS.
- C. J. Serpell, T. G. W. Edwardson, P. Chidchob, K. M. M. Carneiro and H. F. Sleiman, Precision Polymers and 3D DNA Nanostructures: Emergent Assemblies from New Parameter Space, J. Am. Chem. Soc., 2014, 136, 15767–15774 CrossRef CAS.
- P. Chidchob, T. G. W. Edwardson, C. J. Serpell and H. F. Sleiman, Synergy of Two Assembly Languages in DNA Nanostructures: Self-Assembly of Sequence-Defined Polymers on DNA Cages, J. Am. Chem. Soc., 2016, 138, 4416–4425 CrossRef CAS.
- J. J. Fakhoury, T. G. Edwardson, J. W. Conway, T. Trinh, F. Khan, M. Barłóg, H. S. Bazzi and H. F. Sleiman, Antisense precision polymer micelles require less poly(ethylenimine) for efficient gene knockdown, Nanoscale, 2015, 7, 20625–20634 RSC.
- D. Bousmail, L. Amrein, J. J. Fakhoury, H. H. Fakih, J. C. C. Hsu, L. Panasci and H. F. Sleiman, Precision spherical nucleic acids for delivery of anticancer drugs, Chem. Sci., 2017, 8, 6218–6229 RSC.
- M. D. Dore, J. J. Fakhoury, A. Lacroix and H. F. Sleiman, Templated synthesis of spherical RNA nanoparticles with gene silencing activity, Chem. Commun., 2018, 54, 11296–11299 RSC.
- H. H. Fakih, J. J. Fakhoury, D. Bousmail and H. F. Sleiman, Minimalist Design of a Stimuli-Responsive Spherical Nucleic Acid for Conditional Delivery of Oligonucleotide Therapeutics, ACS Appl. Mater. Interfaces, 2019, 11, 13912–13920 CrossRef CAS.
- P. W. K. Rothemund, Folding DNA to create nanoscale shapes and patterns, Nature, 2006, 440, 297–302 CrossRef CAS.
- J. Shi, P. W. Kantoff, R. Wooster and O. C. Farokhzad, Cancer nanomedicine: progress, challenges and opportunities, Nat. Rev. Cancer, 2017, 17, 20–37 CrossRef CAS.
- D. de Rochambeau, Y. Sun, M. Barlog, H. S. Bazzi and H. F. Sleiman, Modular Strategy To Expand the Chemical Diversity of DNA and Sequence-Controlled Polymers, J. Org. Chem., 2018, 83, 9774–9786 CrossRef CAS.
- R. J. Banga, N. Chernyak, S. P. Narayan, S. T. Nguyen and C. A. Mirkin, Liposomal Spherical Nucleic Acids, J. Am. Chem. Soc., 2014, 136, 9866–9869 CrossRef CAS.
- R. Yan, J. Chen, J. Wang, J. Rao, X. Du, Y. Liu, L. Zhang, L. Qiu, B. Liu and Y.-D. Zhao, et al., A NanoFlare-Based Strategy for In Situ Tumor Margin Demarcation and Neoadjuvant Gene/Photothermal Therapy, Small, 2018, 14, 1802745 CrossRef.
- C. Guan, N. Chernyak, D. Dominguez, L. Cole, B. Zhang and C. A. Mirkin, RNA-Based Immunostimulatory Liposomal Spherical Nucleic Acids as Potent TLR7/8 Modulators, Small, 2018, 14, e1803284 CrossRef.
- J. R. Ferrer, J. A. Wertheim and C. A. Mirkin, Dual Toll-Like Receptor Targeting Liposomal Spherical Nucleic Acids, Bioconjugate Chem., 2019, 30, 944–951 CrossRef CAS.
- S. Wang, L. Qin, G. Yamankurt, K. Skakuj, Z. Huang, P.-C. Chen, D. Dominguez, A. Lee, B. Zhang and C. A. Mirkin, Rational vaccinology with spherical nucleic acids, Proc. Natl. Acad. Sci. U. S. A., 2019, 116, 10473–10481 CrossRef CAS.
- S. N. Barnaby, G. A. Perelman, K. L. Kohlstedt, A. B. Chinen, G. C. Schatz and C. A. Mirkin, Design Considerations for RNA Spherical Nucleic Acids (SNAs), Bioconjugate Chem., 2016, 27, 2124–2131 CrossRef CAS.
- C. H. J. Choi, L. Hao, S. P. Narayan, E. Auyeung and C. A. Mirkin, Mechanism for the endocytosis of spherical nucleic acid nanoparticle conjugates, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 7625–7630 CrossRef CAS.
- H. Nemati, M.-H. Ghahramani, R. Faridi-Majidi, B. Izadi, G. Bahrami, S.-H. Madani and G. Tavoosidana, Using siRNA-based spherical nucleic acid nanoparticle conjugates for gene regulation in psoriasis, J. Controlled Release, 2017, 268, 259–268 CrossRef CAS.
- H. H. Fakih, J. J. Fakhoury, D. Bousmail and H. F. Sleiman, Minimalist Design of a Stimuli-Responsive Spherical Nucleic Acid for Conditional Delivery of Oligonucleotide Therapeutics, ACS Appl. Mater. Interfaces, 2019, 11, 13912–13920 CrossRef CAS.
- D. Bousmail, L. Amrein, J. J. Fakhoury, H. H. Fakih, J. C. C. Hsu, L. Panasci and H. F. Sleiman, Precision spherical nucleic acids for delivery of anticancer drugs, Chem. Sci., 2017, 8, 6218–6229 RSC.
- A. Lacroix, E. Vengut-Climent, D. de Rochambeau and H. F. Sleiman, Uptake and Fate of Fluorescently Labeled DNA Nanostructures in Cellular Environments: A Cautionary Tale, ACS Cent. Sci., 2019, 5, 882–891 CAS.
- A. Lacroix, H. H. Fakih and H. F. Sleiman, Detailed cellular assessment of albumin-bound oligonucleotides: Increased stability and lower non-specific cell uptake, J. Controlled Release, 2020, 324, 34–46 CrossRef CAS.
- R. A. Olie, A. P. Simões-Wüst, B. Baumann, S. H. Leech, D. Fabbro, R. A. Stahel and U. Zangemeister-Wittke, A Novel Antisense Oligonucleotide Targeting Survivin Expression Induces Apoptosis and Sensitizes Lung Cancer Cells to Chemotherapy, Cancer Res., 2000, 60, 2805–2809 CAS.
- M. Pennati, M. Folini and N. Zaffaroni, Targeting survivin in cancer therapy, Expert Opin. Ther. Targets, 2008, 12, 463–476 CrossRef CAS.
- D. P. Y. Chan, G. F. Deleavey, S. C. Owen, M. J. Damha and M. S. Shoichet, Click conjugated polymeric immuno-nanoparticles for targeted siRNA and antisense oligonucleotide delivery, Biomaterials, 2013, 34, 8408–8415 CrossRef CAS.
- J. J. Fakhoury, T. G. Edwardson, J. W. Conway, T. Trinh, F. Khan, M. Barlog, H. S. Bazzi and H. F. Sleiman, Antisense precision polymer micelles require less poly(ethylenimine) for efficient gene knockdown, Nanoscale, 2015, 7, 20625–20634 RSC.
- N. L. Rosi, D. A. Giljohann, C. S. Thaxton, A. K. R. Lytton-Jean, M. S. Han and C. A. Mirkin, Oligonucleotide-Modified Gold Nanoparticles for Intracellular Gene Regulation, Science, 2006, 312, 1027–1030 CrossRef CAS.
- J. I. Cutler, E. Auyeung and C. A. Mirkin, Spherical Nucleic Acids, J. Am. Chem. Soc., 2012, 134, 1376–1391 CrossRef CAS.
- C. D. Kusmierz, K. E. Bujold, C. E. Callmann and C. A. Mirkin, Defining the Design Parameters for in Vivo Enzyme Delivery Through Protein Spherical Nucleic Acids, ACS Cent. Sci., 2020, 6, 815–822 CrossRef CAS.
- T. P. Prakash, A. E. Mullick, R. G. Lee, J. Yu, S. T. Yeh, A. Low, A. E. Chappell, M. E. Østergaard, S. Murray and H. J. Gaus, et al., Fatty acid conjugation enhances potency of antisense oligonucleotides in muscle, Nucleic Acids Res., 2019, 47, 6029–6044 CrossRef CAS.
- A. Biscans, J. Caiazzi, S. Davis, N. McHugh, J. Sousa and A. Khvorova, The chemical structure and phosphorothioate content of hydrophobically modified siRNAs impact extrahepatic distribution and efficacy, Nucleic Acids Res., 2020, 48, 7665–7680 CrossRef CAS.
- S. Wada, H. Yasuhara, F. Wada, M. Sawamura, R. Waki, T. Yamamoto, M. Harada-Shiba and S. Obika, Evaluation of the effects of chemically different linkers on hepatic accumulations, cell tropism and gene silencing ability of cholesterol-conjugated antisense oligonucleotides, J. Controlled Release, 2016, 226, 57–65 CrossRef CAS.
- S. Wang, N. Allen, T. P. Prakash, X.-H. Liang and S. T. Crooke, Lipid Conjugates Enhance Endosomal Release of Antisense Oligonucleotides Into Cells, Nucleic Acid Ther., 2019, 29, 245–255 CrossRef CAS.
- Y. Xiao, K. Shi, Y. Qu, B. Chu and Z. Qian, Engineering Nanoparticles for Targeted Delivery of Nucleic Acid Therapeutics in Tumor, Mol. Ther.--Methods Clin. Dev., 2019, 12, 1–18 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0sc06645a |
| This journal is © The Royal Society of Chemistry 2021 |





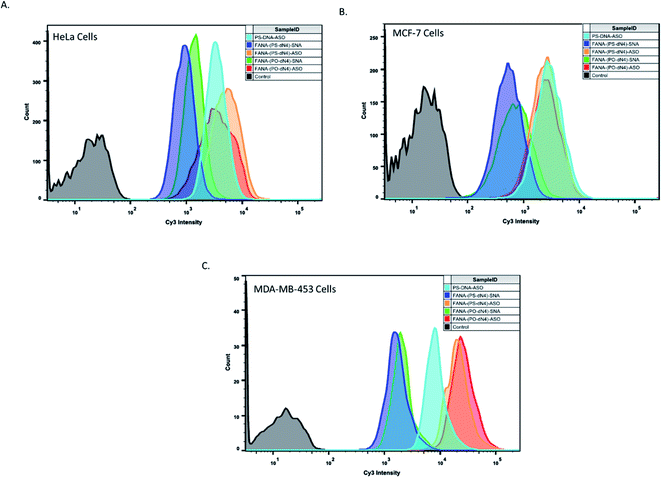
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells count was collected for all samples in HeLa and MCF-7 cells, and 1000 cells count was collected in MDA-453 cells. One duplicate is shown for simplicity.
000 cells count was collected for all samples in HeLa and MCF-7 cells, and 1000 cells count was collected in MDA-453 cells. One duplicate is shown for simplicity.