Open Access Article
Open Access ArticleNature-inspired dimerization as a strategy to modulate neuropeptide pharmacology exemplified with vasopressin and oxytocin†
Zoltan
Dekan‡
 a,
Thomas
Kremsmayr‡
a,
Thomas
Kremsmayr‡
 b,
Peter
Keov
b,
Peter
Keov
 c,
Mathilde
Godin
a,
Ngari
Teakle
a,
Leopold
Dürrauer
b,
Huang
Xiang
d,
Dalia
Gharib
d,
Christian
Bergmayr
d,
Roland
Hellinger
c,
Mathilde
Godin
a,
Ngari
Teakle
a,
Leopold
Dürrauer
b,
Huang
Xiang
d,
Dalia
Gharib
d,
Christian
Bergmayr
d,
Roland
Hellinger
 d,
Marina
Gay
d,
Marina
Gay
 e,
Marta
Vilaseca
e,
Marta
Vilaseca
 e,
Dennis
Kurzbach
e,
Dennis
Kurzbach
 b,
Fernando
Albericio
b,
Fernando
Albericio
 f,
Paul F.
Alewood
f,
Paul F.
Alewood
 a,
Christian W.
Gruber
a,
Christian W.
Gruber
 *cd and
Markus
Muttenthaler
*cd and
Markus
Muttenthaler
 *ab
*ab
aInstitute for Molecular Bioscience, The University of Queensland, Brisbane 4072, Australia. E-mail: m.muttenthaler@uq.edu.au
bInstitute of Biological Chemistry, University of Vienna, Währingerstraße 38, 1090 Vienna, Austria. E-mail: markus.muttenthaler@univie.ac.at
cFaculty of Medicine, School of Biomedical Sciences, The University of Queensland, Brisbane 4072, Australia
dCenter for Physiology and Pharmacology, Medical University of Vienna, Schwarzspanierstraße 17, 1090 Vienna, Austria. E-mail: christian.w.gruber@meduniwien.ac.at
eInstitute for Research in Biomedicine Barcelona, C/ Baldiri Reixac 10, 08028 Barcelona, Spain
fDepartment of Organic Chemistry, University of Barcelona, Barcelona Science Park, Baldiri Reixac 10, 08028 Barcelona, Spain
First published on 4th February 2021
Abstract
Vasopressin (VP) and oxytocin (OT) are cyclic neuropeptides that regulate fundamental physiological functions via four G protein-coupled receptors, V1aR, V1bR, V2R, and OTR. Ligand development remains challenging for these receptors due to complex structure–activity relationships. Here, we investigated dimerization as a strategy for developing ligands with novel pharmacology. We regioselectively synthesised and systematically studied parallel, antiparallel and N- to C-terminal cyclized homo- and heterodimer constructs of VP, OT and dVDAVP (1-deamino-4-valine-8-D-arginine-VP). All disulfide-linked dimers, except for the head-to-tail cyclized constructs, retained nanomolar potency despite the structural implications of dimerization. Our results support a single chain interaction for receptor activation. Dimer orientation had little impact on activity, except for the dVDAVP homodimers, where an antagonist to agonist switch was observed at the V1aR. This study provides novel insights into the structural requirements of VP/OT receptor activation and spotlights dimerization as a strategy to modulate pharmacology, a concept also frequently observed in nature.
Introduction
Vasopressin (VP, CYFQNCPRG) and oxytocin (OT, CYIQNCPLG) are closely-related multifunctional neuropeptides consisting of a six-residue macrocyclic structure that is cyclized by a disulfide bond and a three-residue amidated C-terminal tail. VP and OT act via four receptors (V1aR, V1bR, V2R, OTR) belonging to the rhodopsin-like/class A G protein-coupled receptor (GPCR) family.1–8 VP/OT receptors are drug targets for cardiovascular and bleeding conditions, fluid and electrolyte disorders, anxiety, aggression, stress, depression, autism, schizophrenia, nephrogenic diabetes insipidus, nocturnal enuresis, prevention of preterm labour, pain and cancer.4,6,9–11 The study of these receptors relies on a good structural understanding of the ligand–receptor interactions as well as on molecular probes with defined pharmacology. Despite considerable efforts in probe development,12–15 we still do not have a complete pharmacological toolbox to study this signalling system.16–18 To address this limitation, we have pursued alternative ligand discovery and modification strategies ranging from innovative medicinal chemistry11,18–21 to natural product discovery.22–26 The latter led us to peptide dimerization, a concept frequently observed in nature27–36 and also studied using a variety of conjugation strategies.37–48 Disulfide-linked homo- and heterodimeric peptides are commonly found in venoms, underpinning the evolutionary relevance of this strategy.31,49–56 Dimerization is also integral for human physiology: for instance, β-human atrial natriuretic peptide (β-hANP), a dimer of α-hANP, elicits cardiovascular responses reminiscent of monomeric α-hANP, but with distinct pharmacokinetics and pharmacodynamics, providing slower onset and longer duration.28,57–59 Insulin is another example for a heterodimer controlling glucose homeostasis, with the monomeric A and B chains also able to regulate this function, but to a lesser extent.60–62 We were further encouraged by a naturally occurring antiparallel inotocin (insect version of VP/OT) homodimer that regulates fluid secretion in the migratory locust.63,64We thus set out to systematically study VP/OT dimerization to establish (a) if and how receptor activation occurs, (b) if dimer directionality plays a role, and (c) whether this strategy can be used to modulate pharmacology.
Results and discussion
Dimer design and synthesis
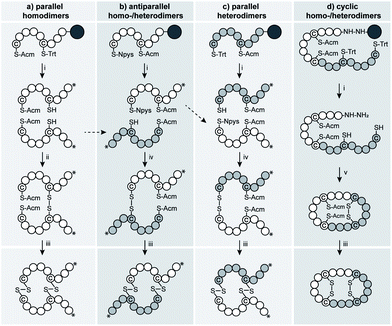
We devised several regioselective synthetic strategies to produce the parallel and antiparallel VP/OT homo- and heterodimers as well as N- to C-terminal cyclized VP/OT dimers (Fig. 1 and S1†). We used Fmoc-SPPS and orthogonal cysteine protecting groups (combinations of S-Trt, S-Acm and S-Npys groups) to access the individual homo- and heterodimers in the desired orientations (parallel and antiparallel) and Fmoc-SPPS in combination with intramolecular native chemical backbone ligation65,66 and directed folding to produce the N- to C-terminal cyclized analogues (Fig. 1).Pharmacological characterization
We evaluated the activity of these dimeric constructs in cells overexpressing each receptor of interest using well-established IP-1 (V1aR, V1bR, OTR) and cAMP (V2R) quantitative second messenger-detecting assays (Fig. 2a–c, S3 and S4†). Interestingly, both parallel and antiparallel VP homodimers activated all four receptors, with only 5–15-fold reduced potency (EC50) compared to VP, yet presented no significant change in selectivity or intrinsic efficacy (Emax). Parallel and antiparallel OT homodimers also activated all four receptors, yet with 10–100-fold reduced potencies compared to OT. No significant differences in selectivity or directionality were observed. The lack of directionality dependence coupled with the potency differences between VP and OT dimers suggested that activation occurs through a single chain of the dimer and that the VP chain is structurally better tolerated than the OT chain. This finding was further supported by the data of the antiparallel OT-VP heterodimer, which was slightly more potent than the OT dimers at OTR and less potent than the VP dimers at the VPRs.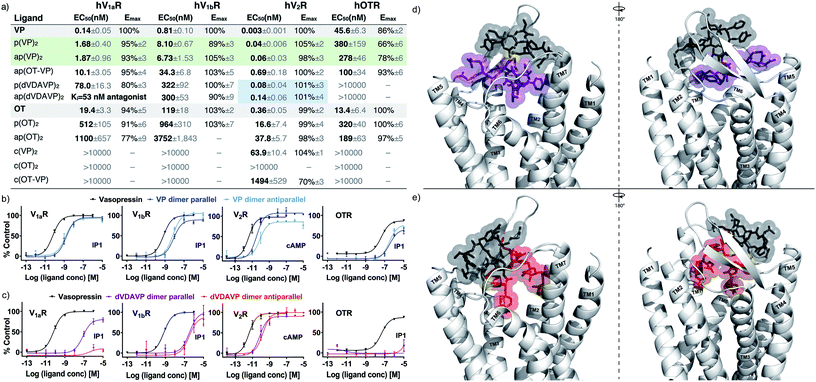 | ||
| Fig. 2 EC50, Emax and representative full-dose response curves of the most potent dimers, (VP)2 and (dVDAVP)2. (a) Table summarising EC50 and Emax; green colour highlighting noteworthy activity (5–15-fold of OT/VP), and blue improved selectivity. Ki determined by radioligand displacement assays (Table S1†). (b) Concentration response curves of VP homodimers with respect to VP across V1aR, V1bR, V2R and OTR. (c) Concentration response curves of dVDAVP dimers with respect to VP across OTR, V1aR, V1bR, V2R and OTR. N = 3 independent experiments; errors presented as standard error of the mean (SEM), some error bars are smaller than the symbol; for concentration response curves of all dimers, please refer to ESI Fig. S3, S4 and S6.† (d) & e) Possible binding mode derived from in silico docking of p(dVDAVP)2 (d) and ap(dVDAVP)2 (e) to hV1aR. Receptor docking was performed using a recently reported homology model of hV1aR. Please refer to the ESI† for details on structure modelling and binding simulation. The individual strands of the dimers are indicted in grey/purple or grey/red, respectively; interstrand disulfide bond connections are highlighted in yellow; the seven transmembrane (TM) V1aR helices are labelled. | ||
The observed pico- to nanomolar potencies were striking considering the switch from a single intramolecular disulfide bond in the monomers to two intermolecular disulfide bonds in the dimers, particularly when put in context with past studies where even minor modifications (e.g., shortening of the OT ring structure by one sulfur atom, or disulfide bond replacement by a dimethylene bridge) led to substantial decrease in activity.18,20,67 We thus ensured that this observation was not due to monomer formation during the assays (confirmed by HPLC before and after, Fig. S5†). Moreover, we determined the EC50 using a fluorescent imaging plate reader (FLIPR) that measured Ca2+ responses within seconds after the addition of freshly prepared dimers, further confirming that signalling was indeed induced by intact dimers (Fig. S6 and Table S2†).
Considering the potent agonism of the VP dimers, we also designed and synthesized N- to C-terminal cyclized VP and OT dimers with the aim of creating metabolically more stable analogues. The cyclic analogues were however largely inactive across the four receptors (Fig. 2a and S4†), underpinning the importance of the N-terminus and the free three-residue C-terminus for activity.
We then expanded this strategy to an improved analogue of the drug desmopressin, namely dVDAVP (dCYFVNCPrG, 1-deamino-4-valine-8-D-arginine-VP), a well-known V2R/V1bR agonist and V1aR antagonist.14,68–72 Interestingly, during the synthesis of monomeric dVDAVP, we observed spontaneous dimerization during folding (60% dimer, 40% monomer), even at a relatively low peptide concentration of 100 μM, which was not observed with VP or OT. MS/MS experiments revealed that the observed product was exclusively the antiparallel homodimer (Fig. S7 and S8†) with no parallel homodimer formed. We also synthesized the dVDAVP homodimers via directed disulfide bond formation to access the parallel homodimer and to confirm the MS/MS experiments via analytical HPLC co-elution study (Fig. S9†). Both dVDAVP homodimers were potent V2R agonists that also displayed some activity at V1bR, but no activity at OTR (Fig. 2a and c). Interestingly, in this case, directionality played a role at V1aR, where the antiparallel homodimer was an antagonist (Ki = 53 nM, similar to dVDAVP monomer:14,68–72Ki = 17 nM, Fig. S2 and Table S1†) whereas the parallel homodimer was a full V1aR agonist (EC50 = 78 nM, Ki = 76 nM). This finding illustrates that dimeric design can lead to significant functional (antagonist/agonist) switches (Fig. 2a, c and S2, Table S1†).
Structural characterization
NMR-based structure determination protocols were not applicable to the dimers due to a lack of long-range nuclear Overhauser effects and assignment ambiguities due to the sequence homology of the dimer chains. Hence, we modelled both, the parallel and antiparallel dVDAVP dimer, by adapting well-established protocols and docked these dVDAVP dimer structures into the hV1aR binding pocket (see ESI† for details on the modelling and docking procedure).22,73–75 The resulting ligand-receptor-complex models supported our experimental in vitro activity data and hypothesis that only a single dimer subunit/chain fits and interacts with the receptor binding pocket (Fig. 2d and e). In qualitative agreement with previously described binding modes of VP and related variants,22,76–78 we observed energetically low-lying complex conformations where Y2 and F3 of the interacting dimer chain, in both parallel and antiparallel (dVDAVP)2, insert into the hydrophobic core of hV1aR.We also carried out circular dichroism (CD) studies (Fig. 3) to provide further insights into overall secondary structures. VP and OT homodimers yielded CD spectra that were highly similar to their corresponding monomeric ligands VP and OT (Fig. 3a and b). The data suggest high flexibility and a structure of intrinsically disordered nature, corresponding well with literature79 and bioactivity data. The OT-VP heterodimer also displayed a CD profile similar to VP and OT (Fig. 3c), while the inactive N- to C-terminal cyclized analogues had a similar, but less pronounced profile (Fig. 3a–c). By contrast, the dVDAVP homodimers displayed considerable differences, not only in comparison with VP and dVDAVP monomers, but also between the parallel and antiparallel orientation (Fig. 3d). We used BeStSel software tool80,81 to estimate the secondary structures and found that the parallel dVDAVP homodimer featured a CD spectrum reminiscent of a β-turn structure (38% structural contribution), while the antiparallel dVDAVP homodimer had a spectrum reminiscent of an antiparallel β-strand (58% structural contribution). The CD data also aligned well with our ligand models as two distinct structures were observed for the parallel and antiparallel dVDAVP homodimer (Fig. 3e and f): The p(dVDAVP)2 model features two stable β-turn motifs between residues F3 and C6 of both subunits, which were less pronounced for ap(dVDAVP)2. These structural differences, might also account for the spontaneous formation of the antiparallel dimer during the undirected folding event, indicating an energetically favoured antiparallel orientation. This was supported by energy calculations for the antiparallel and parallel model structures, which yielded a considerably lower conformational energy (−4501 kJ mol−1) for the antiparallel homodimer versus the parallel dimer (−2364 kJ mol−1).
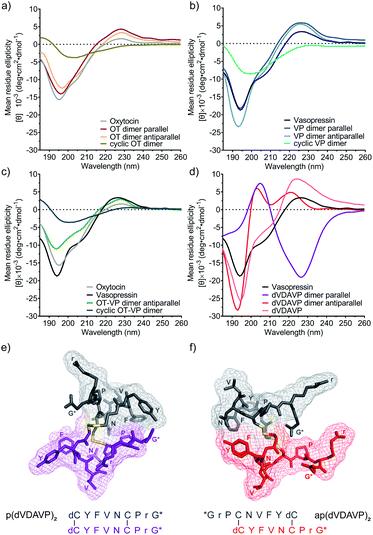 | ||
| Fig. 3 CD spectra of homo and heterodimeric constructs and in silico models of parallel and antiparallel dVDAVP dimers. CD spectra of (a) OT dimers in comparison with OT. (b) VP dimers in comparison with VP. (c) OT-VP antiparallel and cyclic dimers in comparison with OT and VP. (d) dVDAVP dimers in comparison with VP and dVDAVP. CD spectra were recorded in 10 mM aqueous sodium phosphate at pH 7.4 and 25 °C. Ligand concentrations were 50 μM for monomers and 25 μM for dimers. (e) In silico modelled 3D structure of p(dVDAVP)2. (f) In silico modelled 3D structure of ap(dVDAVP)2. Single chains are indicted in different colours; interstrand disulfide bond connections are highlighted in yellow. The structures represent energetically low-lying snapshots of a highly dynamic conformational ensemble. Please refer to the ESI† for details on the structure simulation procedure. | ||
Our work furthermore clarified the mechanism of action of earlier efforts investigating neuropeptide dimers:63,82–86 OT dimers displayed weak but protracted and long-lasting uterotonic activity in rats, which the authors speculated was due to the dimers slowly reverting to monomers.83,86 Here we, however, demonstrated that monomer formation is not required for receptor activation. This also highlights that dimerization can modulate in vivo pharmacokinetics and pharmacodynamics, as observed with the more protracted and longer-lasting uterotonic activity compared to OT.86 As such, dimerization could play a valuable role in optimizing therapeutic effects or in the development of bifunctional heterodimers to generate synergistic effects by targeting two distinct receptors.
Conclusions
We demonstrated that VP and OT homo- and heterodimers retain their abilities to potently activate their receptors despite the structural rearrangement imposed through dimerization. We provided novel structural and mechanistic insights into the concept of dimerization which is commonly observed in nature and adds a new design strategy to the development of ligands with distinct pharmacology. Considering the common activation mechanism across class A GPCRs,1 our findings may also be relevant for other receptors of this family.Author contributions
Z. D. and T. K. contributed equally to this work. Z. D., T. K. and M. G. synthesised the compounds. Z. D. and T. K. performed the CD analysis. P. K., N. T. and L. D. performed the functional assays. L. D., H. X., D. G. and C. B. carried out the radioligand binding assays. R. H., M. G., M. V. conducted the structure determination via mass spectrometry. D. K. and T. K performed the molecular modelling. M. M., C. W. G., P. F. A., and F. A. conceived, funded, and supervised the project. M. M., C. W. G., D. K. and T. K. prepared the manuscript. All authors read the manuscript and provided feedback.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We thank Elena Deliu for her help in drafting this manuscript. This work was supported by the European Research Council under the European Union's Horizon 2020 research and innovation program (grant agreements no 714366 and 801936), by the Australian Research Council Discovery Project (DP190101667) and by the Vienna Science and Technology Fund (WWTF) through project LS18-053. Work in the laboratory of Christian W. Gruber was supported by an Australian Research Council Future Fellowship (FT140100730) and the Austrian Science Fund FWF (I3243 and P32109). T. Kremsmayr was supported by the Austrian Academy of Sciences through a DOC Fellowship (25139).References
- Q. Zhou, D. Yang, M. Wu, Y. Guo, W. Guo, L. Zhong, X. Cai, A. Dai, W. Jang, E. I. Shakhnovich, Z. J. Liu, R. C. Stevens, N. A. Lambert, M. M. Babu, M. W. Wang and S. Zhao, eLife, 2019, 8, e50279 CrossRef.
- B. Jurek and I. D. Neumann, Physiol. Rev., 2018, 98, 1805–1908 CrossRef CAS.
- A. S. Hauser, M. M. Attwood, M. Rask-Andersen, H. B. Schioth and D. E. Gloriam, Nat. Rev. Drug Discovery, 2017, 16, 829–842 CrossRef CAS.
- A. Meyer-Lindenberg, G. Domes, P. Kirsch and M. Heinrichs, Nat. Rev. Neurosci., 2011, 12, 524–538 CrossRef CAS.
- J. A. Salon, D. T. Lodowski and K. Palczewski, Pharmacol. Rev., 2011, 63, 901–937 CrossRef CAS.
- C. W. Gruber, M. Muttenthaler and M. Freissmuth, Curr. Pharm. Des., 2010, 16, 3071–3088 CrossRef CAS.
- Z. R. Donaldson and L. J. Young, Science, 2008, 322, 900–904 CrossRef CAS.
- G. Gimpl and F. Fahrenholz, Physiol. Rev., 2001, 81, 629–683 CrossRef CAS.
- R. Landgraf, Handb. Exp. Pharmacol., 2005, 335–369 CAS.
- G. Leng, R. Pineda, N. Sabatier and M. Ludwig, J. Endocrinol., 2015, 226, T173–T185 CAS.
- A. D. de Araujo, M. Mobli, J. Castro, A. M. Harrington, I. Vetter, Z. Dekan, M. Muttenthaler, J. Wan, R. J. Lewis, G. F. King, S. M. Brierley and P. F. Alewood, Nat. Commun., 2014, 5, 3165 CrossRef.
- K. Jost, M. Lebl and F. Brtnik, CRC handbook of neurohypophyseal hormone analogs, CRC Press Inc., Boca Raton, FL, United States, 1987 Search PubMed.
- M. Manning, S. Stoev, B. Chini, T. Durroux, B. Mouillac and G. Guillon, in Prog. Brain Res., ed. I. D. Neumann and R. Landgraf, Elsevier, 2008, vol. 170, pp. 473–512 Search PubMed.
- M. Manning, A. Misicka, A. Olma, K. Bankowski, S. Stoev, B. Chini, T. Durroux, B. Mouillac, M. Corbani and G. Guillon, J. Neuroendocrinol., 2012, 24, 609–628 CrossRef CAS.
- D. Gulliver, E. Werry, T. A. Reekie, T. A. Katte, W. Jorgensen and M. Kassiou, Trends Pharmacol. Sci., 2019, 40, 22–37 CrossRef CAS.
- B. Chini and M. Manning, Biochem. Soc. Trans., 2007, 35, 737–741 CrossRef CAS.
- B. Chini, M. Manning and G. Guillon, in Prog. Brain Res., ed. I. D. Neumann and R. Landgraf, Elsevier, 2008, vol. 170, pp. 513–517 Search PubMed.
- M. Muttenthaler, A. Andersson, I. Vetter, R. Menon, M. Busnelli, L. Ragnarsson, C. Bergmayr, S. Arrowsmith, J. R. Deuis, H. S. Chiu, N. J. Palpant, M. O’Brien, T. J. Smith, S. Wray, I. D. Neumann, C. W. Gruber, R. J. Lewis and P. F. Alewood, Sci. Signaling, 2017, 10, eaan3398 CrossRef.
- M. Busnelli, G. Kleinau, M. Muttenthaler, S. Stoev, M. Manning, L. Bibic, L. A. Howell, P. J. McCormick, S. Di Lascio, D. Braida, M. Sala, G. E. Rovati, T. Bellini and B. Chini, J. Med. Chem., 2016, 59, 7152–7166 CrossRef CAS.
- M. Muttenthaler, A. Andersson, A. D. de Araujo, Z. Dekan, R. J. Lewis and P. F. Alewood, J. Med. Chem., 2010, 53, 8585–8596 CrossRef CAS.
- J. Wan, M. Mobli, A. Brust, M. Muttenthaler, A. Andersson, L. Ragnarsson, J. Castro, I. Vetter, J. X. Huang, M. Nilsson, S. M. Brierley, M. A. Cooper, R. J. Lewis and P. F. Alewood, Aust. J. Chem., 2017, 70, 162 CrossRef CAS.
- M. G. Di Giglio, M. Muttenthaler, K. Harpsoe, Z. Liutkeviciute, P. Keov, T. Eder, T. Rattei, S. Arrowsmith, S. Wray, A. Marek, T. Elbert, P. F. Alewood, D. E. Gloriam and C. W. Gruber, Sci. Rep., 2017, 7, 41002 CrossRef CAS.
- S. Dutertre, D. Croker, N. L. Daly, A. Andersson, M. Muttenthaler, N. G. Lumsden, D. J. Craik, P. F. Alewood, G. Guillon and R. J. Lewis, J. Biol. Chem., 2008, 283, 7100–7108 CrossRef CAS.
- J. Koehbach, M. O'Brien, M. Muttenthaler, M. Miazzo, M. Akcan, A. G. Elliott, N. L. Daly, P. J. Harvey, S. Arrowsmith, S. Gunasekera, T. J. Smith, S. Wray, U. Goransson, P. E. Dawson, D. J. Craik, M. Freissmuth and C. W. Gruber, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 21183–21188 CrossRef CAS.
- C. W. Gruber and M. Muttenthaler, PLoS One, 2012, 7, e32559 CrossRef CAS.
- C. W. Gruber, J. Koehbach and M. Muttenthaler, Future Med. Chem., 2012, 4, 1791–1798 CrossRef CAS.
- U. Derewenda, Z. Derewenda, G. G. Dodson, R. E. Hubbard and F. Korber, Br. Med. Bull., 1989, 45, 4–18 CrossRef CAS.
- K. Kangawa, A. Fukuda and H. Matsuo, Nature, 1985, 313, 397–400 CrossRef CAS.
- M. W. Hornef, K. Pütsep, J. Karlsson, E. Refai and M. Andersson, Nat. Immunol., 2004, 5, 836–843 CrossRef CAS.
- A. C. Conibear and D. J. Craik, Angew. Chem., Int. Ed., 2014, 53, 10612–10623 CrossRef CAS.
- Z. Dekan, S. J. Headey, M. Scanlon, B. A. Baldo, T. H. Lee, M. I. Aguilar, J. R. Deuis, I. Vetter, A. G. Elliott, M. Amado, M. A. Cooper, D. Alewood and P. F. Alewood, Angew. Chem., Int. Ed., 2017, 56, 8495–8499 CrossRef CAS.
- E. N. Lorenzon, J. P. Piccoli, N. A. Santos-Filho and E. M. Cilli, Protein Pept. Lett., 2019, 26, 98–107 CrossRef CAS.
- S. P. Liu, L. Zhou, R. Lakshminarayanan and R. W. Beuerman, Int. J. Pept. Res. Ther., 2010, 16, 199–213 CrossRef CAS.
- C. V. F. Batista, A. Scaloni, D. J. Rigden, L. R. Silva, A. Rodrigues Romero, R. Dukor, A. Sebben, F. Talamo and C. Bloch, FEBS Lett., 2001, 494, 85–89 CrossRef CAS.
- I. H. Lee, Y. S. Lee, C. H. Kim, C. R. Kim, T. Hong, L. Menzel, L. M. Boo, J. Pohl, M. A. Sherman, A. Waring and R. I. Lehrer, Biochim. Biophys. Acta, Gen. Subj., 2001, 1527, 141–148 CrossRef CAS.
- W. S. Jang, K. N. Kim, Y. S. Lee, M. H. Nam and I. H. Lee, FEBS Lett., 2002, 521, 81–86 CrossRef CAS.
- J. C. Cheronis, E. T. Whalley, K. T. Nguyen, S. R. Eubanks, L. G. Allen, M. J. Duggan, S. D. Loy, K. A. Bonham and J. K. Blodgett, J. Med. Chem., 1992, 35, 1563–1572 CrossRef CAS.
- K. Arai, T. Takei, M. Okumura, S. Watanabe, Y. Amagai, Y. Asahina, L. Moroder, H. Hojo, K. Inaba and M. Iwaoka, Angew. Chem., Int. Ed., 2017, 56, 5522–5526 CrossRef CAS.
- G. M. Williams, K. Lee, X. Li, G. J. Cooper and M. A. Brimble, Org. Biomol. Chem., 2015, 13, 4059–4063 RSC.
- J. W. Kim, F. V. Cochran and J. R. Cochran, J. Am. Chem. Soc., 2015, 137, 6–9 CrossRef CAS.
- G. Mezo, M. Manea, A. Jakab, B. Kapuvari, S. Bosze, G. Schlosser, M. Przybylski and F. Hudecz, J. Pept. Sci., 2004, 10, 701–713 CrossRef.
- J. Karas, F. Shabanpoor, M. A. Hossain, J. Gardiner, F. Separovic, J. D. Wade and D. B. Scanlon, Int. J. Pept., 2013, 2013, 504260 Search PubMed.
- S. J. Mountford, M. Liu, L. Zhang, M. Groenen, H. Herzog, N. D. Holliday and P. E. Thompson, Org. Biomol. Chem., 2014, 12, 3271–3281 RSC.
- K. Conde-Frieboes, S. Andersen and J. Breinholt, Tetrahedron Lett., 2000, 41, 9153–9156 CrossRef CAS.
- J. K. Murray, K. Biswas, J. R. Holder, A. Zou, J. Ligutti, D. Liu, L. Poppe, K. L. Andrews, F. F. Lin, S. Y. Meng, B. D. Moyer, S. I. McDonough and L. P. Miranda, Bioorg. Med. Chem. Lett., 2015, 25, 4866–4871 CrossRef CAS.
- Y. Kambayashi, S. Nakajima, M. Ueda and K. Inouye, FEBS Lett., 1989, 248, 28–34 CrossRef CAS.
- A. Peschel, F. C. Cardoso, A. A. Walker, T. Durek, M. R. L. Stone, N. Braga Emidio, P. E. Dawson, M. Muttenthaler and G. F. King, J. Med. Chem., 2020, 63, 12773–12785 CrossRef CAS.
- J. C. Breger, M. Muttenthaler, J. B. Delehanty, D. A. Thompson, E. Oh, K. Susumu, J. R. Deschamps, G. P. Anderson, L. D. Field, S. A. Walper, P. E. Dawson and I. L. Medintz, Nanoscale, 2017, 9, 10447–10464 RSC.
- D. E. Nolde, A. G. Sobol, K. A. Pluzhnikov, E. V. Grishin and A. S. Arseniev, J. Biomol. NMR, 1995, 5, 1–13 CrossRef CAS.
- S. D. Robinson, A. Mueller, D. Clayton, H. Starobova, B. R. Hamilton, R. J. Payne, I. Vetter, G. F. King and E. A. B. Undheim, Sci. Adv., 2018, 4, eaau4640 CrossRef.
- V. Barasse, A. Touchard, N. Tene, M. Tindo, M. Kenne, C. Klopp, A. Dejean, E. Bonnafe and M. Treilhou, Toxins, 2019, 11, 732 CrossRef CAS.
- A. V. Osipov, I. E. Kasheverov, Y. V. Makarova, V. G. Starkov, O. V. Vorontsova, R. Ziganshin, T. V. Andreeva, M. V. Serebryakova, A. Benoit, R. C. Hogg, D. Bertrand, V. I. Tsetlin and Y. N. Utkin, J. Biol. Chem., 2008, 283, 14571–14580 CrossRef CAS.
- A. H. Jin, M. Muttenthaler, S. Dutertre, S. W. A. Himaya, Q. Kaas, D. J. Craik, R. J. Lewis and P. F. Alewood, Chem. Rev., 2019, 119, 11510–11549 CrossRef CAS.
- A. D. Santos, J. S. Imperial, T. Chaudhary, R. C. Beavis, B. T. Chait, J. P. Hunsperger, B. M. Olivera, M. E. Adams and D. R. Hillyard, J. Biol. Chem., 1992, 267, 20701–20705 CrossRef CAS.
- S. A. Nixon, Z. Dekan, S. D. Robinson, S. Guo, I. Vetter, A. C. Kotze, P. F. Alewood, G. F. King and V. Herzig, Biomedicines, 2020, 8, 185 CrossRef CAS.
- A. Touchard, H. C. Mendel, I. Boulogne, V. Herzig, N. Braga Emidio, G. F. King, M. Triquigneaux, L. Jaquillard, R. Beroud, M. De Waard, O. Delalande, A. Dejean, M. Muttenthaler and C. Duplais, ACS Pharmacol. Transl. Sci., 2020, 3, 1211–1224 CrossRef CAS.
- N. Chino, K. Yoshizawa-Kumagaye, Y. Noda, T. X. Watanabe, T. Kimura and S. Sakakibara, Biochem. Biophys. Res. Commun., 1986, 141, 665–672 CrossRef CAS.
- K. Kangawa and H. Matsuo, Biochem. Biophys. Res. Commun., 1984, 118, 131–139 CrossRef CAS.
- C. Nagai-Okatani, K. Kangawa and N. Minamino, J. Pept. Sci., 2017, 23, 486–495 CrossRef CAS.
- A. Marglin and S. W. Cushman, Biochem. Biophys. Res. Commun., 1967, 29, 710–716 CrossRef CAS.
- J. C. Meek and R. E. Bolinger, Biochemistry, 1966, 5, 3198–3203 CrossRef CAS.
- P. Volvin, A. M. Chambaut, D. Eboue Bonis, H. Clauser, O. Brinkhoff, H. Bremer, J. Meienhofer and H. Zahn, Nature, 1964, 203, 408–409 CrossRef CAS.
- J. P. Proux, C. A. Miller, J. P. Li, R. L. Carney, A. Girardie, M. Delaage and D. A. Schooley, Biochem. Biophys. Res. Commun., 1987, 149, 180–186 CrossRef CAS.
- G. M. Coast, R. C. Rayne, T. K. Hayes, A. I. Mallet, K. S. Thompson and J. P. Bacon, J. Exp. Biol., 1993, 175, 1–14 CAS.
- J. S. Zheng, S. Tang, Y. Guo, H. N. Chang and L. Liu, Chembiochem, 2012, 13, 542–546 CrossRef CAS.
- J. S. Zheng, S. Tang, Y. K. Qi, Z. P. Wang and L. Liu, Nat. Protoc., 2013, 8, 2483–2495 CrossRef CAS.
- J. L. Stymiest, B. F. Mitchell, S. Wong and C. Vederas John, J. Org. Chem., 2005, 70, 7799–7809 CrossRef CAS.
- M. Manning, L. Balaspiri, M. Acosta and W. H. Sawyer, J. Med. Chem., 1973, 16, 975–978 CrossRef CAS.
- W. H. Sawyer, M. Acosta, L. Balaspiri, J. Judd and M. Manning, Endocrinology, 1974, 94, 1106–1115 CrossRef CAS.
- W. H. Sawyer, M. Acosta and M. Manning, Endocrinology, 1974, 95, 140–149 CrossRef CAS.
- M. Kruszynski, B. Lammek, M. Manning, J. Seto, J. Haldar and W. H. Sawyer, J. Med. Chem., 1980, 23, 364–368 CrossRef CAS.
- S. Derick, L. L. Cheng, M. J. Voirol, S. Stoev, M. Giacomini, N. C. Wo, H. H. Szeto, M. Ben Mimoun, M. Andres, R. C. Gaillard, G. Guillon and M. Manning, Endocrinology, 2002, 143, 4655–4664 CrossRef CAS.
- Y. Shen, J. Maupetit, P. Derreumaux and P. Tufféry, J. Chem. Theory Comput., 2014, 10, 4745–4758 CrossRef CAS.
- J. Copps, R. F. Murphy and S. Lovas, Methods Mol. Biol., 2008, 494, 115–126 CrossRef CAS.
- H. Geng, F. Chen, J. Ye and F. Jiang, Comput. Struct. Biotechnol. J., 2019, 17, 1162–1170 CrossRef CAS.
- B. Mouillac, B. Chini, M.-N. Balestre, J. Elands, S. Trumpp-Kallmeyer, J. Hoflack, M. Hibert, S. Jard and C. Barberis, J. Biol. Chem., 1995, 270, 25771–25777 CrossRef CAS.
- M. Thibonnier, P. Coles, A. Thibonnier and M. Shoham, in Prog. Brain Res., Elsevier, 2002, vol. 139, pp. 179–196 Search PubMed.
- M. J. Ślusarz, E. Sikorska, R. Ślusarz and J. Ciarkowski, J. Med. Chem., 2006, 49, 2463–2469 CrossRef.
- V. S. Ananthanarayanan and K. S. Brimble, Biopolymers, 1996, 40, 433–443 CrossRef CAS.
- A. Micsonai, F. Wien, L. Kernya, Y.-H. Lee, Y. Goto, M. Refregiers and J. Kardos, Proc. Natl. Acad. Sci. U. S. A., 2015, 112, E3095–E3103 CrossRef CAS.
- A. Micsonai, F. Wien, E. Bulyaki, J. Kun, E. Moussong, Y. H. Lee, Y. Goto, M. Refregiers and J. Kardos, Nucleic Acids Res., 2018, 46, W315–W322 CrossRef CAS.
- H. L. Aanning and D. Yamashiro, J. Am. Chem. Soc., 1970, 92, 5214–5216 CrossRef CAS.
- D. Yamashiro, D. B. Hope and V. Du Vigneaud, J. Am. Chem. Soc., 1968, 90, 3857–3860 CrossRef CAS.
- A. V. Schally and R. Guillemin, J. Biol. Chem., 1964, 239, 1038–1041 CrossRef CAS.
- M. C. Munson, M. Lebl, J. Slaninova and G. Barany, Pept. Res., 1993, 6, 155–159 CAS.
- L. Chen, H. Bauerova, J. Slaninova and G. Barany, Pept. Res., 1996, 9, 114–121 CAS.
Footnotes |
| † Electronic supplementary information (ESI) available: Experimental procedures, analytical characterization data (HPLC traces, mass spectra), pharmacological characterization data, structural characterization via MS/MS, structural modelling and receptor docking procedures. See DOI: 10.1039/d0sc05501h |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2021 |