DOI:
10.1039/D1RA07658B
(Paper)
RSC Adv., 2021,
11, 39994-40010
Exploring the charge transfer dynamics of hydrogen bonded crystals of 2-methyl-8-quinolinol and chloranilic acid: synthesis, spectrophotometric, single-crystal, DFT/PCM analysis, antimicrobial, and DNA binding studies†
Received
16th October 2021
, Accepted 19th November 2021
First published on 16th December 2021
Abstract
The new chemistry of the hydrogen-bonded charge and proton transfer complex (HB CT) between electron-donor 2-methyl-8-quinolinol (2 MQ) and electron-acceptor chloranilic acid (CHLA) has been studied using electronic absorption spectroscopy in acetonitrile (ACN), methanol (MeOH), and ethanol (EtOH) polar media at room temperature. The stoichiometric proportion of the HB CT complex was observed to be 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 from the Job data and photometric titration process. The association constant (KCT) and molar absorptivity (εCT) of the HB CT complex were determined by using the modified Benesi–Hildebrand equation in three polarities. Other spectroscopic physical parameters like the energy of interaction (ECT), ionization potential (ID), resonance energy (RN), standard free energy change (ΔG°), oscillator strength (f), and transition dipole moment (μ) were also evaluated. The HB CT complex structure was confirmed by different characterization techniques, such as FT-IR, NMR, TGA-DTA, and SEM-EDX analysis. Powder XRD and single-crystal XRD were used to determine the nature and structure of the synthesized HB CT complex. DNA binding studies for the HB CT complex produced a good binding constant value of 2.25 × 104 L mol−1 in UV-visible and 1.17 × 104 L mol−1 in fluorescence spectroscopy. The biological activity of the HB CT complex was also tested in vitro against the growth of bacteria and fungi, and the results indicated remarkable activity for the HB CT complex compared to the standard drugs, ampicillin and clindamycin. Hence, the abovementioned biological results of the synthesized HB CT complex show it could be used as a pharmaceutical drug in the future. Computational analysis was carried out by DFT studies using the B3LYP function with a basis set of 6-31G(d,p) in the gas phase and PCM analysis. The computational studies further supported the experimental results by confirming the charge and proton transfer complex.
1 from the Job data and photometric titration process. The association constant (KCT) and molar absorptivity (εCT) of the HB CT complex were determined by using the modified Benesi–Hildebrand equation in three polarities. Other spectroscopic physical parameters like the energy of interaction (ECT), ionization potential (ID), resonance energy (RN), standard free energy change (ΔG°), oscillator strength (f), and transition dipole moment (μ) were also evaluated. The HB CT complex structure was confirmed by different characterization techniques, such as FT-IR, NMR, TGA-DTA, and SEM-EDX analysis. Powder XRD and single-crystal XRD were used to determine the nature and structure of the synthesized HB CT complex. DNA binding studies for the HB CT complex produced a good binding constant value of 2.25 × 104 L mol−1 in UV-visible and 1.17 × 104 L mol−1 in fluorescence spectroscopy. The biological activity of the HB CT complex was also tested in vitro against the growth of bacteria and fungi, and the results indicated remarkable activity for the HB CT complex compared to the standard drugs, ampicillin and clindamycin. Hence, the abovementioned biological results of the synthesized HB CT complex show it could be used as a pharmaceutical drug in the future. Computational analysis was carried out by DFT studies using the B3LYP function with a basis set of 6-31G(d,p) in the gas phase and PCM analysis. The computational studies further supported the experimental results by confirming the charge and proton transfer complex.
1. Introduction
The chemistry of charge transfer (CT) complexes has become important in materials and biological science in recent years.1,2 To produce a charge transfer complex, the sharing of electronic charge between two appropriate molecular entities, one of which can donate and the other of which can accept the electrons, has already been investigated. The CT complex mechanism was initially exposed by Mullikan and further developed by Foster.3,4 CT complexes have been broadly investigated and explored for extensive use in several areas, like photocatalysis,2,5 solar energy, organic semiconductors,6 electrical properties,7 DNA binding,8,9 antimicrobial studies,10–13 and drug-receptor binding mechanisms.14 CT complexes show a vital characteristic in the area of electric and optical properties.15–17,68 The proton transfer hydrogen bonding phenomenon was predicted by Pauling and other researchers.18,19 Hydrogen-bonded CT complexes are used in areas such as solar energy batteries,20 surface chemistry, and also in many organic fields and analytical chemistry.
Nitrogen-containing heterocyclic compounds act as electron donors in CT complex formation. The donor 2-methyl-8-quinolinol (2 MQ) is a methyl-substituted quinolinol derivative and it has various applications in the fields of biological and material chemistry.21,22 In general, quinoline derivatives have generated a lot of interest as biomolecules due to their exciting pharmacological properties.23 Quinoline derivatives are used as anti-asthmatic, antibacterial, antifungal, antimalarial, antiviral, and anti-inflammatory agents.24 Researchers have reported that chloranilic acid (CHLA) acts as an electron acceptor in the formation of CT complexes with 2,3-diaminopyridine,25 5,6-dimethyl benzimidazole,26 p-nitroaniline,27 2-amino-4-methylthiazole,19 4-aminoquinoline,18 ethyl carbamate,28 pyrazole,29 and its derivative 3-amino-1,5-dimethyl pyrazole.30
A literature survey has revealed that hetero atoms containing CT complexes have recently received a lot of interest for their pharmacological properties. CT complexes may play a significant role in antimicrobial and DNA binding studies.25,31–34 Based on the preceding discussion, and owing to the importance of 2 MQ from a chemistry point of view and to the wide range of biological activities and pharmacological applications of 2 MQ, this work focuses on exploring antimicrobial and DNA binding studies. The experimental work will take in the use of different spectroscopic techniques such as UV-vis, FT-IR, 1H and 13C NMR spectroscopies; SEM-EDX; single-crystal X-ray diffraction (XRD); powder XRD; and TGA-DTA analysis to characterize the produced HB CT complex in different solvents: ACN, MeOH, and EtOH. The present investigation in this article involves antimicrobial studies, DNA binding studies, and computational analysis. Hence, the DNA binding and biological results of the synthesized HB CT complex show it could be used as a pharmaceutical drug in the future. Theoretical analysis of DFT gas phase/PCM estimations explores the CT complex structure. The experimental results are well supported by the theoretical analysis of the charge and proton transfer hydrogen-bonded complex.
2. Experimental section
2.1. Materials required
The donor 2-methyl-8-quinolinol (2 MQ), acceptor chloranilic acid (CHLA), and CT-DNA were acquired from Sigma-Aldrich USA (98%). Sodium chloride and tris buffer (99.5%) were procured from SD Fine Chemicals Limited. The solvents acetonitrile (Finar 99.5%), methanol (Finar 99.5%), and ethanol (Hayman 100%) were used at a high analytical grade. Safety glasses, hand covers, and dust masks were used during all experiments.
2.2. Preparation of standard stock solutions
To prepare standard stock solutions of 2-methyl-8-quinolinol (0.01 M) and chloranilic acid (0.01 M), 0.015918 g of donor and 0.0208 g of acceptor were added to separate standard volumetric flasks (10 mL) using acetonitrile, methanol, and ethanol. The concentrations of 2 MQ and CHLA (0.0005 M) were separately prepared in a standard 25 mL volumetric flask by diluting the standard stock solutions. The CHLA concentration was kept constant and varying concentrations of 2 MQ were made by diluting the standard stock solution (0.0001 M, 0.00015 M, 0.0002 M, 0.00025 M, 0.0003 M, and 0.00035 M, 0.0004 M, 0.00045 M, 0.0005 M) in each separate 25 mL standard volumetric flask.
2.3. Synthesis of the HB CT complex
The HB CT complex was synthesized by the mixing a 5 mmol saturated methanol solution of 2 MQ (0.795 g) and CHLA (1.04 g) in an isolated round bottom flask. A violet-colored solution was formed upon mixing, which was then stirred continuously for 90 min and allowed to evaporate slowly at 25 °C. The solid was collected, washed several times with methanol solvent to eliminate any unreacted components, and then dried under vacuum over anhydrous CaCl2. The chemical formula of the HB CT complex is C16H11Cl2NO5+ (m wt: 368.17) and the elemental compositions is C, 52.20%, H, 3.01%, Cl, 19.26%, N, 3.80%, and O, 21.73%.
2.4. Single-crystal growth
The solid compound was dissolved in methanol and allowed to slowly evaporate to obtain the crystal. The suspended impurities were separated from the solution with Whatman: 41 filter paper and the collective filtrate was allowed to remain in a dust-free environment for 12 days at room temperature. After 12 days, we obtained good-quality violet rod-shaped crystals and studied them on a Rigaku diffractometer at room temperature; the CCDC no of which is 2070105.
2.5. Instrumentational analysis
2.5.1. UV-visible absorption spectra. The UV-visible absorption spectra were recorded in the region of 200–800 nm using a Shimadzu UV-2600 spectrophotometer containing a quartz cell of path length 1.0 cm.
2.5.2. Infrared spectra. The FT-IR spectra of the reacting molecules and HB CT complex were recorded in the region from 550 to 4000 cm−1 in a PerkinElmer infrared spectrometer.
2.5.3. 1H NMR spectra. The 1H NMR spectra of the reacting molecules and corresponding HB CT complex were recorded on a Bruker 400 MHz NMR instrument using DMSO-d6 as the solvent and TMS as the reference.
2.5.4. SEM-EDX elemental composition. The elemental composition and surface morphology of the SEM-EDX were estimated using a scanning electron microscope (SEM, Quanta FEG 250). An acceleration voltage of 20 kV was used to power the device.
2.5.5. Thermal analysis. The thermal stability was verified in a temperature range of 20–800 °C under a N2 flow (50 mL min−1) at a static heat rate of 10 °C min−1 using a DTG-60H Thermal Analyzer (Shimadzu).
2.5.6. Powder XRD analysis. Powder X-ray diffraction analysis was performed with a Rigaku MiniFlex 600 X-ray diffractometer with Cu-Kα radiation (λ = 1.5406 Å) at a step size of 0.02° and a scan step time of 0.15 s in the 2θ range 10–80°.
2.5.7. Single-crystal XRD. Single-crystal XRD for an HB CT crystal was carried out on a Rigaku Oxford Diffraction CCD single-crystal diffractometer furnished with a graphite monochromator and Mo Kα radiation (0.71073 Å) at 293 K. Unit cell measurement, data collection, integration, scaling, and absorption corrections were executed using Rigaku Oxford Diffraction. Multiscan absorption corrections were carried out using SADABS-2014. The crystal structure was solved by direct methods with SHELXS-97 and refined using the full matrix least squares process using the SHELXL-2014 stream in the OLEX2 Software. Aromatic and H(N) hydrogen positions were derived from geometrical considerations, methyl and H(O) ones from difference maps. All H-atoms were then refined at idealized positions riding on the parent atoms with isotropic displacement parameters Uiso(H) = 1.2U ∼ eq ∼ (C/N) or 1.5U ∼ eq ∼ (–CH ∼ 3∼ and –OH hydrogens). All methyl and OH hydrogen atoms were allowed to rotate but not to tip (AFIX 43, 137, 147). The packing figure of the crystal was produced using Mercury 3.8 software.
2.6. Antimicrobial studies
2.6.1. Antibacterial activity. The antibacterial activity of the HB CT complex was screened for selected bacterial strains: for two Gram-positive bacteria Staphylococcus aureus (MTCC 96) and Bacillus subtilis (MTCC 441), and two Gram-negative bacteria Escherichia coli (MTCC 43) and Klebsiella pneumoniae (MTCC 530), by the cup-plate agar diffusion method. Experiments were performed on 5 mm sample discs made from Whatman filter paper that was sterilized at 140 °C for 1 h. The above-mentioned cultures were incubated in autoclaved nutrient broth media inoculation, for 24 h at 37 °C in a vibrator for bacterial growth. 0.3 mL of bacterial culture was taken and protected using a diffuser on newly made autoclaved agar plates. DMSO solvent was used as a control and the concentration of the HB CT complex was kept at 1 mg mL−1. A reference drug, ampicillin, was used to compare the activity of the CT complex. The observed zone of inhibition after 24 h was measured using a metal scale. The experiments for each sample were measured in triplicate for accuracy.
2.6.2. Antifungal activity. The antifungal activity of the HB CT complex was examined using the dilution technique. The activity was tested against Fusarium oxysporum, Sclerotium rolfsii, and Phytophthora strains. An agar plug of vigorously growing fungal culture was positioned in the center of the plate. At that time, compound discs were placed at a distance of 2 cm from the center where the fungus was placed, and changed aliquots were incubated at 30 ± 2 °C for 36 to 96 h. Clindamycin and DMSO were used as a reference drug and a control, respectively. The experiments for each sample were measured in triplicate for accuracy.
2.7. Preparation of DNA stock solution
A calf thymus-DNA (CT-DNA) stock solution was made using 5 mM Tris–HCl and 50 mM NaCl at 25 °C in double-distilled water. The solution mixture was stirred overnight to form a clear solution. The pH was adjusted to 7.2 with HCl added to the homogenous clear buffer solution. The purity of the CT-DNA solution was calculated from the absorbance ratio at A260/A280 nm, giving a value of 1.8–1.9. This confirms that no impurity protein was present in the CT-DNA. UV-visible electronic spectroscopy was used to calculate the concentration of CT-DNA at absorbance wavelength 260 nm by a molar absorptivity of 6600 M−1 cm−1. The prepared solutions were kept at a lower temperature (3–4 °C) and used for 3–4 days. The HB CT complex was dissolved using a 50% acetonitrile and 50% Tris–HCl buffer solution throughout the examination. Binding constant titrations were completed through a static concentration of the HB CT complex by changing the concentrations of CT-DNA (0–10 μM).
2.8. Fluorescence study
An ethidium bromide (EB) displacement assay was carried out with quartz cuvettes with a 1 cm path length on a HORIBA FluoroMax spectrofluorometer. Fluorescence spectroscopy was used to investigate the binding ability of the HB CT complex using an EB assay. The HB CT complex fluorescence emission spectra were measured in the region of 500–750 nm. At room temperature, increasing concentrations of HB CT complex (0–80 μM) were titrated using a buffer solution containing CT-DNA (20 μM) and EB (5 μM).
2.9. Theoretical analysis
The theoretical investigation was carried out by DFT studies using the B3LYP function with basis set 6-31G(d,p) in the gas phase, and IEF-PCM (integrated integral equation formalism for the polarizable continuum model). The 2 MQ, CHLA, and HB CT complex were all estimated with the Gaussian 09 program. The optimized geometrical structural parameters, such as bond lengths, bond angles, Mulliken atomic charges, MEP maps, FMO surfaces, and reactivity parameters of the HB CT complex were estimated.
3. Results and discussion
3.1. Electronic absorption spectra
3.1.1. Observation of charge transfer band. The UV-visible electronic absorption spectra of 2 MQ, CHLA, and HB CT complex were recorded in acetonitrile (ACN), methanol (MeOH), and ethanol (EtOH) solvents. CT bands were observed at 516.5, 523.5, and 523 nm (Fig. 1), which are not shown in the free 2 MQ and CHLA absorption spectra. The new CT bands facilitate the formation of the HB CT complex between 2 MQ and CHLA with its representative absorption band shifted to longer wavelengths compared with the individual molecules. Among the solvents, acetonitrile is aprotic while methanol and ethanol are protic, and these protic solvents support the intermolecular hydrogen bonding between the OH group of methanol and ethanol with 2 MQ, to attain the stability of the HB CT complex. The mixing of the donor (0.0005 M) and acceptor (0.0005 M) produces an instant violet color35,36 (Scheme 1), which seems to produce the proton transfer hydrogen bonded CT complex in all three solvents, and the probable mechanism is shown in Scheme 2. Moreover, the absorbance of the HB CT complex increases with increasing donor concentration without changing the λmax of the complex, suggesting the formation of a stable HB CT complex.18
 |
| | Fig. 1 Electronic absorption spectra of 2 MQ (A), CHLA (B), and CTC (C) in acetonitrile, methanol, and ethanol, respectively, at room temperature. | |
 |
| | Scheme 1 Naked eye color of donor 2 MQ, acceptor CHLA, and HB CT complex in methanol solvent. | |
 |
| | Scheme 2 Probable mechanism of the HB CT complex. | |
3.1.2. Stoichiometric proportion. The stoichiometric proportion of the HB CT complex was estimated from Job process data. In this method, a spectrum was plotted between absorbance versus mole fraction in all solvents and is depicted in Fig. S1.† The maximum absorption is shown at the 0.5 mole fraction in Fig. S1.† This result indicates that the HB CT complex has a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 molar ratio. Photometric titration calculations were used to estimate the ratio of the formed HBCT complex, where two lines were formed which intersected, which gives a 1
1 molar ratio. Photometric titration calculations were used to estimate the ratio of the formed HBCT complex, where two lines were formed which intersected, which gives a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 molar ratio in all solvents.37 A graph was plotted of absorbance against the D/A ratio and is depicted in Fig. S2.† Subsequently, one can confirm that the solvent polarity does not affect the HBCT complex molecular composition.
1 molar ratio in all solvents.37 A graph was plotted of absorbance against the D/A ratio and is depicted in Fig. S2.† Subsequently, one can confirm that the solvent polarity does not affect the HBCT complex molecular composition.
3.1.3. Effect of time for the HB CT complex. The HB CT complex spectra depend on time and, in this complex, the donor concentration (5 × 10−4 M) and acceptor concentration (5 × 10−4 M) are both mixed at a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio at room temperature.35,36 The absorption band of the CT complex increases slowly with time, and it reveals that the formation of HB CT is strong in all solvents. The HB CT band λmax for all three solutions is not changed, but the HB CT band absorbance slowly increases with time and is presented in Fig. S3.†
1 ratio at room temperature.35,36 The absorption band of the CT complex increases slowly with time, and it reveals that the formation of HB CT is strong in all solvents. The HB CT band λmax for all three solutions is not changed, but the HB CT band absorbance slowly increases with time and is presented in Fig. S3.†
3.2. Estimation of the spectroscopic physical parameters
3.2.1. Energy. The energy (ECT) of the HB CT complex39 is calculated from the following equation and these results are presented in Table 2 in the three polar media.| | |
ECT = 1243.667/λCT nm
| (2) |
in this calculation, λCT is the wavelength of the HB CT complex. These energy results show that the maximum value of ECT is observed in acetonitrile, where there is an accumulation of acetonitrile molecules with the HB CT complex, leading to steric hindrance, which causes charge transmission from 2 MQ to CHLA due to the lower value of the association constant. Lower energy was noticed in methanol for the HB CT complex due to increased stability.
Table 2 Spectroscopic physical parameters of HB CT complex in three different polarities
| Solvent |
ECT (eV) |
ΔG° × 103 (kJ mol−1) |
ID (eV) |
W (eV) |
f |
μ (Debye) |
RN (eV) |
| Acetonitrile |
2.40 |
−14.16 |
8.70 |
5.2 |
4.39 |
21.95 |
0.197 |
| Methanol |
2.375 |
−25.00 |
8.663 |
5.188 |
0.662 |
2.67 |
0.041 |
| Ethanol |
2.377 |
−24.66 |
8.666 |
5.189 |
0.458 |
7.13 |
0.027 |
3.2.2. Standard Gibbs free energy (ΔG°). The standard Gibbs free energy change of the HB CT complex was evaluated with the following equation40 in different polar solvents.| |
ΔG° = −RT![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ln ln![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) KCT KCT
| (3) |
in this equation, ΔG° is the free energy change of the HB CT complex, R is the universal gas constant (8.314 J mol−1 K), T is the absolute temperature (Kelvin) at 25 °C, and KCT is the association constant of the HB CT complex (L mol−1). The values for the HB CT complex in the three polar solvents are depicted in Table 2. The negative results reveal that formation of the HB CT complex is spontaneous and exothermic. The reaction between the 2 MQ and CHLA molecules shows a strong interaction between them, which can be taken from the higher negative value of free energy. The maximum standard Gibbs free energy was noted in methanol and it shows the high stability of the HB CT complex.
3.2.3. Estimation of ionization energy. The ionization energy (IE) of the HB CT complex was calculated with the following equation in the three polar solvents, which is associated with Aloisi and Pignataro.41| | |
ID = 5.76 + 1.52 × 10−4 υCT
| (4) |
in this equation, υCT and ID are the wavenumber in cm−1 and the ionization energy of the donor molecule. The electron-contributing power of 2 MQ is estimated from its ionization energy, which is the energy needed to emit an electron from the HOMO of the 2 MQ molecule in different polarities. These values are presented in Table 2.
3.2.4. Dissociation energy (W). The dissociation energy36 (W) of the HB CT complex was evaluated by using the energy (ECT), the IE of the donor (ID), and electron affinity (EA) of the acceptor from the following equation in different polarities. These values are depicted in Table 2.
3.2.5. Estimation of oscillator strength (f) and transition dipole moment (μEN). The HB CT complex has an electronic absorption spectrum in which the oscillator strength (f) is a dimensionless quantity that is used for stating the transition probability of the CT bands in the three polar solvents and can be determined from the following equation.18,23| | |
f = 4.32 × 10−9[εmaxΔυ1/2]
| (6) |
in this equation, Δυ1/2 denotes the half bandwidth and εmax is the molar extinction coefficient. The oscillator strength (f) results show a strong interaction between 2 MQ and CHLA moieties in different polarities. The calculated results for the oscillator strength (f) of the HB CT complex are depicted in Table 2.The transition dipole moment of the HB CT complex in different polarities was evaluated, which related to the molar absorptivity which has been estimated from the following equation.22,36
| |
 | (7) |
in this expression,
υmax is the wavenumber corresponding to the highest absorption of the HB CT complex band and the results are depicted in
Table 2. The transition dipole moment is used for the evaluation of the acceptable changes between the HOMO of the 2 MQ and the LUMO of the CHLA.
3.2.6. Calculation of resonance energy (RN). The resonance energy (RN) of the HB CT complex was evaluated by following the principle of Briegleb and Czekalla in the three polar solvents.35,42| | |
εCT = 7.7 × 104/[hυCT/|RN| − 3.5]
| (8) |
in this equation, υCT and εCT are the frequency and molar absorptivity of the CT band, respectively. The resonance energy values are presented in Table 2. These results show the higher stability of the resultant HB CT complex in the three polar solvents.
3.3. Spectral data
3.3.1. IR spectra. The IR spectra of 2 MQ, and CHLA, along with the HB CT complex, were performed as KBr discs from 4000 to 550 cm−1. This examination is seemingly recognized to cause the changes in electronic density as an outcome of the interaction of the proton with CT within the reactants to produce the HB CT complex.43,44 Changes in the frequencies and band intensities were noted in the HB CT complex spectrum related to 2 MQ and CHLA. This results in the making of new charge and proton transfers upon complexation. These spectra are depicted in Fig. 3 and the band assignments are given in Table S1.† In the HB CT spectral data, an important observation is the nonappearance of the OH group of CHLA at 3229 cm−1, signifying its proton transfer to the basic center of the donor 2 MQ, to produce proton transfer H-bonding between the OH of CHLA and the pyridine ring nitrogen of 2 MQ. The existence of H-bonding in the produced complex is shown by the presence of a broad absorption band at 2970 cm−1 for O–H⋯N. From Fig. 3, it is concluded that the OH group of 2 MQ does not participate in a proton transfer interaction with CHLA, and the shift in the NH group stretching frequencies confirms the charge transfer from 2 MQ to CHLA. The asymmetric stretching vibration of CH3 in 2 MQ is detected at 2865 cm−1 in the HB CT complex. It merges with the broad peak, and the bending and rocking C–H vibrations of the methyl group exhibit weak signals at 1384 and 1044 cm−1. In the CHLA moiety, the appearance of band frequencies was noted before and after complex formation, ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) are 1663 cm−1 and 1635 cm−1. The ν(C–Cl) band frequencies are 836, and 745 cm−1 in the HB CT complex compared to 845, and 752 cm−1 for CHLA itself, and ν(C
O) are 1663 cm−1 and 1635 cm−1. The ν(C–Cl) band frequencies are 836, and 745 cm−1 in the HB CT complex compared to 845, and 752 cm−1 for CHLA itself, and ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C) has a band frequency of 1628 cm−1 compared to 1599 cm−1 for free CHLA. Hence, the occurrence of proton transfer besides charge transfer was confirmed from the IR spectrum of the HB CT complex.
C) has a band frequency of 1628 cm−1 compared to 1599 cm−1 for free CHLA. Hence, the occurrence of proton transfer besides charge transfer was confirmed from the IR spectrum of the HB CT complex.
 |
| | Fig. 3 FT-IR spectra of 2MQ, CHLA, and HBCT complex. | |
3.3.2. 1H NMR spectra. The 1H NMR spectra of 2 MQ and HB CT complex were recorded in DMSO-d6 solvent and are shown in Fig. 4.16,26 The donor 2 MQ, acceptor CHLA, and HB CT complex add additional validation to the discovery of the manner of bonding between 2 MQ and CHLA. The 1H NMR spectrum of 2 MQ shows two doublet signals at 8.20 and 7.07 ppm between one triplet signal at 7.43 ppm which are assigned to the three aromatic protons of 2 MQ (phenyl ring). Two other doublet signals appear at 8.18 and 7.41 ppm due to another two aromatic protons of 2 MQ (pyridine ring). The OH group proton phenyl ring of 2 MQ gives rise to a signal at 9.39 ppm and the pyridine ring methyl protons causes a signal at 2.50 ppm which is a singlet. The main objective of the 1H NMR spectrum is to show the occurrence of electron charges and proton transport interactions for the HB CT complex. A new peak appears for the +N–H proton at 3.35 ppm in the HB CT complex and that for the O–H proton of free CHLA disappears. In the HB CT complex, proton peaks of the 2 MQ (phenyl ring) moiety are detected at 7.27, 6.94–6.81, and 6.44 ppm, and there are peaks for the two protons of the (pyridine ring) at 7.57 and 7.41–7.35 ppm. The CH3 protons of 2 MQ give rise to a single peak at 3.17 ppm and the OH peak of the 2 MQ moiety is detected at 11.19 ppm. Hence, the 1H NMR spectra predict the occurrence of charge transfer by the transport of the proton, which confirms its structure.
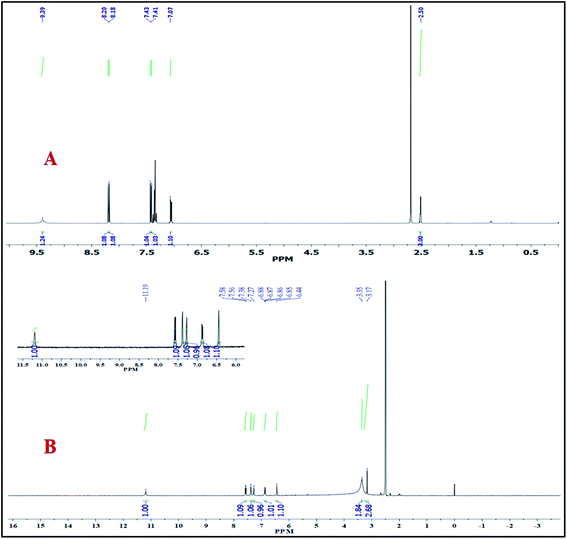 |
| | Fig. 4 1H NMR spectra of (A) 2 MQ and (B) HBCT complex in DMSO-d6. | |
3.3.3. 13C NMR spectra. The HB CT complex was further analyzed using 13C NMR spectra.25,26 The donor 2 MQ, acceptor CHLA and formed HB CT complex are recorded in DMSO-d6 solvent at 25 °C and the obtained spectra are depicted in Fig. S4.† From Fig. S4,† observation of the CHLA spectrum alone showed two signals at 165.75 and 109.87 ppm, which are attributed to a carbonyl carbon and a carbon connected to the OH and Cl groups of CHLA. CHLA participates in HB CT complex formation, and the carbonyl carbon signal recorded a small downfield shift of 167.71 ppm, whereas those attached to the OH and Cl groups showed an upfield shift of 105.49 ppm, confirming the charge transfer from 2 MQ to CHLA. This outcome could be described based on the contribution of the OH groups and the carbonyl of CHLA being involved in hydrogen bond formation. For the 2 MQ carbons, the upfield shift of a few carbon signals involved in complex formation was detected compared with 2 MQ itself. HB CT complex C(6) is noted at 149.73 ppm compared with 152.97 ppm for 2 MQ itself. Similarly, C(2) was observed at 131.93 ppm in the HB CT complex compared with 138.27 ppm for 2 MQ itself. This condition is powerfully associated with the proton transfer contact between 2 MQ and CHLA when the proton transfer proceeds and NH+ is produced. Furthermore, electron transfer from the OH and methyl groups produces shielding of the adjacent carbon atoms. Therefore, from the 13C NMR spectra, along with the 1H NMR and vibrational spectrum, the presence of both charge and proton transfer in the HB CT complex is fully evidenced.
3.4. SEM-EDX analysis
The surface morphology45 and chemical composition of the HB CT complex were studied through SEM-EDX analysis. The corresponding results are presented in Fig. 5. The surface morphology of the HB CT complex shows a sheet-like microstructure. The chemical composition of the HB CT complex produced with 2 MQ and CHLA was confirmed from energy dispersive X-ray (EDX) spectra, which show C, N, O, and Cl elemental peaks, and these data are shown in Table S2.† This composition exactly matches the experimental analysis of the HB CT complex.
 |
| | Fig. 5 SEM images and EDX spectrum for HB CT complex. | |
3.5. Thermal analysis
The thermal stability of the HB CT complex was determined by TGA/DTA analysis.46 The TGA/DTA examination was executed under N2 gas flowing at 50 mL min−1 in the temperature range of 20–800 °C at a heating system rate of 10 °C min−1. In this investigation, 8.5 mg of the HB CT complex was used and the TGA/DTA curvatures of the HB CT complex are presented in Fig. S5.† From Fig. S5,† the thermal behavior of the HB CT complex was shown to be stable up to 148.45 °C and the HB CT complex shows a major two-step decomposition. According to Fig. S5,† two weight loss portions of 34.525% and 7.005% (ΔH = 2101.67 mJ g−1, ΔH = 3.34 × 102 mJ g−1) were observed in the two steps at 148.45 °C and 229.02 °C, respectively. The other HB CT complex decomposed with a weight loss of 58.47%. The TGA-DTA outcomes show the excellent thermal stability of the synthesized HB CT complex.
3.6. Powder X-ray diffraction studies
The powder X-ray studies of 2 MQ, CHLA, and HB CT complex are presented in Fig. 6. Some sharp and strong peaks were detected in all three samples.44,46 High-intensity peaks were detected at diffraction angles 2θ of 22.61°, 24.03° and 15.24° for 2 MQ, CHLA, and HB CT complex, respectively. It can be seen that the HB CT complex sample shows the highest intensity diffraction peaks, which confirms its highly crystalline nature. Moreover, the diffraction peak, which is detected in 2 MQ and CHLA, is not detected in the HB CT complex. This is in good agreement with the HB CT complex having a crystalline nature. The proton transfer between the OH group of CHLA and the N atom of 2 MQ also plays a key part in the crystalline nature of the produced HB CT complex. The crystalline size of the HB CT complex was evaluated from the Debye–Scherrer equation using the high-intensity peak.
D = 0.94λ/β![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) cos cos![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) θ θ |
in this equation, λ is the wavelength of the X-rays used (0.15406 nm), D is the crystalline size in nm, 0.94 is the Scherrer constant, and θ and β are the position and the full width at half maximum (FWHM), respectively, of the particular diffraction peak. The calculated crystalline sizes for 2 MQ, CHLA, and HB CT complex are 23.99, 24.34, and 45.14 nm, respectively. The XRD spectral data is presented in Table 3.
 |
| | Fig. 6 Powder XRD patterns show the crystalline nature of 2 MQ, CHLA, and HB CT complex. | |
Table 3 Powdered XRD spectral data for the 2 MQ, CHLA, and HB CT complex
| Compound |
Bragg angle, 2θ |
FWHM (β) |
Crystalline size (nm) |
| 2 MQ |
22.61° |
0.15274 |
23.99 |
| CHLA |
24.03° |
0.14898 |
24.34 |
| HB CT |
15.24° |
0.08497 |
45.14 |
3.7. Single-crystal XRD
Single-crystal XRD analysis was carried out to acquire structural evidence for the HB CT complex.47–50 The crystallography study reveals it has one unit acceptor moiety and one donor moiety unit, demonstrating that complexation occurs in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio. The empirical formula of the HB CT complex crystal structure is C16H11Cl2NO5. Single-crystal X-ray determined the formation of proton transfer hydrogen bonding between the OH of CHLA and the 2 MQ pyridine ring nitrogen. The intermolecular hydrogen bond N1–H⋯O3 with H⋯O of 1.93 Å and the intramolecular hydrogen bond O5–H⋯O4 with H⋯O of 2.13 Å formed, and the molecular structure of the HB CT complex with anisotropic displacement ellipsoids is presented in Fig. 7. The crystal system was found to be orthorhombic with space group Pna21 and Z = 4. The lattice parameters are a = 22.9722(6) Å, b = 13.0888(3) Å, and c = 5.0930(1) Å and other crystallographic data and refinement parameters for the HB CT complex are summarized in Table 4.51–53 The bonding network of crystal packing is presented in Fig. 8. The HB CT complex hydrogen bonding bond lengths are fully correlated to the DFT analysis.
1 ratio. The empirical formula of the HB CT complex crystal structure is C16H11Cl2NO5. Single-crystal X-ray determined the formation of proton transfer hydrogen bonding between the OH of CHLA and the 2 MQ pyridine ring nitrogen. The intermolecular hydrogen bond N1–H⋯O3 with H⋯O of 1.93 Å and the intramolecular hydrogen bond O5–H⋯O4 with H⋯O of 2.13 Å formed, and the molecular structure of the HB CT complex with anisotropic displacement ellipsoids is presented in Fig. 7. The crystal system was found to be orthorhombic with space group Pna21 and Z = 4. The lattice parameters are a = 22.9722(6) Å, b = 13.0888(3) Å, and c = 5.0930(1) Å and other crystallographic data and refinement parameters for the HB CT complex are summarized in Table 4.51–53 The bonding network of crystal packing is presented in Fig. 8. The HB CT complex hydrogen bonding bond lengths are fully correlated to the DFT analysis.
 |
| | Fig. 7 (A) ORTEP view of the HB CT complex. (B) Hydrogen bonding interaction between donor and acceptor in the HB CT complex with anisotropic displacement ellipsoids drawn at the 50% probability level. | |
Table 4 Crystal data and structural refinement for the synthesized crystal of HB CT complex
| System |
HB CT crystal |
| Sum formula |
C16H11Cl2NO5 |
| Formula weight |
368.16 |
| CCDC no. |
2070105 |
| Crystal system |
Orthorhombic |
| Temperature |
293(2) K |
| Wavelength |
0.71073 |
| Space group |
Pna21 |
| a [Å] |
22.9722(6) |
| b [Å] |
13.0888(3) |
| c [Å] |
5.0930(1) |
| V [Å3] |
1531.36(6) |
| Z |
4 |
| Dcalcd [g cm−3] |
1.597 |
| Abs. coeff. change [mm−1] |
0.452 |
| F(000) |
752 |
| h, k, l max |
−27/28, ±15, ±6 |
| Theta (max) |
25.679 |
| Rflns collect |
2909 |
| Unique rflns |
2716 |
| Obsd rflns |
2370 |
| Parameters |
220 |
| R1 |
0.0408 |
| wR2 |
0.1128 |
| Parameters refined |
220 |
| Data completeness |
1.67/0.93 |
| GoF (F2) |
1.007 |
| Min delta F |
2.36 |
| Max delta F |
24.99 |
| Rint |
0.0889 |
 |
| | Fig. 8 (A and B) Crystal packing of the HB CT complex. (Red, grey, blue, green, and white colors correspond to oxygen, carbon, nitrogen, chlorine, and hydrogen atoms, respectively.) | |
3.8. Biological studies
Charge transfer complexes are known to have promising biological activities that include anti-bacterial and anti-fungal effects. In the present investigation, the synthesized CT complex was tested for its anti-bacterial and anti-fungal activities against selected microbial strains. The HB CT complex was solubilized in DMSO to get a 1 mg mL−1 stock solution. The zone of inhibition was calculated, which displays the growth inhibition of the screened microorganism. The HB CT complex showed remarkable antimicrobial activity against bacterial and fungal strains.
3.8.1. Antibacterial activity. Antibacterial activity was examined against four infective microorganisms: two Gram-negative bacteria, Escherichia coli and Klebsiella pneumoniae, and two Gram-positive bacteria, Staphylococcus aureus and Bacillus subtilis.54–57 The newly synthesized HB CT complex showed crucial suppressing activity against the growth of bacterial strains, as presented in Fig. S6.† The standard drug ampicillin was used for the evaluation of the bacterial results and the zone of inhibition (cm) is shown in Table 5. These results were observed at a concentration of 1 mg mL−1 in the case of Gram-negative and Gram-positive bacteria. The maximum zone of inhibition was detected for Klebsiella pneumoniae and then for Bacillus subtilis. The other bacterial strains, Escherichia coli and Staphylococcus aureus, showed a relatively lower zone of inhibition. The presence of polar hydroxyl groups and a nitrogen atom facilitates the passage of small molecules into the microbial cells and affects their growth inhibition. The hydroxyl groups improve the permeability of the CT complex by allowing it to dissolve in the aqueous phase of the cell membrane of microorganisms.
Table 5 Antibacterial activity of HB CT complex
| Bacterial strains |
Ampicillin |
Control (DMSO) |
HB CT complex |
| Bacillus subtilis |
0.8 ± 0.02 cm |
0 cm |
1.2 ± 0.02 cm |
| Staphylococcus aureus |
1.1 ± 0.03 cm |
0 cm |
0.8 ± 0.01 cm |
| Escherichia coli |
1.4 ± 0.02 cm |
0 cm |
0.9 ± 0.03 cm |
| Klebsiella pneumoniae |
1.1 ± 0.01 cm |
0 cm |
1.6 ± 0.05 cm |
3.8.2. Antifungal activity. The antifungal activity was tested against three different fungal strains, and clindamycin was used as a standard drug.31 A concentration of 1 mg mL−1 of CT complex was treated against Fusarium oxysporum, Sclerotium rolfsii, and Phytophthora strains, and the result of the HB CT complex on the growth of fungal strains is depicted in Fig. S7.† The activity was estimated by measuring the zone of inhibition (in cm) and the data is presented in Table 6. The fungal strains were found to be more sensitive towards treatment by the synthesized complex. From the results, it is evident that the complex exhibited remarkable antifungal properties as compared to the standard drug, clindamycin.
Table 6 Antifungal activity of HB CT complex
| Fungal strains |
Control (DMSO) |
Clindamycin |
HB CT complex |
| Measurement |
% |
Measurement |
% |
Measurement |
% |
| Sclerotium rolfsii |
0 cm |
0 |
1.4 ± 0.08 cm |
72 |
2.3 ± 0.02 cm |
54 |
| Fusarium oxysporum |
0 cm |
0 |
2.6 ± 0.10 cm |
48 |
2 ± 0.06 cm |
60 |
| Phytophthora strains |
0 cm |
0 |
1.9 ± 0.09 cm |
62 |
2 ± 0.02 cm |
60 |
3.9. DNA binding studies
The interaction between a drug and DNA is the most vital characteristic in biotic research used to identify antiproliferative agents. In the cellular system, deoxyribonucleic acid (DNA) is one of the most important hereditary materials.58 Up to now, exploration for new DNA targeted agents has developed into a significant range of studies. Due to the variety in DNA systems, the examination of explicit DNA binding for a system is an intriguing subject for investigation. In general, the binding mode of DNA can occur in a covalent manner or a noncovalent manner. Of these two binding manners, the noncovalent manner is safer due to low toxicity. The noncovalent binding molecules may bind by groove binding, intercalation, and electrostatic modes.
3.9.1. DNA binding analysis from UV-visible spectroscopy. In investigations of CT-DNA binding, analysis of CT complexes through UV absorption spectroscopy is a prominent method. After adding the DNA solution to the complex, the binding interaction of the HB CT complex through DNA helices is defined by changes in absorbance and wavelength.59,60 The binding affinity was evaluated in this current study by observing the absorbance change of the CT complex when the CT-DNA concentration increased as well as when the CT complex concentration was kept constant. The absorption spectra of the CT-DNA binding with HB CT complex are presented in Fig. 9. The spectra display a redshift and are hypochromic, expressing the intercalation binding mode between CT-DNA and the HB CT complex. A change in the absorbance spectrum of the HB CT complex was seen by increasing the concentration of CT-DNA, when the wavelength shifted to a higher wavelength (redshift) as a result of strong associations between contiguous base pairs of DNA and the aromatic chromosphere of the HB CT complex via intercalation. The HB CT complex had a strong interaction with the DNA helix via an intercalation mode, which shows the extent of the hypochromic behavior generally associated with the intercalation binding mode. The intrinsic binding constant (Kb) was calculated using the Wolfe–Shimmer equation based on the absorption data.| | |
[DNA]/(εa − εf) = [DNA]/(εb − εf) + 1/Kb(εb − εf)
| (9) |
in this equation, [DNA] is the concentration of CT-DNA, εa is the apparent coefficient, εf and εb correspond to the extinction coefficient of free HB CT and HB CT fully bound to DNA molecules, and Kb is the intrinsic binding constant. A graph was plotted between [DNA]/(εa − εf) and [DNA], and the binding constant Kb was calculated by determining the gradient to intercept ratio. The Kb value for the HB CT complex is 2.25 × 104 L mol−1, which shows a strong binding affinity.61–63
 |
| | Fig. 9 UV-visible absorption spectra of HB CT complex with increasing concentration of CT-DNA from 0 to 10 μM and static concentration of HB CT complex. The arrow (↓) displays changes in absorption intensity upon increasing DNA concentration. Inset: linear plot for the calculation of the intrinsic binding constant, Kb. | |
3.9.2. DNA binding analysis from fluorescence spectroscopy. Competitive displacement of ethidium bromide by the HB CT complex was examined by a fluorescence spectral technique and this enables us to understand the binding modes of the HB CT complex with target CT-DNA.59,63,64 The interaction or binding mode of the HB CT complex with CT-DNA was studied using an EB-bound CT-DNA solution in 5 mM Tris–HCl/NaCl buffer (pH = 7.2). Due to tenable intercalation between adjacent DNA base pairs, EB emits intense fluorescence at about 580–620 nm in the presence of CT-DNA. The intensity of absorption in a solution containing EB and CT-DNA gradually decreased as the concentration of HB CT complex increased and the spectra are presented in Fig. 10. The absorption changes clearly show that the HB CT complex competes to replace EB from CT-DNA, indicating the complex is intercalatively binding to CT-DNA. Displacement of CT-DNA-bound EB occurs when changes to the secondary structure of CT-DNA take place. Fig. 10 depicts the replacement of intercalatively bound EB by the HB CT complex. The data were evaluated using the Stern–Volmer equation by observing a decrease in fluorescence intensities.where Io and I are the fluorescence intensities in the absence and presence of quencher, KSV is the Stern–Volmer constant, and [Q] is the molar concentration of quencher. The extent to which the emission intensity is quenched is a measure of the affinity of the interacting molecule for binding to CT-DNA. The Stern–Volmer quenching constant value KSV was calculated from the slope of Io/I vs. Q[HB CT complex] and found to be 1.17 × 104 M−1. Our findings reveal that the HB CT complex seems to have an excellent binding affinity to CT-DNA, which is one of the most important molecular targets in the development of bioactive materials.
 |
| | Fig. 10 Fluorescence quenching curve of EB bound to CT-DNA by the HB CT complex. The arrow shows the intensity changes on increasing the concentration of the complex. Inset: plot of I0/I vs. [HB CT complex]. | |
3.10. Computational analysis
3.10.1. Estimation of bond lengths and bond angles. The optimized structures of the 2 MQ, CHLA, and HB CT complex in the gas phase are shown in Fig. 11. PCM analysis16,36 of the HB CT complex was undertaken in acetonitrile, methanol, and ethanol (IEF PCM), and is shown in Fig. S8.† The calculations of bond lengths and bond angles in the gas phase and PCM analysis are given in Table S3.† The HB CT complex has two kinds of hydrogen bonds: one is intermolecular and the other is an intramolecular hydrogen bond. These bonds are supportive and helpful in the progression of charge transfer contact between 2 MQ and CHLA. In an intermolecular hydrogen bond, a proton has transferred from O11–H13 to N17. The bond length has a value of 1.4673 Å in the gas phase and PCM analysis has shown that the values are 1.7236, 1.7233, and 1.726 Å when dissolved in ACN, MeOH, and EtOH compared to free CHLA, which has bond lengths of 0.9899 Å in the gas phase and 0.98996, 0.9896, and 0.9896 Å in PCM analysis, confirming the transfer of a proton to the N of the 2 MQ ring. In PCM analysis all three solvents gave nearly similar values; hence no solvent effect was present in the bond lengths of the HB CT complex. From Table S3,† the bond length of C4–O11 decreases by 1.2723 Å in the gas phase, and 1.2741, 1.2741, and 1.2742 Å in ACN, MEOH, and ETOH, respectively, in PCM analysis. Free CHLA in the gas phase has bond length reductions of 1.3526 Å and 1.349, 1.349 and 1.3491 Å in PCM analysis. These results confirm the formation of proton transfer hydrogen bonding at O11⋯H13 + N17. It can also be noted that the change in C5–Cl9 and C2–Cl10 bond lengths after HB CT complex formation in the gas phase and the difference in PCM analysis compared to free CHLA are due to the growing electron density on the Cls of the acceptor which supports the contribution of 2 MQ in HB CT development along with CHLA. An intramolecular hydrogen bond of O12–H14 after HB CT complex formation increases its bond length in gas and PCM analysis compared to free CHLA in the gas phase and PCM analysis. The above bond length results strongly support the HB CT and proton transfer interaction between CHLA and 2 MQ.
 |
| | Fig. 11 1Optimized structures of donor 2 MQ, acceptor CHLA, and HB CT complex in the gas phase. | |
From Table S4,† the bond angle C(4)–O(11)–H(13) is increased upon HB CT complex formation compared to free CHLA in the gas phase and PCM analysis. C(3)–C(4)–O(11) and C(5)–C(4)–O(11) also see their bond angles increase, which supports the intermolecular hydrogen bond. It can be noticed the bond angle of C(1)–O(12)–H(14) decreases compared to free CHLA in gas and PCM analysis. These results indicate the formation of intramolecular hydrogen bonds. From donor 2 MQ the C(4)–N(17)–C(12) bond angle increases in the HB CT complex compared to free 2 MQ in gas and PCM analysis. These results support proton transfer hydrogen bonding taking place between 2 MQ and CHLA. There is also no solvent effect on the bond angle.
3.10.2. Mulliken atomic charge distribution. The Mullikan color range of 2 MQ, CHLA, and the HB CT complex in the gas phase and PCM analysis60 are depicted in Fig. S9 and S10.† The main changes are noticed before and after the complexation of the reacting molecules and these estimated Mulliken atomic charges are given in Table S5.† The acceptor CHLA oxygen atoms of O11 and O12 are increased to −0.589653 a.u. and −0.199742 a.u. in the HB CT complex gas phase compared to free CHLA values of −0.154863 a.u. and −0.138610 a.u. in the gas phase. Moreover, N17 is reduced to −0.460791 a.u. in the HB CT complex compared with −0.509682 a.u. in free 2 MQ. The two chlorines (Cl9 and Cl10), and their attached carbons (C2 and C5) displayed an increase in Mullikan atomic charges: −0.369019 a.u. and −0.340620 a.u. compared to free CHLA in gas phase −0.293884 a.u. and −0.321396 a.u. Furthermore, the Mullikan charges of the two chlorines decrease (0.037488 a.u. and −0.128462 a.u.) after complexation compared to free CHLA (−0.193577 a.u. and −0.157256 a.u.) in the gas phase. It is concluded that two chlorines and their involved carbon atoms are contributors to the accepting capacity of CHLA. Hence these results give strong support to the charge transfer and proton transfer contact between CHLA and 2 MQ. From Table S5,† a similar conclusion was also drawn in PCM analysis in the three different polarities.
3.10.3. Electrostatic potential maps (ESP). The electrostatic potential (ESP) surface is the most suitable graphic demonstration for showing the charge density distribution.36,65 This describes the nucleophilic and electrophilic areas at the molecular level. Electrostatic potential (ESP) surface maps were estimated by DFT in the gas phase and PCM analysis. Molecular electrostatic potential surface maps (gas and PCM) are depicted in Fig. 12 and S11.† Molecular electrostatic potential surface maps have a positive region (electrophile), a negative region (nucleophile), and neutral electrostatic potential regions characterized by the association of color. In CHLA two electrophilic regions are observed, one for OH protons and the other for the CHLA ring (blue color), whereas the negative (nucleophile) area is placed on the aromatic rings of 2 MQ N atom (red color). In 2 MQ, the methyl and OH group are supportive of their high electron-donating capacity. Subsequently, the –OH group of CHLA can act as an H-donor and the π system of CHLA can act as the acceptor. In CHLA a positive region is located on the CHLA moiety at the center of the ring (0.0390004 a.u. to 0.0188502 a.u.) and OH proton (0.0786752 a.u. to 0.0511666 a.u.) in the gas phase. Consideration of the 2 MQ ESP plot shows a negative region that is mostly positioned at the N moiety of the ring (−0.101808 a.u. to −0.0337212 a.u.). In the HB CT complex, the MEP surface map shows the value on 2 MQ N is −0.00517464 a.u. to 0.00955661 a.u., the CHLA moiety at the center of the ring is −0.0347706 a.u. to −0.0262269 a.u. and the CHLA OH proton is 0.0263817 a.u. to 0.0159533 a.u. These results clearly show that charge transfer takes place from 2 MQ to CHLA. Similar results are shown in PCM analysis for the three different solvents. Hence, the ESP map surfaces show good agreement with the experimental results.
 |
| | Fig. 12 Electrostatic potential surface maps of 2 MQ, CHLA, and HB CT complex in the gas phase. | |
3.10.4. Estimation of the FMOs. The outcomes of an FMO (frontier molecular orbital) study show that the molecular orbitals are usually composed of p-atomic orbitals and the most reactive condition is for π-electronic systems. The HOMO and LUMO surfaces behave as an electron donor and an electron acceptor, respectively. The n, π-molecular orbitals are detected as HOMOs and the π* molecular orbital are LUMOs; therefore, the charge transfer can be assigned as n–π* and π–π*. This yields information about the reactivity and appropriate contacts of atoms in the charge transfer phenomenon.16,36,66,67 The HOMO–LUMO surface plots are presented in Fig. S12,† and the results are given in Table S6.† From Fig. S12,† the spatial plots of HOMO and HOMO−3 are distributed only on the 2 MQ moiety and HOMO−1, HOMO−2, HOMO−4, and HOMO−5 are distributed on the CHLA moiety, while the LUMOs are mostly distributed on the CHLA moiety as well as LUMO+2 and LUMO+3, which are subsidized by 2 MQ in the gas phase. In PCM analysis, the three different polarities also show a similar trend. The spatial plots and energy gaps of the HB CT complex in the gas phase and PCM analysis are displayed in Fig. 13. The electronic energy gap of the HOMO to LUMO HB CT complex (gas and PCM) is very similar to the experimental calculated energy.
 |
| | Fig. 13 FMO surfaces of the HB CT complex in the PCM analysis. | |
The FMO energy results for HOMO, HOMO−n, LUMO, and LUMO+n, where n = 1–5 are shown in Table S6,† where the HOMO energy level of the HB CT complex in the gas and PCM analysis were observed to be 0.20977 Ha, −0.22423 Ha, −0.22413 Ha and −0.22375 Ha, compared to 2 MQ in gas and PCM analysis which had values of −0.21071 Ha, −0.21737 Ha, −0.21733 Ha and −0.21720 Ha. While the LUMO in gas and PCM analysis had values of −0.10913 Ha, −0.11851 Ha, −0.11843 Ha and −0.11812 Ha compared with values for CHLA alone in gas and PCM analysis of −0.05129 Ha, −0.05355 Ha, −0.05353 Ha and −0.05348 Ha. These values are close to each other; hence it can be concluded that orbital interaction energy increases mostly due to the electronic charge transition from high occupied MOs to unoccupied MOs. The above results fully correlate with those of the experimental proton transfer hydrogen bonded complex.
3.10.5. Reactivity descriptors. Various reactivity descriptors, such as ionization potential (Ip), electron affinity (A), chemical potential (μ), hardness (η), and softness (σ) as well as electrophilicity index (ω), all calculated from the HOMO–LUMO surfaces, give insightful characteristics into the reactivity related to chemical reactions.26 A explanation of these descriptors is given by eqn (11)–(16).| | |
Ionization potential (Ip) = −EHOMO
| (11) |
| | |
Electron affinity (A) = −ELUMO
| (12) |
| | |
Hardness (η) = (Ip − A)/2
| (13) |
| | |
Chemical potential (μ) = (Ip + A)/2
| (14) |
| | |
Electrophilicity index (ω) = μ2/2η
| (15) |
Descriptors for the electronic interaction of 2 MQ and CHLA with the formed HB CT complex are given in Table S7.† From this table, the electronic nature of CHLA and 2 MQ molecules can be concluded. When deriving the HOMO–LUMO energies of a molecule, a maximum EHOMO makes it a good electron donor, while a minimum ELUMO makes for a good electron acceptor. Here CHLA has a lower ELUMO than 2 MQ, so it is considered an electron acceptor; on the other hand, 2 MQ has a greater EHOMO than the CHLA molecule in gas and PCM analysis, so it is considered an electron donor. Additionally, the chemical potential is a potential index that specifies the path of the flow of electrons between molecules. The flow of electrons arises from a structure with a maximum chemical potential to one with a minimum chemical potential. In this interpretation, 2 MQ has a greater maximum chemical potential than CHLA. Furthermore, the electrophilicity index (ω) is an estimate of the electrophilicity of the molecule. Subsequently, CHLA has greater values of ω than 2 MQ, so the former is a better electrophile than the latter. The softness ‘σ’ values and these results additionally show that 2 MQ is an electron donor while CHLA is an electron acceptor in gas and PCM analysis.
4. Conclusions
In this paper, UV-visible electronic spectroscopy was used to investigate a new organic charge and proton transfer hydrogen bonded complex between an electron donor (proton acceptor) 2 MQ and electron acceptor (proton donor) CHLA in acetonitrile, methanol, and ethanol media. The stoichiometry of compound HB CT is a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 molar ratio, confirmed by Job's and photometric methods. Spectroscopic physical parameters, such as KCT, εCT, ECT, ID, RN, f, W, ΔG°, and μEN for HB CT were evaluated using spectral data. It is concluded that the stability of the produced HB CT complex depends on the polarity of the solvent. The compound HB CT showed higher stability in the less polar solvents methanol and ethanol compared to acetonitrile, due to the formation of an intermolecular hydrogen bond between the OH group of methanol and ethanol with the N moiety of 2 MQ. The synthesis of the HB CT complex was confirmed through different techniques like FT-IR and NMR. The crystal nature and structure of the HB CT complex were confirmed by powdered XRD and single crystal XRD analysis. The surface morphology and elemental analysis were studied by carrying out SEM EDX spectra. The thermal stability was studied by TGA-DTA analysis. The HB CT complex has remarkable pharmacological properties, as it shows excellent antimicrobial activity and interacts with CT-DNA intercalatively. A theoretical analysis of the HB CT complex was conducted by the DFT method, and the function B3LYP/6-31G(d,p) basis set was employed for optimization in the gas phase and PCM analysis. The bond lengths, bond angles, Mulliken atomic charges, ESP maps, reactivity parameters, and FMO surfaces were also calculated. The experimental results are supported well by the theoretical analysis of the charge and proton transfer hydrogen bonded complex. The remarkable antimicrobial activity and CT-DNA interactions give scope for the development of a pharmaceutical drug.
1 molar ratio, confirmed by Job's and photometric methods. Spectroscopic physical parameters, such as KCT, εCT, ECT, ID, RN, f, W, ΔG°, and μEN for HB CT were evaluated using spectral data. It is concluded that the stability of the produced HB CT complex depends on the polarity of the solvent. The compound HB CT showed higher stability in the less polar solvents methanol and ethanol compared to acetonitrile, due to the formation of an intermolecular hydrogen bond between the OH group of methanol and ethanol with the N moiety of 2 MQ. The synthesis of the HB CT complex was confirmed through different techniques like FT-IR and NMR. The crystal nature and structure of the HB CT complex were confirmed by powdered XRD and single crystal XRD analysis. The surface morphology and elemental analysis were studied by carrying out SEM EDX spectra. The thermal stability was studied by TGA-DTA analysis. The HB CT complex has remarkable pharmacological properties, as it shows excellent antimicrobial activity and interacts with CT-DNA intercalatively. A theoretical analysis of the HB CT complex was conducted by the DFT method, and the function B3LYP/6-31G(d,p) basis set was employed for optimization in the gas phase and PCM analysis. The bond lengths, bond angles, Mulliken atomic charges, ESP maps, reactivity parameters, and FMO surfaces were also calculated. The experimental results are supported well by the theoretical analysis of the charge and proton transfer hydrogen bonded complex. The remarkable antimicrobial activity and CT-DNA interactions give scope for the development of a pharmaceutical drug.
Conflicts of interest
The authors state that they have no known competing financial interests or personal relationships that could have influenced the research presented in this study.
Acknowledgements
The author PMK gives special thanks to the Head, Department of Chemistry, Osmania University, Hyderabad, for providing facilities for the research work and is thankful to UGC, New Delhi, India for financial support through CSIR-UGC-SRF.
References
- K. P. Goetz, D. Vermeulen, M. E. Payne, C. Kloc, L. E. McNeil and O. D. Jurchescu, J. Mater. Chem. C, 2014, 2, 3065 RSC.
- M. C. Grossel and S. C. Weston, Chem. Mater., 1996, 8, 977–980 CrossRef CAS.
- R. S. Mulliken, J. Am. Chem. Soc., 1952, 74, 811 CrossRef CAS.
- R. Foster, Organic Charge-Transfer Complexes, Academic Press, New York, 1970 Search PubMed.
- I. M. Khan, K. Alam and M. Alam, J. Mol. Liq., 2020, 310, 113213 CrossRef CAS.
- A. Eychmüller and A. L. Rogach, Pure Appl. Chem., 2000, 72, 179–188 Search PubMed.
- H. Sakurai, A. Izuoka and T. Sugawara, Mol. Cryst. Liq. Cryst., 1997, 306, 415–421 CrossRef CAS.
- G. Vadivelan, M. Saravanabhavan, V. Murugesan, G. Gohulvani, M. Sekar and B. Babu, Mol. Cryst. Liq. Cryst., 2017, 652, 242–254 CrossRef CAS.
- M. Mohammad, H. Al Rasid Gazi, K. Pandav, P. Pandya and M. M. Islam, ACS Omega, 2021, 6, 2613–2625 CrossRef CAS PubMed.
- A. S. Gaballa and A. S. Amin, Spectrochim. Acta, Part A, 2015, 145, 302–312 CrossRef CAS PubMed.
- L. H. Abdel-Rahman, A. M. Abu-Dief, H. Moustafa and S. K. Hamdan, Appl. Organomet. Chem., 2017, 31, e3555 CrossRef.
- S. Ray, R. Mohan, J. K. Singh, M. K. Samantaray, M. M. Shaikh, D. Panda and P. Ghosh, J. Am. Chem. Soc., 2007, 129, 15042–15053 CrossRef CAS PubMed.
- O. R. Chittepu, Grain Oil Sci. Technol., 2019, 2, 15–20 CrossRef.
- A. Dozal, H. Keyzer, H. Kim and W. Wang, Int. J. Antimicrob. Agents, 2000, 14, 261–265 CrossRef CAS PubMed.
- K. Sathya, P. Dhamodharan and M. Dhandapani, Opt. Laser Technol., 2018, 101, 328–340 CrossRef CAS.
- K. M. Al-Ahmary, M. M. Habeeb and S. H. Aljahdali, J. Mol. Liq., 2019, 277, 453–470 CrossRef CAS.
- H. Liu, Q. Zhao, Y. Li, Y. Liu, F. Lu, J. Zhuang, S. Wang, L. Jiang, D. Zhu and D. Yu, J. Am. Chem. Soc., 2005, 127, 1120–1121 CrossRef CAS PubMed.
- K. M. Al-Ahmary, M. S. Alenezi and M. M. Habeeb, J. Mol. Liq., 2016, 220, 166–182 CrossRef CAS.
- L. Miyan and A. Ahmad, J. Mol. Liq., 2018, 262, 514–526 CrossRef CAS.
- G. Zhang, G. Kim and W. Choi, Energy Environ. Sci., 2014, 7, 954–966 RSC.
- B.-Q. Zou, Q.-P. Qin, Y.-X. Bai, Q.-Q. Cao, Y. Zhang, Y.-C. Liu, Z.-F. Chen and H. J. M. Liang, J. Med. Chem., 2017, 8, 633–639 CAS.
- E. T. Seo, T. L. Riechel and D. T. Sawyer, Inorg. Chem., 1977, 16, 734–738 CrossRef CAS.
- J.-H. Jeon, C.-H. Lee and H.-S. Lee, J. Korean Soc. Appl. Biol. Chem., 2009, 52(2), 202–205 CrossRef CAS.
- Y. Song, H. Xu, W. Chen, P. Zhan and X. Li, MedChemComm, 2015, 6, 61–74 RSC.
- K. M. Al-Ahmary, M. M. Habeeb and A. H. Al-Obidan, Spectrochim. Acta, Part A, 2018, 196, 247–255 CrossRef CAS PubMed.
- N. Singh, I. M. Khan, A. Ahmad and S. Javed, J. Mol. Liq., 2016, 221, 1111–1120 CrossRef CAS.
- Zulkarnain, I. M. Khan, A. Ahmad, L. Miyan, M. Ahmad and N. Aziz, J. Mol. Struct., 2017, 1141, 687–697 CrossRef CAS.
- A. M. A. Adam, M. S. Refat and H. A. Saad, J. Mol. Struct., 2013, 1037, 376–392 CrossRef CAS.
- I. M. Khan, M. Islam, S. Shakya, K. Alam, N. Alam and M. Shahid, Bioorg. Chem., 2020, 99, 103779 CrossRef CAS PubMed.
- K. M. Al-Ahmary, M. M. Habeeb and S. H. Aljahdali, J. Mol. Struct., 2019, 1181, 48–60 CrossRef CAS.
- I. M. Khan, S. Shakya, R. Akhtar, K. Alam, M. Islam and N. Alam, Bioorg. Chem., 2020, 100, 103872 CrossRef CAS PubMed.
- Zulkarnain, L. Miyan, A. Ahmad, Md. Fazle Alam and H. Younus, J. Photochem. Photobiol., B, 2017, 174, 195–208 CrossRef CAS PubMed.
- H. Al Rabiah, H. A. Abdel-Aziz and G. A. E. Mostafa, J. Mol. Liq., 2019, 286, 110754 CrossRef.
- I. M. Khan, A. Ahmad and M. F. Ullah, J. Photochem. Photobiol., B, 2011, 103, 42–49 CrossRef CAS PubMed.
- P. Manojkumar, B. Naveen and T. Parthasarathy, J. Solution Chem., 2018, 47, 975–992 CrossRef.
- N. Venkatesh, B. Naveen, A. Venugopal, G. Suresh, V. Mahipal, P. Manojkumar and T. Parthasarathy, J. Mol. Struct., 2019, 1196, 462–477 CrossRef CAS.
- G. Suresh, N. Venkatesh, B. Naveen, V. Mahipal, M. Madhavi and T. Parthasarathy, J. Solution Chem., 2020, 49, 777–797 CrossRef CAS.
- H. A. Benesi and J. H. J. Hildebrand, J. Am. Chem. Soc., 1949, 71, 2703–2707 CrossRef CAS.
- I. M. Khan, K. Alam, M. J. Alam and M. Ahmad, New J. Chem., 2019, 43, 9039–9051 RSC.
- N. Singh, I. M. Khan, A. Ahmad and S. Javed, J. Mol. Liq., 2016, 221, 1111–1120 CrossRef CAS.
- G. G. Aloisi and S. Pignataro, J. Chem. Soc., Faraday Trans. 1, 1973, 69, 534–539 RSC.
- G. Briegleb, Elektronen–Donator–Acceptor–Komplexe, Springer-Verlag, 2013 Search PubMed.
- K. M. Al-Ahmary, M. S. Alenezi and M. M. Habeeb, J. Mol. Struct., 2015, 1098, 377–392 CrossRef CAS.
- M. T. Basha, R. M. Alghanmi, S. M. Soliman and W. J. Alharby, J. Mol. Liq., 2020, 309, 113210 CrossRef CAS.
- F. A. Al-Saif, A. A. El-Habeeb, M. S. Refat, A. M. A. Adam, H. A. Saad, A. I. El-Shenawy and H. Fetooh, J. Mol. Liq., 2019, 287, 110981 CrossRef CAS.
- K. Alam and I. M. Khan, J. Electron., 2018, 63, 7–22 CAS.
- I. M. Khan, S. Shakya and N. Singh, J. Mol. Liq., 2019, 287, 110981 CrossRef.
- B. Zhang, H. Wu, Z. Wang, A. Qin and B. Z. Tang, J. Mater. Chem. C, 2020, 8, 4754–4762 RSC.
- P. W. Doheny, J. K. Clegg, F. Tuna, D. Collison, C. J. Kepert and D. M. D'Alessandro, Chem. Sci., 2020, 11, 5213–5220 RSC.
- J. Gao, H. Zhai, P. Hu and H. Jiang, Crystals, 2020, 10, 993 CrossRef CAS.
- U. Neupane, M. Singh, P. Pandey and R. Rai, J. Mol. Struct., 2019, 1195, 131–139 CrossRef CAS.
- O. Kataeva, M. Khrizanforov, Y. Budnikova, D. Islamov, T. Burganov, A. Vandyukov, K. Lyssenko, B. Mahns, M. Nohr and S. Hampel, Cryst. Growth Des., 2016, 16, 331–338 CrossRef CAS.
- J. I. Lachowicz, G. Dalla Torre, R. Cappai, E. Randaccio, V. M. Nurchi, R. Bachor, Z. Szewczuk, L. Jaremko, M. Jaremko and M. B. Pisano, Dalton Trans., 2020, 49, 2862–2879 RSC.
- A. Karmakar, P. Bandyopadhyay, S. Banerjee, N. C. Mandal and B. Singh, J. Mol. Liq., 2020, 299, 112217 CrossRef CAS.
- I. M. Khan, A. Ahmad, M. M. Oves and B. Spectroscopy, Spectrochim. Acta, Part A, 2010, 77, 1059–1064 CrossRef PubMed.
- R. Kumar, S. Obrai, A. Kaur, M. S. Hundal, H. Meehnian and A. K. Jana, New J. Chem., 2014, 38, 1186–1198 RSC.
- H. Ke, F. Hu, L. Meng, Q.-H. Chen, Q.-S. Lai, Z.-C. Li, Z.-L. Huang, J.-Z. Liao, J.-D. Qiu and C.-Z. Lu, Chem. Commun., 2020, 56, 14353–14356 RSC.
- D. Mahendiran, G. Vinitha, S. Shobana, V. Viswanathan, D. Velmurugan and A. K. J. R. a. Rahiman, RSC Adv., 2016, 6, 60336–60348 RSC.
- I. M. Khan, A. Ahmad and M. F. Ullah, Spectrochim. Acta, Part A, 2013, 102, 82–87 CrossRef CAS PubMed.
- P. Manojkumar, N. Venkatesh, G. Suresh, V. Mahipal, M. Ramesh and T. Parthasarathy, Chem. Data Collect., 2020, 29, 100493 CrossRef CAS.
- B. V. Emma and T. Gunnlaugsson, J. Org. Chem., 2010, 75, 5513–5525 CrossRef PubMed.
- S. G. Aziz, S. A. Elroby, A. Jedidi, B. A. Babgi, N. S. Alshehri and M. A. Hussien, J. Mol. Struct., 2020, 1215, 128283 CrossRef CAS.
- D. Sreenu, A. Rambabu, N. Vamsikrishna, G. Nirmala and S. Raj, J. Coord. Chem., 2019, 72, 1973–1993 CrossRef.
- B. Pradip, A. Abhishek, P. Brandao, M. Sunil Kumar, I. Bhattacharyya, G. Mondal, J. Abhimanyu, S. Ananyakumari and B. Pulakesh, New J. Chem., 2021, 45, 11999–12015 RSC.
- V. Mahipal, N. Venkatesh, B. Naveen, G. Suresh, V. Manaiah and T. Parthasarathy, Chem. Data Collect., 2020, 28, 100474 CrossRef CAS.
- R. M. Alghanmi, M. T. Basha, S. M. Soliman and R. K. Alsaeedi, J. Mol. Liq., 2021, 326, 115199 CrossRef CAS.
- M. Sun, J. Chem. Phys., 2006, 124, 054903 CrossRef PubMed.
- L. Cui, S. Zhu and M. Sun, Phys. E, 2021, 134, 114840 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. CCDC 2070105. For ESI and crystallographic data in CIF or other electronic format see DOI: 10.1039/d1ra07658b |
|
| This journal is © The Royal Society of Chemistry 2021 |
Click here to see how this site uses Cookies. View our privacy policy here.  Open Access Article
Open Access Article *a
*a
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 from the Job data and photometric titration process. The association constant (KCT) and molar absorptivity (εCT) of the HB CT complex were determined by using the modified Benesi–Hildebrand equation in three polarities. Other spectroscopic physical parameters like the energy of interaction (ECT), ionization potential (ID), resonance energy (RN), standard free energy change (ΔG°), oscillator strength (f), and transition dipole moment (μ) were also evaluated. The HB CT complex structure was confirmed by different characterization techniques, such as FT-IR, NMR, TGA-DTA, and SEM-EDX analysis. Powder XRD and single-crystal XRD were used to determine the nature and structure of the synthesized HB CT complex. DNA binding studies for the HB CT complex produced a good binding constant value of 2.25 × 104 L mol−1 in UV-visible and 1.17 × 104 L mol−1 in fluorescence spectroscopy. The biological activity of the HB CT complex was also tested in vitro against the growth of bacteria and fungi, and the results indicated remarkable activity for the HB CT complex compared to the standard drugs, ampicillin and clindamycin. Hence, the abovementioned biological results of the synthesized HB CT complex show it could be used as a pharmaceutical drug in the future. Computational analysis was carried out by DFT studies using the B3LYP function with a basis set of 6-31G(d,p) in the gas phase and PCM analysis. The computational studies further supported the experimental results by confirming the charge and proton transfer complex.
1 from the Job data and photometric titration process. The association constant (KCT) and molar absorptivity (εCT) of the HB CT complex were determined by using the modified Benesi–Hildebrand equation in three polarities. Other spectroscopic physical parameters like the energy of interaction (ECT), ionization potential (ID), resonance energy (RN), standard free energy change (ΔG°), oscillator strength (f), and transition dipole moment (μ) were also evaluated. The HB CT complex structure was confirmed by different characterization techniques, such as FT-IR, NMR, TGA-DTA, and SEM-EDX analysis. Powder XRD and single-crystal XRD were used to determine the nature and structure of the synthesized HB CT complex. DNA binding studies for the HB CT complex produced a good binding constant value of 2.25 × 104 L mol−1 in UV-visible and 1.17 × 104 L mol−1 in fluorescence spectroscopy. The biological activity of the HB CT complex was also tested in vitro against the growth of bacteria and fungi, and the results indicated remarkable activity for the HB CT complex compared to the standard drugs, ampicillin and clindamycin. Hence, the abovementioned biological results of the synthesized HB CT complex show it could be used as a pharmaceutical drug in the future. Computational analysis was carried out by DFT studies using the B3LYP function with a basis set of 6-31G(d,p) in the gas phase and PCM analysis. The computational studies further supported the experimental results by confirming the charge and proton transfer complex.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 molar ratio. Photometric titration calculations were used to estimate the ratio of the formed HBCT complex, where two lines were formed which intersected, which gives a 1
1 molar ratio. Photometric titration calculations were used to estimate the ratio of the formed HBCT complex, where two lines were formed which intersected, which gives a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 molar ratio in all solvents.37 A graph was plotted of absorbance against the D/A ratio and is depicted in Fig. S2.† Subsequently, one can confirm that the solvent polarity does not affect the HBCT complex molecular composition.
1 molar ratio in all solvents.37 A graph was plotted of absorbance against the D/A ratio and is depicted in Fig. S2.† Subsequently, one can confirm that the solvent polarity does not affect the HBCT complex molecular composition.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio at room temperature.35,36 The absorption band of the CT complex increases slowly with time, and it reveals that the formation of HB CT is strong in all solvents. The HB CT band λmax for all three solutions is not changed, but the HB CT band absorbance slowly increases with time and is presented in Fig. S3.†
1 ratio at room temperature.35,36 The absorption band of the CT complex increases slowly with time, and it reveals that the formation of HB CT is strong in all solvents. The HB CT band λmax for all three solutions is not changed, but the HB CT band absorbance slowly increases with time and is presented in Fig. S3.†![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 Benesi–Hildebrand principle38 at 25 °C in the three polar solvents.
1 Benesi–Hildebrand principle38 at 25 °C in the three polar solvents.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 HB CT complex in the three polarities. They are shown in Fig. 2. We calculated the gradient and intercept (1/ε and 1/KCTε) values from Fig. 2. These values are collected for the different polarities and shown in Table 1.
1 HB CT complex in the three polarities. They are shown in Fig. 2. We calculated the gradient and intercept (1/ε and 1/KCTε) values from Fig. 2. These values are collected for the different polarities and shown in Table 1.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif)
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif)
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ln
ln![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) KCT
KCT

![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) are 1663 cm−1 and 1635 cm−1. The ν(C–Cl) band frequencies are 836, and 745 cm−1 in the HB CT complex compared to 845, and 752 cm−1 for CHLA itself, and ν(C
O) are 1663 cm−1 and 1635 cm−1. The ν(C–Cl) band frequencies are 836, and 745 cm−1 in the HB CT complex compared to 845, and 752 cm−1 for CHLA itself, and ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C) has a band frequency of 1628 cm−1 compared to 1599 cm−1 for free CHLA. Hence, the occurrence of proton transfer besides charge transfer was confirmed from the IR spectrum of the HB CT complex.
C) has a band frequency of 1628 cm−1 compared to 1599 cm−1 for free CHLA. Hence, the occurrence of proton transfer besides charge transfer was confirmed from the IR spectrum of the HB CT complex.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) cos
cos![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) θ
θ![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio. The empirical formula of the HB CT complex crystal structure is C16H11Cl2NO5. Single-crystal X-ray determined the formation of proton transfer hydrogen bonding between the OH of CHLA and the 2 MQ pyridine ring nitrogen. The intermolecular hydrogen bond N1–H⋯O3 with H⋯O of 1.93 Å and the intramolecular hydrogen bond O5–H⋯O4 with H⋯O of 2.13 Å formed, and the molecular structure of the HB CT complex with anisotropic displacement ellipsoids is presented in Fig. 7. The crystal system was found to be orthorhombic with space group Pna21 and Z = 4. The lattice parameters are a = 22.9722(6) Å, b = 13.0888(3) Å, and c = 5.0930(1) Å and other crystallographic data and refinement parameters for the HB CT complex are summarized in Table 4.51–53 The bonding network of crystal packing is presented in Fig. 8. The HB CT complex hydrogen bonding bond lengths are fully correlated to the DFT analysis.
1 ratio. The empirical formula of the HB CT complex crystal structure is C16H11Cl2NO5. Single-crystal X-ray determined the formation of proton transfer hydrogen bonding between the OH of CHLA and the 2 MQ pyridine ring nitrogen. The intermolecular hydrogen bond N1–H⋯O3 with H⋯O of 1.93 Å and the intramolecular hydrogen bond O5–H⋯O4 with H⋯O of 2.13 Å formed, and the molecular structure of the HB CT complex with anisotropic displacement ellipsoids is presented in Fig. 7. The crystal system was found to be orthorhombic with space group Pna21 and Z = 4. The lattice parameters are a = 22.9722(6) Å, b = 13.0888(3) Å, and c = 5.0930(1) Å and other crystallographic data and refinement parameters for the HB CT complex are summarized in Table 4.51–53 The bonding network of crystal packing is presented in Fig. 8. The HB CT complex hydrogen bonding bond lengths are fully correlated to the DFT analysis.

![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 molar ratio, confirmed by Job's and photometric methods. Spectroscopic physical parameters, such as KCT, εCT, ECT, ID, RN, f, W, ΔG°, and μEN for HB CT were evaluated using spectral data. It is concluded that the stability of the produced HB CT complex depends on the polarity of the solvent. The compound HB CT showed higher stability in the less polar solvents methanol and ethanol compared to acetonitrile, due to the formation of an intermolecular hydrogen bond between the OH group of methanol and ethanol with the N moiety of 2 MQ. The synthesis of the HB CT complex was confirmed through different techniques like FT-IR and NMR. The crystal nature and structure of the HB CT complex were confirmed by powdered XRD and single crystal XRD analysis. The surface morphology and elemental analysis were studied by carrying out SEM EDX spectra. The thermal stability was studied by TGA-DTA analysis. The HB CT complex has remarkable pharmacological properties, as it shows excellent antimicrobial activity and interacts with CT-DNA intercalatively. A theoretical analysis of the HB CT complex was conducted by the DFT method, and the function B3LYP/6-31G(d,p) basis set was employed for optimization in the gas phase and PCM analysis. The bond lengths, bond angles, Mulliken atomic charges, ESP maps, reactivity parameters, and FMO surfaces were also calculated. The experimental results are supported well by the theoretical analysis of the charge and proton transfer hydrogen bonded complex. The remarkable antimicrobial activity and CT-DNA interactions give scope for the development of a pharmaceutical drug.
1 molar ratio, confirmed by Job's and photometric methods. Spectroscopic physical parameters, such as KCT, εCT, ECT, ID, RN, f, W, ΔG°, and μEN for HB CT were evaluated using spectral data. It is concluded that the stability of the produced HB CT complex depends on the polarity of the solvent. The compound HB CT showed higher stability in the less polar solvents methanol and ethanol compared to acetonitrile, due to the formation of an intermolecular hydrogen bond between the OH group of methanol and ethanol with the N moiety of 2 MQ. The synthesis of the HB CT complex was confirmed through different techniques like FT-IR and NMR. The crystal nature and structure of the HB CT complex were confirmed by powdered XRD and single crystal XRD analysis. The surface morphology and elemental analysis were studied by carrying out SEM EDX spectra. The thermal stability was studied by TGA-DTA analysis. The HB CT complex has remarkable pharmacological properties, as it shows excellent antimicrobial activity and interacts with CT-DNA intercalatively. A theoretical analysis of the HB CT complex was conducted by the DFT method, and the function B3LYP/6-31G(d,p) basis set was employed for optimization in the gas phase and PCM analysis. The bond lengths, bond angles, Mulliken atomic charges, ESP maps, reactivity parameters, and FMO surfaces were also calculated. The experimental results are supported well by the theoretical analysis of the charge and proton transfer hydrogen bonded complex. The remarkable antimicrobial activity and CT-DNA interactions give scope for the development of a pharmaceutical drug.













