Open Access Article
Open Access ArticlePd(II) and Pt(II) complexes of tridentate ligands with selective toxicity against Cryptococcus neoformans and Candida albicans†
Ahmed M. Mansour *
*
Department of Chemistry, Faculty of Science, Cairo University, Gamma Street, Giza, Cairo 12613, Egypt. E-mail: mansour@sci.cu.edu.eg; inorganic_am@yahoo.com
First published on 14th December 2021
Abstract
Novel Pd(II) and Pt(II) complexes of the tridentate 2,6-bis(1-ethyl-benzimidazol-2′-yl)pyridine (LBZ), and 4′-(2-pyridyl)-2,2′:6′,2′′-terpyridine (LPY) ligands were synthesized, characterized using a variety of analytical and spectroscopic tools, and screened for their potential antimicrobial properties against some bacterial and fungal strains as well as cytotoxicity against healthy human embryonic kidney (HEK293) cells. The electronic structures of the complexes were investigated by time-dependent density functional theory calculations. The free ligand LPY and benzimidazole complexes exhibited selective toxicity against Cryptococcus neoformans and Candida albicans, while displaying no cytotoxicity against HEK293. In the case of Cryptococcus neoformans, the antifungal activities of the benzimidazole-based complexes (MIC = 1.58–2.62 μM) are higher than those of the reference drug fluconazole (26.1 μM).
Introduction
Antimicrobial resistance may be an emerging public health emergency, and it seems that organic chemistry alone is not adequate to supply the world with new effective antimicrobial agents.1 Metal-based compounds are relatively uncommon drugs compared with organic compounds, having made their clinical entrance in 1978 with the endorsement of cisplatin. While some metal complexes are currently used in clinical applications such as anticancer therapies,2,3 little attention has been given to their capability as antimicrobial agents. With the current drug pipeline being very scarce, essentially comprising of some antibiotic derivatives, modern classes of antimicrobials are compulsorily required. In fact, a few reports have examined the antimicrobial properties of Pd(II) and Pt(II) compounds.4–11 These two elements are fairly rare and expensive, restricting their use as clinical antibiotics if substantial doses are necessary. In 2014, Lippard and co-workers reported the effect of a monofunctional Pt(II) agent, phenanthriplatin (Fig. 1a), on Escherichia coli and bacteriophage λ lysogens.4 The ability of phenanthriplatin to initiate bacterial filamentation and lysis in lysogenic bacteria proves that the toxicity of this compound is mediated by its interaction with DNA.4 Mixed-ligand platinum(II) complexes with 1,10-phenanthroline and 1,2-diaminoethane (or 1S,2S- or 1R,2R-diaminocyclohexane) (Fig. 1b) exhibit higher selective toxicity towards Escherichia coli (MIC = 10–20 μM) than those of their Cu(II) analogues (MIC ≥10–20 μM).5 Trans-Pt(II)(salicylaldimine)(pyridine)·BF4 and trans-(Pt(II)(salicylaldimine))2(4,4-bipyridine)·2BF4 (Fig. 1c) induce significant retardation in the growth of Escherichia coli compared to the control and cisplatin by changing the morphology of the bacterial cells.6 In 2018, we reported on the antimicrobial activity and protein binding affinities of four Pd(II) and Pt(II) cisplatin analogues bearing N,N-pyridylbenzimidazole derivatives (Fig. 1d) decorated with an alkylated phosphonium or sulfonate side chain.7 Both Pd(II) sulfonate and the Pt(II) phosphonium complexes displayed higher toxicity against Cryptococcus neoformans (MIC = 5–30 μM) and Candida albicans (MIC = 2–30 μM) than the reference drug fluconazole (MIC = 26.1 μM), while displaying no antibacterial activity against the tested bacterial strains and healthy HEK293 cells.7 The simplest Pt(II) complex of 1,5-cyclooctadiene (COD) (Fig. 1e) exhibited promising selective antibacterial activity against a board of Gram-positive bacteria, including vancomycin- and methicillin-resistant Staphylococcus aureus, while demonstrating no toxicity against HEK293 cells or haemolytic properties at the highest tested concentrations.1 A series of terpene-derived chiral palladium(II) complexes exhibited interesting antimicrobial activity against the Gram-positive pathogens, C. albicans and C. neoformans var. grubii with MIC ≤0.25–16 μg mL−1.8 Among the examined Pd(II) terpene complexes, the amine-linked Pd(II) derivative (Fig. 1f) displayed promising antifungal activity (MIC ≤0.25 μg mL−1) and possessed an exceptional selectivity index with respect to C. neoformans var. grubii (>128 against HEK293).8 Besides, the Pd(II) complex of 1-quinolin-8-yliminomethyl-naphthalen-2-ol exhibited antibacterial activity against S. marcescent, E. coli, and M. luteus microbes with MIC of 2.25–3.75 μM.10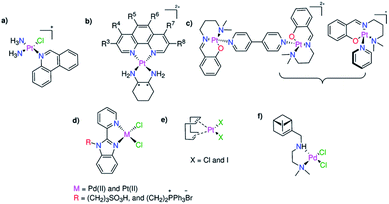 | ||
| Fig. 1 Previously reported Pd(II)- and Pt(II)-based complexes (a–f) exhibiting antimicrobial activity. | ||
In addition, a review has reported a sudden high hit rate for some classes of metal complexes (9.9%) compared with some organic compounds (0.87%) submitted to the same examination.11 Of the 63 examined platinum-based compounds, 27 complexes exhibited promising activity against at least one of the tested bacterial and fungal strains.11 Further examination of the antimicrobial activity of the Pt complexes revealed that a total of 18 compounds displayed no cytotoxic effects toward healthy mammalian cells and induced no haemoglobin release at the tested concentration. This encouraged me to synthesize new Pd(II) and Pt(II) complexes of 2,6-bis(1-ethyl-benzimidazol-2′-yl)pyridine (LBZ), and 4′-(2-pyridyl)-2,2′:6′,2′′-terpyridine (LPY) (Scheme 1), and evaluate their potential antimicrobial activity against five bacterial strains (Staphylococcus aureus, Klebsiella pneumoniae, Pseudomonas aeruginosa, Escherichia coli, and Acinetobacter baumannii) and two fungal (Candida albicans and Cryptococcus neoformans) pathogens. The cell viability of healthy HEK293 cells treated with the synthesized complexes was determined to assess if the toxicity was limited to the tested microbes or extended to mammalian cells as well. Their compatibility with human red blood cells (RBCs) was evaluated by determining the HC10 and HC50 (the concentrations at which 10% and 50% haemolysis occur). After the administration of the biologically active metal complexes, there is a chance for them to be tackled by the surface accessible histidyl proteins, which will affect the biodistribution and pharmacokinetics. Thus, it is essential to investigate the stability of the compounds after designing and before developing a new effective drug, specifically in presence of some target and/or model proteins. Thus, the stability of the Pd(II) and Pt(II) complexes was evaluated in the presence of hen white egg lysozyme (HEWL) by mass spectrometry.
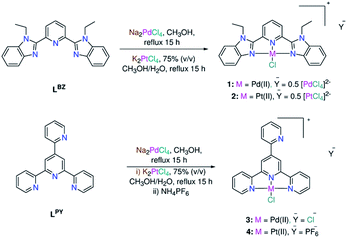 | ||
| Scheme 1 Synthesis of square-planar Pd(II) and Pt(II) complexes coordinated by tridentate ligands (1–4). | ||
Results and discussion
Synthesis and characterization
Benzimidazole LBZ was synthesized via the selective alkylation of 2,6-bis(benzimidazole-2′-yl)pyridine12 using iodoethane and a mixture of fresh sodium hydride and DMF.13 In a strongly basic medium (potassium tert-butoxide and ammonia), the reaction of 2-pyridine carboxaldehyde with two equivalents of 2-acetyl-pyridine gave 4′-(2-pyridyl)-2,2′:6′,2′′-terpyridine (LPY) with a good yield and in pure form.14 The reactions between the ligands and Na2PdCl4 in methanol afforded the square-planar complexes 1 and 3. The IR spectra are presented as ESI (Fig. S1†). The ESI mass spectra, in the positive mode, displayed two fragments at m/z = 510.0516 (calcd 510.0515) (1) and 452.9917 (calcd 452.9940) (3) corresponding to {PdClL}+ (L = LBZ and LPY), respectively (Fig. S2†). A literature search showed that L coordinated with metal ions in either a tridentate fashion via all the three nitrogen donor atoms,13,15 bidentate mode k2–N1,1′, in which one of the two outer heterocyclic rings is bent out of the mean plane of the other two coordinated centers,14,16,17 or monodentate k1–N1 binding mode in few examples.18 The 1H NMR spectra of the free ligands and complex 1 were compared to each other (Fig. S3†). The most striking difference between the 1H NMR spectra of LBZ and complex 1 occurred at the triplet signal of the pyridyl ring and the doublets of benzimidazole- H4/H4′, which lay closest to the metal center and chloride ion. For example, the triplet signal experienced a downfield shift from 8.22 to 8.60 ppm as a result of complex formation. In the case of 1, the observation of the doublet and triplet signals of the pyridine ring in a 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio (Fig. S4–S7†) is consistent with the tridentate coordination mode of LBZ. For 3, the presence of a singlet 1H NMR signal (Fig. S8–S11†) for the central pyridine-H3/H5 confirmed that all the three close nitrogen donor atoms of LPY were involved in complex formation.15 The chloro Pt(II) analogues (2 and 4) were prepared by heating to reflux a mixture of K2PtCl4 and L in a 75% (v/v) methanol/water mixture for 15 h. The shift of ν(CN)/ν(CC) from 1610 (LBZ) and 1616 cm−1 (LPY) in the spectra of free ligands to ≈1604 cm−1 in those of 1–4 may be attributed to the participation of the pyridine and benzimidazole nitrogen atoms in complex formation (Fig. S1†). The ESI mass spectra of 2 and 4 (Fig. S12†) showed the {PtClL}+ peaks at 597.1133 (calcd 597.1128) (2) and 540.0546 (calcd 540.0549) (4). As expected, the NMR spectra of 1 and 2 were typical (Fig. S13–S16†), revealing to the same mode of coordination of LBZ irrespective of changing the metal ion. Unlike 1–3, complex 4 had poor solubility in most organic solvents, including DMSO and DMF. To address the solubility problem, the counterion was replaced by the hexafluorophosphate ion. However, the new form of 4 also showed limited solubility in most organic solvents, such as DMF, and decomposed in DMSO (Fig. S17†). In DMF-d7, the 1H NMR pattern of LPY in 4 was consistent with the tridentate mode of chelation as in 3 (Fig. S18†). The PF6− counterion was tracked by observing the doublet 19F NMR (Fig. S19†) and septet 31P NMR signals (Fig. S20†) at −71.8 (1JPF = 709 Hz) and −141.8 (1JPF = 709 Hz), respectively. Compound 4 was also analysed by solid-state NMR (13C{1H}, 19F and 31P) (Fig. S21–S23†). The PF6− signals were observed at −70.1 and −141.4 (1JPF = 710 Hz) in the 19F and 31P NMR spectra, respectively.
1 ratio (Fig. S4–S7†) is consistent with the tridentate coordination mode of LBZ. For 3, the presence of a singlet 1H NMR signal (Fig. S8–S11†) for the central pyridine-H3/H5 confirmed that all the three close nitrogen donor atoms of LPY were involved in complex formation.15 The chloro Pt(II) analogues (2 and 4) were prepared by heating to reflux a mixture of K2PtCl4 and L in a 75% (v/v) methanol/water mixture for 15 h. The shift of ν(CN)/ν(CC) from 1610 (LBZ) and 1616 cm−1 (LPY) in the spectra of free ligands to ≈1604 cm−1 in those of 1–4 may be attributed to the participation of the pyridine and benzimidazole nitrogen atoms in complex formation (Fig. S1†). The ESI mass spectra of 2 and 4 (Fig. S12†) showed the {PtClL}+ peaks at 597.1133 (calcd 597.1128) (2) and 540.0546 (calcd 540.0549) (4). As expected, the NMR spectra of 1 and 2 were typical (Fig. S13–S16†), revealing to the same mode of coordination of LBZ irrespective of changing the metal ion. Unlike 1–3, complex 4 had poor solubility in most organic solvents, including DMSO and DMF. To address the solubility problem, the counterion was replaced by the hexafluorophosphate ion. However, the new form of 4 also showed limited solubility in most organic solvents, such as DMF, and decomposed in DMSO (Fig. S17†). In DMF-d7, the 1H NMR pattern of LPY in 4 was consistent with the tridentate mode of chelation as in 3 (Fig. S18†). The PF6− counterion was tracked by observing the doublet 19F NMR (Fig. S19†) and septet 31P NMR signals (Fig. S20†) at −71.8 (1JPF = 709 Hz) and −141.8 (1JPF = 709 Hz), respectively. Compound 4 was also analysed by solid-state NMR (13C{1H}, 19F and 31P) (Fig. S21–S23†). The PF6− signals were observed at −70.1 and −141.4 (1JPF = 710 Hz) in the 19F and 31P NMR spectra, respectively.
Density functional theory calculations
The local minimum structures of 1–4 (Fig. 2) were obtained by optimizing the models representing their molecular structures ([MClL]+ (M = Pd(II) and Pt(II))) and analysing the harmonic frequencies using the Becke 3-parameter (exchange) Lee–Yang–Parr (B3LYP)19 functional and the effective core potentials (ECP) of the Hady and Wadt, LANL2DZ basis set.20,21 Because of the absence of the imaginary frequencies, the obtained geometries were labelled as local minima. The geometries around 1–4 could be described as square-planar and were defined by the tridentate ligand L (L = LBZ and LPY) and the chloride ion. Selected bond lengths and angles are given in Table 1. The trans-M–N bonds in 1–4 are equal [PdN2 = PdN12 = 2.047 Å (1), PtN2 = PtN12 = 2.033 Å (2), PdN37 = PdN38 = 2.055 Å (3) and PtN37 = PtN38 = 2.041 Å (4)]. Although the PdCl bond lengths in 1 and 3 are equal, the trans-M–N bonds of 1 are shorter than those of 3 by 0.008 Å. The distance from the Pd(II) ion to the central pyridine N in 1 is longer than the corresponding distance in 3 by 0.035 Å. The Pd–N bonds of 1 and 3 are shorter than the equivalent bonds in 2 and 4. The corresponding cis-angles in the four complexes are nearly equal.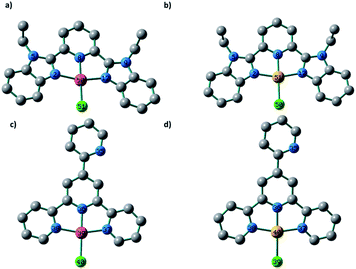 | ||
| Fig. 2 Local minimum structures of the cationic part of (a) 1, (b) 2, (c) 3 and (d) 4 obtained at the B3LYP/LANL2DZ level of theory (H atoms are omitted for clarity). | ||
| 1 | 2 | 3 | 4 |
|---|---|---|---|
| PdN2 = 2.047 | PtN2 = 2.033 | PdN36 = 1.970 | PtN36 = 1.960 |
| PdN8 = 2.005 | PtN8 = 1.986 | PdN37 = 2.055 | PtN37 = 2.041 |
| PdN12 = 2.047 | PtN12 = 2.033 | PdN38 = 2.056 | PtN38 = 2.041 |
| PdCl = 2.365 | PtCl = 2.395 | PdCl = 2.363 | PtCl = 2.396 |
| N2PdN8 = 80.0 | N2PtN8 = 80.2 | N36PdN37 = 80.8 | N36PtN37 = 81.1 |
| ClPdN2 = 100.0 | ClPtN2 = 99.8 | ClPdN37 = 99.3 | ClPtN37 = 98.9 |
| N8PdN12 = 80.0 | N8PtN12 = 80.2 | N36PdN38 = 80.6 | N36PtN38 = 80.9 |
| ClPdN12 = 100.0 | ClPtN12 = 99.8 | ClPdN38 = 99.3 | ClPtN38 = 99.0 |
To get an insight into the electronic structures and transitions of 1–4, time-dependent density functional theory calculations were executed using their local minimum structures. In the singlet state, the lowest 30 spin-allowed excitation states were calculated using the B3LYP/LANL2DZ level of theory and the SMD solvation model.22 The electronic spectra were simulated using GaussSum. Each excited state was included by a Gaussian convolution with the full-width at half-maximum (FWHM) value of 3000. The electronic absorption spectrum (Fig. S24†) of 1 is characterized by three electronic transitions at 317, 370, and 431 nm corresponding to HOMO−1 → LUMO+1, HOMO−1 → LUMO and HOMO → LUMO/LUMO+2, respectively. As shown in Fig. 3, HOMO is mainly composed of π(Cl), LUMO is on π*(LBZ) and LUMO+2 is contained to d(Pd)/π(Cl)/p(N donors).
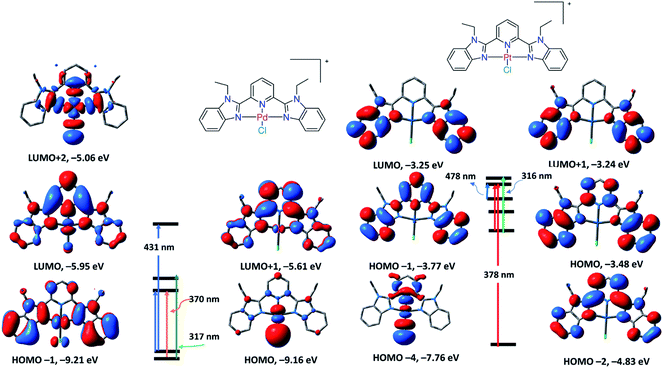 | ||
| Fig. 3 Selected frontier molecular orbitals of 1 (left) and 2 (right) obtained by the B3LYP/LANL2DZ method. The electronic transitions were obtained at the same level of theory. | ||
Thus, the transition at 431 nm is assigned to LLCT/LMCT, while the other transitions at 370 and 317 nm are assigned to π–π* within the ligand framework. The TDDFT spectrum of 2 displayed three electronic transitions at 316, 378 and 478 nm with oscillator strengths of 0.3256, 0.3244, and 0.0036, respectively. As shown in Fig. 3, the longest wavelength transition at 478 nm (HOMO → LUMO) is that of π–π* within LBZ, while the main band at 378 nm (HOMO−4/HOMO−2 → LUMO) represents a mixture of π–π*/LLCT. Experimentally, the electronic absorption of 2 demonstrated four electronic transitions in DMSO at 318, 340, 372, and 460 nm, which are in agreement with the calculated electronic spectrum, disclosing the suitability of this calculation method in the case of Pd(II) and Pt(II) complexes. The calculated spectrum of 3 (Fig. S24†) characterized three main transitions at 312, 330 and 370 nm with oscillator strengths of 0.3204, 0.0175 and 0.1455 in that order, which could be attributed to HOMO → LUMO, HOMO → LUMO+1 and HOMO−6 → LUMO, respectively. The TDDFT spectrum of 4 showed three main transitions at 405, 356 and 280 nm related to HOMO → LUMO, HOMO → LUMO+1 and HOMO−6 → LUMO+1, respectively. The descriptions of the frontier MOs and the relocation of the electron density of 3 and 4 are depicted in Fig. 4. Similar to 1 and 2, the transitions at 370 and 405 nm in the spectra of 3 and 4 could be assigned to LLCT, LMCT or π–π* within the terpyridine ligand framework.
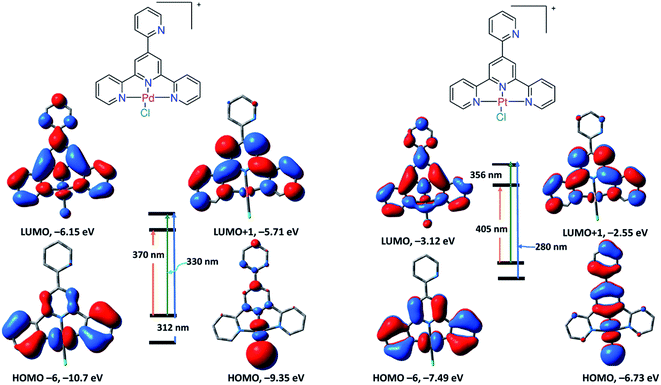 | ||
| Fig. 4 Selected frontier molecular orbitals of 3 (left) and 4 (right) obtained by the B3LYP/LANL2DZ method. The electronic transitions were obtained at the same level of theory. | ||
Stability
The stability of 1–3 in DMSO and 2% (v/v) DMSO–PBS mixture was assessed over two days by UV/vis spectroscopy. The electronic absorption spectra (Fig. S25–S30†) collected in both media as a function of time were nearly superimposable, revealing the good stability of 1–3 under these experimental conditions. In the case of 2, the slight spectral fluctuation may be credited to the exchange of Cl− with the solvent molecules. Since complex 4 displayed poor solubility and weak stability in most organic solvents, it was not further examined for potential antimicrobial properties.Reactivity towards lysozyme and imidazole
Studying the interactions of platinum-based drugs with proteins is vital since it offers valuable information about metallodrug recognition and alteration of the pharmacological profile, as well as biodistribution.23,24 The acquired toxicity or resistance of some Pt(II) based drugs towards certain diseases may be understood by investigating their interactions with some model and/or target proteins. For example, while the mechanism of action of cisplatin and carboplatin is mainly based on direct binding with the double helix,25,26 Lippard group showed that oxaliplatin kills cells by inducing ribosome biogenesis stress.27 On the other hand, some proteins are used as biocompatible carriers.28,29 Thus, it is essential to determine the stability of the biologically active species in the presence of some target and model proteins to select the appropriate biocompatible carrier. In this regard, Merlino and co-workers briefly described and comparatively examined the molecular structures of some adducts obtained upon the reaction of selected Pt-based drugs with proteins.30 Data disclosed that the attached ligands play a substantial role in driving the recognition process, as well as the non-covalent interactions that happen in the early stages of the Pt complex/protein recognition process, plays a critical role in determining the nature of the final Pt–protein adduct.27 For this purpose, the stability of 3, as an illustrative example, in the presence of hen egg white lysozyme, a model protein, was assessed by electrospray ionization mass spectrometry (ESI MS). Compound 3 was dissolved in DMSO and then mixed with the HEWL aqueous solution in a 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio. The reaction mixture was directly injected into the instrument. The ESI MS spectrum (Fig. S13†) displayed a peak corresponding to the free protein at m/z 1590.4299 Da, indicating the incompleteness of the recognition process. Five Pd-containing protein fragments obviously showed up at m/z 1601.9728, 1606.3034, 1613.4102, 1636.9846 and 1647.9731 Da, which were assigned to HEWL/Pd2+, HEWL/Pd(OH2)22+, HEWL/2 × Pd2+, HEWL/[Pd(LPY)]2+ and HEWL/([Pd(LPY)]2+ + Pd2+), respectively. The examination of the latter HEWL adducts revealed a coordinative covalent mode of binding. Besides, the observation of these fragments indicates that the reaction of 3 with lysozyme started with the loss of the chloride ion and ended with the departure of the terpyridine-based ligand. The mass spectrometry data indicated the existence of at least two Pd binding sites on the HEWL surface.
1 ratio. The reaction mixture was directly injected into the instrument. The ESI MS spectrum (Fig. S13†) displayed a peak corresponding to the free protein at m/z 1590.4299 Da, indicating the incompleteness of the recognition process. Five Pd-containing protein fragments obviously showed up at m/z 1601.9728, 1606.3034, 1613.4102, 1636.9846 and 1647.9731 Da, which were assigned to HEWL/Pd2+, HEWL/Pd(OH2)22+, HEWL/2 × Pd2+, HEWL/[Pd(LPY)]2+ and HEWL/([Pd(LPY)]2+ + Pd2+), respectively. The examination of the latter HEWL adducts revealed a coordinative covalent mode of binding. Besides, the observation of these fragments indicates that the reaction of 3 with lysozyme started with the loss of the chloride ion and ended with the departure of the terpyridine-based ligand. The mass spectrometry data indicated the existence of at least two Pd binding sites on the HEWL surface.
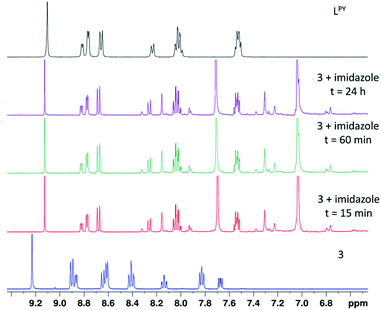
The His15 side chain is a recognized metal-binding site of lysozyme.31,32 Similar to the observation during the binding of the pyridylbenzimidazole Pd(II) and Pt(II) complexes with some target and model proteins,33 it is likely that compound 3 may be able to bind covalently via the imidazole ring of the His15 side chain or maybe decomposed because of the action of the His15 side chain. To authenticate the interaction between 3 and the His15 site, the reaction between 3 and two equivalents of imidazole was investigated by 1H NMR in DMSO-d6 (Fig. 5). The assessment of the reaction product was achieved by comparison with the parent complex and free terpyridine LPY.
As shown in Fig. 5, the reaction between 3 and imidazole was instantaneous and ended within a few minutes with no additional changes until the end of the experiment (24 h). The 1H NMR signals of the imidazole product coincided with those of the free ligand, indicating the decomposition of the complex and the departure of the terpyridine ligand, in agreement with the results obtained in the case of the model protein. To conclude, compound 3 may be degraded in the presence of proteins containing free histidyl side chains.
DNA binding study
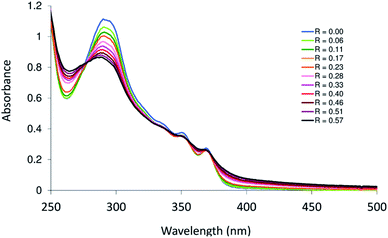
Platinum-based drugs are broadly used in clinics for cancer therapy. The mechanism of action of some of these drugs relies on their binding to DNA.34,35 The coordination of Pt complexes to DNA causes a deformation that interferes with replication and transcription and ultimately leads to cell death via apoptosis.36 In general, Pt compounds interact with a double helix through covalent and/or noncovalent modes. Intercalation, groove (minor and major) and electrostatic binding represent the noncovalent modes of DNA binding. Groove modes depend on the size of the biologically active molecule and the strength of hydrogen bonds, which enable the molecules to occupy a specific groove. Intercalation modes (intercalator and insertors) are influenced by the planarity of the molecules. The stacking of Pt(II) compounds of heterocyclic ligands between the strands of DNA stiffens and lengthens the double helix. Experimentally, it is simply to assess the binding of heterocyclic intercalators to DNA by following the electronic spectral changes upon successive addition of DNA to a fixed amount of the tested compound. The intensity of the charge transfer band of the complex may be altered upon interaction with DNA. Groove induces hyperchromism, while intercalation brings a hypochromic shift to the CT band. The DNA titration plot of 3, which was tested as the representative compound, is shown in Fig. 6. The absorption spectrum of 3 (in 2% (v/v) DMSO–H2O) displays three main transitions at 292, 351 and 370 nm. The addition of calf thymus DNA (CT-DNA) to complex 3 caused hypochromism in the band at 292 nm, suggesting the intercalation mode of binding.36,37 To verify the magnitude of interaction of 3 with CT-DNA, the intrinsic binding constant (Kb)38 was calculated and compared to those of closely related structures. The Kb value of 3 was (3.5 ± 0.1) × 104 M−1. The obtained Kb value of 3 is similar to that of other reported Pd(II) complexes based on terpyridine-like ligands, such as 4′-(4-(2-(piperidin-1-yl)ethoxy)phenyl)-2,2′:6′,2′′-terpyridine (Kb = 1.06 ± × 104 M−1),39 and is lower than that of 4′-(4-hydroxyphenyl)-2,2′,6′,2′′-terpyridine (Kb = 1.4 ± × 105 M−1).40Antimicrobial activity
Primary screening of the free ligands and their Pd(II) and Pt(II) complexes (1–3) for potential antimicrobial properties was executed at 32 μg mL−1 concentration against key bacterial (Staphylococcus aureus ATCC 43300, Pseudomonas aeruginosa ATCC 27853, Escherichia coli ATCC 25922, Klebsiella pneumoniae ATCC 700603 and Acinetobacter baumannii ATCC 19606) and fungal (Cryptococcus neoformans and Candida albicans) pathogens. A follow-up dose-responsive toxicity assay against the same strains was performed for the compounds displaying marked inhibition at 32 μg mL−1. The minimum inhibitory concentration (MIC) was determined as the lowest concentration that inhibited 80% and 70% growth of C. albicans and C. neoformans, respectively. The benzimidazole ligand (LBZ) was inactive against all the seven tested microbes. The terpyridine ligand (LPY) and complexes 1–3 were effective at inhibiting the drug-susceptible yeast reference strains C. albicans and C. neoformans, while they exhibited no antibacterial activity. The antimicrobial activity of compound 4 could not be determined because it is decomposed in DMSO.Compared to fluconazole (Flu), the antifungal activity of the tested ligands against C. albicans was in the following order: Flu (0.40 μM) > 1 (0.78 μM) > 2 (1.31 μM) > 3 (66.0 μM) > LPY (103.2 μM). In the case of C. neoformans, the order of antifungal activity was as follows: 1 (1.58 μM) > 2 (2.62 μM) > Flu (26.1 μM) > 3 (33.0 μM) > LPY (103.2 μM) (Table 2).
Several biologically active compounds display general toxicity toward both microbes and mammalian cells. To verify this, the cell viability of normal HEK293 cells treated with LPY and 1–3 was determined to inspect if their toxicity was limited to the fungal strains or had a similar effect on mammalian cells. Media and cells without inhibitors were used, respectively as the negative and positive controls. While LPY and compounds (1 and 2) were safe for HEK293 with CC50 > 32 μg mL−1, complex 3 displayed slight toxicity toward these cells with a CC50 value of 25.91 μg mL−1. On the other hand, the haemolytic properties of the synthesized compounds towards human red blood cells (RBCs) were evaluated by determining the concentrations at which 10% and 50% haemolysis occurred (HC10 and HC50). None of the compounds induced haemolysis up to 32 μg mL−1.
On comparing with some of the previously published complexes of LPY and LBZ, complexes 1 and 2 displayed higher antifungal activities than triazolate–Mn(CO)3 of LPY, which was potent against C. albicans and C. neoformans (MIC = 6.0 μM).41 Besides, compounds 1 and 2 exhibited higher potency against the same two pathogens than the Pd(II) sulfonate and Pt(II) phosphonium complexes of benzimidazole ligands (MIC = 2–30 μM) we reported earlier7 and a morpholine-based Pd(II) complex (MIC = 32–64 μM),42 while displaying comparable antifungal activity to other benzimidazole Pd(II) complexes (MIC = 500–2 μg mL−1),43,44 an oxime Schiff-base (MIC = 500 μg mL−1),45 and thiosalicylic acid (MIC = 1000–62 μg mL−1).46 Based on the interesting antifungal properties of 1 and 2 against C. neoformans, further evaluation of these two compounds and other closely related structures as antifungal agents is highly recommended (Table 2).
| E. coli | K. pneumoniae | A. baumanni | P. aeruginosa | ||
|---|---|---|---|---|---|
| a Full names: Staphylococcus aureus ATCC 43300, Escherichia coli ATCC 25922, Klebsiella pneumoniae ATCC 700603, Acinetobacter baumannii ATCC 19606, Pseudomonas aeruginosa ATCC, Candida albicans ATCC 90028 and Cryptococcus neoformans var. grubii H99; ATCC 208821.b Data of ligands published previously.12,13c Not determined.d Fluconazole. | |||||
| LBZ | >32 | >32 | >32 | >32 | |
| LPy | >32 | >32 | >32 | >32 | |
| 1 | >32 | >32 | >32 | >32 | |
| 2 | >32 | >32 | >32 | >32 | |
| 3 | >32 | >32 | >32 | >32 | |
| 4 | n.dc | n.d | n.d | n.d | |
| Flud | — | — | — | — | |
| S. aureus | C. albicans | C. neoformans | ||
|---|---|---|---|---|
| LBZ | >32 | >32 | >32 | |
| LPy | >32 | 32 | 32 | |
| 1 | >32 | 0.5 | 1 | |
| 2 | >32 | 1 | 2 | |
| 3 | >32 | 32 | 16 | |
| 4 | n.d | n.d | n.d | |
| Flud | — | 0.125 | 8 |
Conclusion
As reported in the literature, only a few reports have examined the antimicrobial properties of Pd(II) and Pt(II) complexes owing to the cost of these two elements, which restricts their use as clinical antibiotics when substantial doses are necessary. In this context, novel Pd(II) and Pt(II) complexes of two tridentate ligands (2,6-bis(1-ethyl-benzimidazol-2′-yl)pyridine, and 4′-(2-pyridyl)-2,2′:6′,2′′-terpyridine) were synthesized and fully characterized using a variety of analytical and spectroscopic tools. The metal complexes displayed good stability in DMSO and 2% (v/v) DMSO–H2O except for the Pt(II) complex of terpyridine, which had poor solubility in most organic solvents and decomposed in DMSO. After administration, there is a high probability that the biologically active species bind to the surface-accessible histidyls of proteins, and consequently, we investigated the stability of the Pd(II) terpyridine complex, as an illustrated example, in the presence of a model protein (hen white egg lysozyme) by electrospray ionization mass spectrometry. The results showed that the decomposition of the complex occurred in a stepwise manner, starting with the loss of the highly reactive coordinated chloride ion and ending with the departure of the terpyridine ligand. The mass spectrometry data indicated the existence of at least two Pd binding sites on the HEWL surface. As it is likely that the Pd(II) and Pt(II) complexes bind to proteins covalently via the imidazole ring of the His15 side chain or maybe decomposed because of the action of the His15 side chain, the reaction between the Pd(II) terpyridine complex and imidazole was investigated by 1H NMR. The reaction was instant, and the NMR signals of the reaction product coincided with those of the free ligand, suggesting the decomposition of the complex. Primary screening of the free ligands and their Pd(II) and Pt(II) complexes for potential antimicrobial properties indicated that the free terpyridine ligand and the benzimidazole-based complexes were effective at inhibiting the drug-susceptible reference yeast strains C. albicans and C. neoformans, while they displayed no antibacterial activity. The Pd(II) and Pt(II) complexes (MIC = 1.58–2.62 μM) of 2,6-bis(1-ethyl-benzimidazol-2′-yl)- pyridine exhibited higher toxicity against C. neoformans than fluconazole (MIC = 26.1 μM) and displayed no cytotoxicity towards normal mammalian cells, as well as exhibited good compatibility with RBCs. Therefore, further evaluation of these two complexes and other closely related benzimidazole-based complexes as antifungal agents is highly recommended.Experimental section
Materials and instruments
Na2PdCl4 and K2PtCl4 were purchased from Strem and used as received. 2,6-Bis(1-ethyl-benzimidazol-2′-yl)pyridine (LBZ),12 and 4′-(2-pyridyl)-2,2′:6′,2′′-terpyridine (LPY)13 were synthesized according to the published procedures. Elemental micro-analysis was achieved with a Vario Micro Cube analyzer from Elementar Analysensysteme or an EA 3000 elemental analyser from HEKtech. The electrospray mass spectra were registered with a ThermoFisher Exactive Plus instrument with an Orbitrap mass analyzer at a resolution of R = 70.000 and a solvent flow rate of 50 μL min−1. The electronic absorption spectra were recorded on a Specord 210 Plus spectrophotometer. The ATR IR spectra of the solid-state complexes were recorded on a Brucker Alpha-E instrument. The 1H, 13C, 19F and 31P NMR spectra were recorded with Bruker-Avance 500 (1H, 500.13 MHz; 13C{1H}, 125.77 MHz; 19F, 470.59 MHz; 31P, 202.46 MHz) and Bruker-Avance 400 (1H, 400.40 MHz; 13C{1H}, 100.68 MHz; 31P, 162.08 MHz) spectrometers. The assignment of NMR signals was performed with the aid of two-dimensional NMR methods, {1H, 1H} COS90 and {1H, 13C} HSQC.Synthesis
1: To a round-bottom flask charged with LBZ (0.5 mmol; 183 mg) and sodium tetrachloropalladate (1.0 mmol; 294 mg), methanol (40 mL) was added, and then the reaction mixture was heated to reflux for 15 h. A yellow precipitate was collected by filtration, washed with methanol and diethyl ether, and dried in vacuo. Yield: 67% (210 mg, 0.33 mmol): IR (ATR): ν = 3099 (w, CH), 3026 (w, CH), 2973 (w, CH), 1604 (m, CC/CN), 1525, 1483, 1441, 1340, 1131, 973, 751 cm−1. 1H NMR (DMSO-d6, 400.40 MHz): δ = 8.60 (t, 3JH,H = 8.3 Hz, 1H, py-H4), 8.40 (d, 3JH,H = 8.2 Hz, 2H, py-H3/H5), 7.64 (d, 3JH,H = 7.9 Hz, 2H, bim-H4/H4′), 7.46 (d, 3JH,H = 8.1 Hz, 2H, bim-H7/H7′), 7.24 (t, 3JH,H = 7.1 Hz, 2H, bim-H5/H5′), 7.19 (t, 3JH,H = 7.2 Hz, 2H, bim-H6/H6′), 4.48 (q, 3JH,H = 7.2 Hz, 4H, NCH2), 1.39 (t, 3JH,H = 7.11 Hz, 6H, NCH2CH3) ppm. 13C-NMR (DMSO-d6, 100.68 MHz): δ = 149.7 (py-C2/C6), 146.8 (bim-C2/C2′), 144.1 (py-C4), 138.1 (bim-C3a/C3a′), 133.1 (bim-C7a/C7a′), 126.1 (bim-C6/C6′), 126.0 (bim-C5/C5′), 124.8 (py-C3/C5), 116.6 (bim-C7/C7′), 111.9 (bim-C4/C4′), 40.6 (CH2/CH2′), 15.0 (CH3/CH3′) ppm. ESI-MS (positive mode, methanol): m/z = 510.0516 {M–X}+. C23H21ClN5Pd·0.5(PdCl4)·2H2O: C 41.26, H 3.76, N 10.46, found C 41.09, H 3.43, N 10.14.
2: To the methanolic solution (25 mL) of LBZ (0.5 mmol; 183 mg), 5 mL aqueous solution of potassium tetrachloroplatinate(II) (1.0 mmol; 415 mg) was added. The reaction mixture was heated to reflux for 15 h, whereupon a grey precipitate of complex 2 formed. It was filtered off, washed with water, methanol, and diethyl ether and then dried in vacuo. Yield: 54% (207 mg, 0.27 mmol): IR (ATR): ν = 3098 (w, CH), 3063 (w, CH), 1605 (m, CC/CN), 1525, 1482, 1442, 1341, 1156, 1079, 749 cm−1. 1H NMR (DMSO-d6, 400.40 MHz): δ = 8.58 (t, 3JH,H = 8.2 Hz, 1H, py-H4), 8.41 (d, 3JH,H = 8.1 Hz, 2H, py-H3/H5), 7.84 (d, 3JH,H = 8.00 Hz, 2H, bim-H4/H4′), 7.60 (d, 3JH,H = 8.1 Hz, 2H, bim-H7/H7′), 7.39 (t, 3JH,H = 6.9 Hz, 2H, bim-H5/H5′), 7.33 (t, 3JH,H = 7.7 Hz, 2H, bim-H6/H6′), 4.56 (q, 3JH,H = 7.0 Hz, 4H, NCH2), 1.42 (t, 3JH,H = 6.9 Hz, 6H, NCH2CH3) ppm. 13C-NMR (DMSO-d6, 100.68 MHz): δ = 152.4 (py-C2/C6), 147.3 (bim-C2/C2′), 143.1 (py-C4), 138.6 (bim-C3a/C3a′), 133.5 (bim-C7a/C7a′), 126.7 (bim-C6/C6′), 126.0 (bim-C5/C5′), 124.4 (py-C3/C5), 116.2 (bim-C7/C7′), 112.4 (bim-C4/C4′), 40.6 (CH2/CH2′), 15.0 (CH3/CH3′) ppm. ESI-MS (positive mode, methanol): m/z = 597.1133 {M–X}+. C23H21ClN5Pt·0.5(PtCl4)·1.5H2O: C 34.82, H 3.05, N 8.83, found C 34.67, H 2.69, N 8.54.
3: To a round flask charged with the terpyridine ligand (1 mmol; 310 mg) and Na2PdCl4 (0.85 mmol; 249 mg), methanol (50 mL) was added. The reaction mixture was heated to reflux for 15 h. A yellow precipitate was collected by filtration, washed with methanol, chloroform, and diethyl ether and then dried under vacuum. Yield: 42% (176 mg, 0.32 mmol). IR (ATR): ν = 3474 (br, OH2), 3028 (w, CH), 3001 (w, CH), 1604 (s, CC/CN), 1558, 1475, 1407, 1245, 1162, 1028, 891, 780 cm−1. 1H NMR (DMSO-d6, 400.40 MHz): δ = 9.21 (s, 2H, H3/H5), 8.88 (m, 3H, H6′/H6′′/H6′′′), 8.69 (d, 2H, 3JH,H = 5.2 Hz, H3′/H3′′), 8.56 (d, 1H, 3JH,H = 8.2 Hz, H3′′′), 8.45 (td, 2H, JH,H = 7.9 Hz, JH,H = 1.6 Hz, H5′/H5′′), 8.17 (td, 1H, JH,H = 7.8 Hz, 3JH,H = 1.7 Hz, H4′′′), 7.86 (m, 2H, H4′/H4′′), 7.68 (t, 1H, 3JH,H = Hz, H5′′′) ppm. 13C-NMR (DMSO-d6, 100.70 MHz): δ = 157.8, 154.9, 152.0, 151.7, 150.8, 150.3, 142.5, 138.2, 128.9, 126.2, 125.7, 123.0, 121.2 ppm. ESI-MS (positive mode, 20% (v/v) DMSO/methanol): m/z = 452.9917 {M–Cl}+. C20H14Cl2N4Pd·3H2O: C 44.34, H 3.72, N 10.34, found C 44.53, H 3.34, N 10.68.
4: To a round-bottom flask charged with LPY (1.0 mmol; 300 mg) and 25 mL methanol, 5 mL aqueous solution of K2PtCl4 (1.0 mmol; 415 mg) was added. The reaction mixture was heated to reflux for 15 h, whereupon a brown precipitate formed. Filtration was done. The product was treated with NH4PF6 (1.0 mmol; 163 mg) in a 75–25% acetone/water mixture and stirred overnight. The formed brown precipitate was collected by filtration, washed with water and diethyl ether, and dried in vacuo. Both forms of the complexes, with and without PF6, were subjected to analysis. Before the addition of NH4PF6, the results are as follows: IR (ATR): ν = 3496 (m, OH2), 3051 (s, CH), 1605 (s, CC/CN), 1554, 1470, 1409, 1302, 1254, 1163, 1032, 990, 891, 778, 715 cm−1. ESI-MS (positive mode, 20% (v/v) DMSO/methanol): m/z = 540.0546 {M–X}+. C20H14Cl2N4Pt·0.5PtCl4·2H2O. C 33.03, H 2.22, N 7.70, found C 33.06, H 2.10, N 7.57. After the addition of NH4PF6, the results are as follows: colour: brown powder. IR (ATR): ν = 3476 (br, OH2), 3050 (w, CH), 1605 (s, CC/CN), 1474, 1411, 1249, 1163, 1034, 839, 776 cm−1. The complex decomposed in DMSO-d6 (ESI†) and had poor solubility in DMF-d7. 1H NMR (DMF-d7, 400.40 MHz): δ = 9.16 (s, 2H, H3/H5), 9.31 (d, 3JH,H = 6.0 Hz, 1H), 9.25 (d, 3JH,H = 8.1 Hz, 2H), 9.12 (d, 3JH,H = 4.1 Hz, 1H), 8.88 (m, 3H), 8.38 (t, 3JH,H = 6.8 Hz, 1H), 8.30 (m, 2H), 7.91 (m, 1H) (one proton signal overlapped with the solvent residual). Because of its poor solubility, it was difficult to record the 13C{1H} NMR data. 31P-NMR (DMF-d7, 161.94 MHz): δ = −141.8 (sept, 1JPF = 709 Hz, PF6−). 19F-NMR (DMF-d7, 396.5 MHz): δ = −71.8 (doublet, 1JPF = 709 Hz, PF6−) ppm. Solid-state NMR analyses were done. 13C-NMR (solid-state, 100.68 MHz): δ = 155.9, 152.9, 151.2, 149.8, 149.0, 143.5, 138.7, 131.1, 126.8, 125.3, 123.5, 120.2, 117.4 ppm. 31P-NMR (solid-state, 161.94 MHz): δ = −141.4 ppm (sept, 1JPF = 710 Hz, PF6−). 19F-NMR (solid-state, 396.5 MHz): δ = −70.1 (br, PF6−) ppm. ESI-MS (positive mode, 20% (v/v) DMSO/methanol): m/z = 540.0546 {M–PF6}+. C20H14Cl2F6N4PPt·2H2O: C 33.28, H 2.51, N 7.76, found C 33.41, H 2.42, N 7.61.
DNA binding studies
Initially, the CT-DNA solution was prepared at a proper concentration so that the intensity ratio of the transitions at 260 and 280 nm is nearly 1.9 to ensure that the double helix is free of protein. The standardization of CT-DNA was performed by using the molar absorptivity value at 260 nm (ε260 = 6600 L mol−1 cm−1). The reactivity of complex 3 towards calf-thymus DNA was assessed by following the electronic absorption changes upon the successive addition of CT-DNA to a fixed amount of the complex at room temperature.47 Following the addition of CT-DNA, the solution was agitated, and after 10 min, the spectrum was registered. The procedure was repeated until no further significant change was observed.Lysozyme affinity
An aqueous solution of hen white egg lysozyme was mixed with five equivalents of complex 3, and then the reaction mixture was immediately injected into the mass spectrometer. The positive mode ESI MS spectrum was registered by the direct introduction of the sample at a flow rate of 10 μL min−1. The working conditions were as follows: spray voltage 3.80 kV, capillary voltage 45 V, and capillary temperature 320 °C. For spectral acquisition, Thermo Xcalibur qual was used.Biological activity testing
The evaluation of the antimicrobial properties of the free ligands and metal complexes was achieved by CO-ADD (The Community for Antimicrobial Drug Discovery), and this was financed by Wellcome Trust (UK) and University of Queensland (Australia). The susceptibility of the seven treated bacterial and fungal strains to the active compounds was tested at 32 μg mL−1 using standard broth microdilution assays.11 For compounds that showed toxicity, a follow-up confirmation of toxicity was effected by means of a dose-response assay against the same microbial strain. No animals were used in this study. The cell lines (microbial and mammalian) were obtained from the American Type Culture Collection (ATCC). Human blood was obtained from the Australian Red Cross Blood Service with informed consent, and its use in haemolysis assays was approved by the University of Queensland Institutional Human Research Ethics Committee, approval number 2014000031.Density functional theory calculations
The local minimum structures and vibrational spectra of 1–4 were obtained at the B3LYP (ref. 18)/LANL2DZ (ref. 19 and 20) level of theory using GaussView03.48 Starting from the coordinates of the optimized structures, the time dependent density functional theory calculations were executed using the same level of theory. The SMD solvation model was used in the TDDFT calculations.21 The visualization of the local minimum structures, and the vibrational and electronic spectra, as well as Frontier molecular orbitals, was obtained by GaussView03.49Conflicts of interest
There are no conflicts to declare.Acknowledgements
A. Mansour thanks Dr Rüdiger Bertermann for solid-state NMR analysis, Dr Ola Shehab for mass spectrometry measurements and Mrs Rabaa Khaled for doing the stability tests. Antimicrobial screening was performed by CO-ADD (The Community for Antimicrobial Drug Discovery), funded by the Wellcome Trust (UK) and The University of Queensland (Australia).References
- A. Frei, S. Ramu, G. J. Lowe, H. Dinh, L. Semenec, A. G. Elliott, J. Zuegg, A. Deckers, N. Jung, S. Braese, A. Cain and M. A. Blaskovich, Platinum cyclooctadiene complexes with activity against Gram-positive bacteria, ChemMedChem, 2021, 16(20), 3165–3171 CrossRef CAS PubMed.
- T. C. Johnstone, K. Suntharalingam and S. J. Lippard, The next generation of platinum drugs: targeted Pt(II) agents, nanoparticle delivery and Pt(IV) prodrugs, Chem. Rev., 2016, 116, 3436–3486 CrossRef CAS PubMed.
- R. G. Kenny and C. J. Marmion, Toward multi-targeted platinum and ruthenium drugs-a new paradigm in cancer drug treatment regimens, Chem. Rev., 2019, 119, 1058–1137 CrossRef CAS PubMed.
- T. C. Johnstone, S. M. Alexander, W. Lin and S. J. Lippard, Effects of monofunctional platinum agents on bacterial growth: A retrospective study, J. Am. Chem. Soc., 2014, 136, 116–118 CrossRef CAS PubMed.
- N. S. Ng, P. Leverett, D. E. Hibbs, Q. Yang, J. C. Bulanadi, M. J. Wu and J. R. Aldrich-Wright, The antimicrobial properties of some copper(II) and platinum(II) 1,10-phenanthroline complexes, Dalton Trans., 2013, 42, 3196–3209 RSC.
- F.-U. Rahman, A. Ali, I. U. Khan, M. Z. Bhatti, M. Petroselli, H.-Q. Duong, J. Martì-Rujas, Z.-T. Li, H. Wang and D.-W. Zhang, Monofunctional supramolecular Pt(II) complexes: Synthesis, single crystal structure, anticancer activity, E. coli growth retardation and DNA interaction study, Inorg. Chem. Commun., 2019, 102, 95–103 CrossRef CAS.
- A. M. Mansour and O. R. Shehab, Lysozyme and DNA binding affinity of Pd(II) and Pt(II) complexes bearing charged N,N-pyridylbenzimidazole bidentate ligands, Dalton Trans., 2018, 47, 3459–3468 RSC.
- O. Zalevskaya, Y. Gureva, A. Kutchin and K. A. Hansford, Antimicrobial and antifungal activities of Terpene-Derived Palladium complexes, Antibiotics, 2020, 9, 277 CrossRef CAS PubMed.
- E. H. Avdović, Ž. B. Milanović, M. N. Živanović, D. S. Seklić, I. D. Radojević, L. R. Čomić, S. R. Trifunović, A. Amić and Z. S. Marković, Synthesis, spectroscopic characterization, biological activity, DFT and molecular docking study of novel 4-hydroxycoumarine derivatives and corresponding palladium(II) complexes, Inorg. Chim. Acta, 2020, 504, 119465 CrossRef.
- F. S. Aljohani, A. M. Abu-Dief, R. M. El-Khatib, H. A. Al-Abdulkarium, A. Alharbi, A. Mahran, M. E. Khalifa and N. M. El-Metwaly, J. Mol. Struct., 2021, 1246, 131139 CrossRef CAS.
- A. Frei, J. Zuegg, A. G. Elliott, M. Baker, S. Braese, C. Brown, F. Chen, C. G. Dowson, G. Dujardin, N. Jung, A. P. King, A. M. Mansour, M. Massi, J. Moat, H. A. Mohamed, A. K. Renfrew, P. J. Rutledge, P. J. Sadler, M. W. Todd, C. E. Willans, J. J. Wilson, M. A. Cooper and M. A. T. Blaskovich, Metal complexes as a promising source for new antibiotics, Chem. Sci., 2020, 11, 2627–2639 RSC.
- A. W. Addison and P. J. Burke, Synthesis of some imidazole- and pyrazole-derived chelating agents, J. Heterocycl. Chem., 1981, 18, 803–807 CrossRef CAS.
- A. M. Mansour and O. R. Shehab, Photoactivatable CO-releasing properties of {Ru(CO)2}-core pyridylbenzimidazole complexes and reactivity towards lysozyme, Eur. J. Inorg. Chem., 2017, 4299–4310 CrossRef CAS.
- A. M. Mansour and O. R. Shehab, {Ru(CO)x}-core terpyridine complexes: lysozyme binding affinity, DNA and photoinduced carbon monoxide releasing properties, J. Photochem. Photobiol., A, 2018, 364, 406–414 CrossRef CAS.
- A. M. Mansour, Terpyridine Zn(II) azide compounds: Spectroscopic and DFT calculations, J. Mol. Struct., 2021, 1242, 130737 CrossRef CAS.
- A. M. Mansour, K. Radacki and O. R. Shehab, Role of the ancillary ligand in controlling the lysozyme binding affinity and electronic properties of terpyridine fac-Re(CO)3 complexes, Dalton Trans., 2021, 50, 1197–1201 RSC.
- A. M. Mansour and A. Friedrich, The CO release properties of k2–N1,N2 Mn(I) tricarbonyl PhotoCORMs with tridentate benzimidazole coligands, Inorg. Chem. Front., 2017, 4, 1517–1524 RSC.
- A. Hildebrandt, N. Wetzold, P. Ecorchard, B. Walfort, T. Rüffer and H. Lang, Synthesis and Reaction Chemistry of Heterodi-and Heterotrimetallic Transition-Metal Complexes Based on 1-(Diphenylphosphanyl)-1′-terpyridylferrocene, Eur. J. Inorg. Chem., 2010, 3615–3627 CrossRef CAS.
- A. D. Becke, Density-functional thermochemistry. III. The role of exact exchange, J. Chem. Phys., 1993, 98, 5648–5652 CrossRef CAS.
- P. J. Hay and W. R. Wadt, Ab initio effective core potentials for molecular calculations. Potentials for the transition metal atoms Sc to Hg, J. Chem. Phys., 1985, 82, 270–283 CrossRef CAS.
- P. J. Hay and W. R. Wadt, Ab initio effective core potentials for molecular calculations. Potentials for main group elements Na to Bi, J. Chem. Phys., 1985, 82, 284–298 CrossRef.
- A. V. Marenich, C. J. Cramer and D. G. Truhlar, Universal Solvation Model Based on Solute Electron Density and on a Continuum Model of the Solvent Defined by the Bulk Dielectric Constant and Atomic Surface Tensions, J. Phys. Chem. B, 2009, 113, 6378–6396 CrossRef CAS PubMed.
- L. Messori and A. Merlino, Cisplatin binding to proteins: A structural perspective, Coord. Chem. Rev., 2016, 513, 67–89 CrossRef.
- F. Arnesano and G. Natile, Platinum on the road: Interactions of antitumoral cisplatin with proteins, Pure Appl. Chem., 2008, 80(12), 2715–2725 CAS.
- S. E. Sherman, D. Gibson, A. H. Wang and S. J. Lippard, X-ray structure of the major adduct of the anticancer drug cisplatin with DNA: cis-[Pt(NH3)2(d(pGpG))], Science, 1985, 230(4724), 412–417 CrossRef CAS PubMed.
- Y. Jung and S. J. Lippard, Direct cellular responses to platinum induced DNA damage, Chem. Rev., 2007, 107(5), 1387–1407 CrossRef CAS PubMed.
- P. M. Bruno, Y. Liu, G. Y. Park, J. Murai, C. E. Koch, T. J. Eisen, J. R. Pritchard, Y. Pommier, S. J. Lippard and M. T. Hemann, A subset of platinum-containing chemotherapeutic agents kills cells by inducing ribosome biogenesis stress, Nat. Med., 2017, 23(4), 461–471 CrossRef CAS PubMed.
- M. ChavesFerreira, I. S. Albuquerque, D. Matak-Vinkovic, A. C. Coelho, S. M. Carvalho, L. M. Saraiva, C. C. Romao and G. J. L. Bernardes, Spontaneous CO Release from RuII(CO)2–Protein Complexes in Aqueous Solution, Cells, and Mice, Angew. Chem., Int. Ed., 2015, 54, 1172–1175 CrossRef CAS PubMed.
- H. Tabe, K. Fujita, S. Abe, M. Tsujimoto, T. Kuchimaru, S. Kizaka-Kondoh, M. Takano, S. Kitagawa and T. Ueno, Preparation of a cross-linked porous protein crystal containing Ru carbonyl complexes as a CO-releasing extracellular scaffold, Inorg. Chem., 2015, 54, 215–220 CrossRef CAS PubMed.
- G. Ferraro, D. Loreto and A. Merlino, Interaction of platinum drugs with proteins: An overview of representative crystallographic studies, Curr. Top. Med. Chem., 2021, 21, 6–27 CrossRef CAS PubMed.
- D. Loreto and A. Merlino, The interaction of rhodium compounds with proteins: A structural overview, Coord. Chem. Rev., 2021, 442, 213999 CrossRef CAS.
- A. M. Mansour, K. Radacki and O. R. Shehab, Sulfonate improves water solubility and cell selective toxicity and alters the lysozyme binding activity of half sandwich Rh(III) complexes, Dalton Trans., 2021, 50, 10701 RSC.
- G. Ferraro, A. M. Mansour and A. Merlino, Exploring the interactions between model proteins and Pd(II) or Pt(II) compounds bearing charged N,N-pyridylbenzimidazole bidentate ligands by X-ray crystallography, Dalton Trans., 2018, 47, 10130–10138 RSC.
- S. E. Sherman, D. Gibson, A. H. Wang and S. J. Lippard, X-ray structure of the major adduct of the anticancer drug cisplatin with DNA: cis-[Pt(NH3)2(d(pGpG))], Science, 1985, 230(4724), 412–417 CrossRef CAS PubMed.
- Y. Jung and S. J. Lippard, Direct cellular responses to platinum induced DNA damage, Chem. Rev., 2007, 107(5), 1387–1407 CrossRef CAS PubMed.
- Q. L. Zhang, J. G. Liu, H. Chao, G. Q. Xue and L. N. Ji, DNA-binding and photocleavage studies of cobalt(III) polypyridyl complexes:[Co(phen)2IP]3+ and [Co(phen)2PIP]3+, J. Inorg. Biochem., 2001, 83, 49–55 CrossRef CAS PubMed.
- Z. C. Liu, B. D. Wang, B. Li, Q. Wang, Z. Y. Yang, T. R. Li and Y. Li, Crystal structures, DNA-binding and cytotoxic activities studies of Cu(II) complexes with 2-oxo-quinoline-3-carbaldehyde Schiff-bases, Eur. J. Med. Chem., 2010, 45, 5353 CrossRef CAS PubMed.
- A. M. Pyle, J. P. Rehmann, R. Meshoyrer, C. V. Kumar, N. J. Turro and J. K. Barton, Mixed-ligand complexes of ruthenium(II): factors governing binding to DNA, J. Am. Chem. Soc., 1989, 111, 3051–3058 CrossRef CAS.
- W. Chu, Y. Wang, S. Liu, X. Yang, S. Wang, S. Li, G. Zhou, X. Qin, C. Zhou and J. Zhang, Synthesis, cytotoxicity and DNA-binding properties of Pd(II), Cu(II) and Zn(II) complexes with 4′-(4-(2-(piperidin-1-yl)ethoxy)phenyl)-2,2′:6′,2′′-terpyridine, Bioorg. Med. Chem. Lett., 2013, 23, 5187–5191 CrossRef CAS PubMed.
- F. Darabi, H. Hadadzadeh, J. Simpson and A. Shahpiri, A water-soluble Pd(II) complex with a terpyridine ligand: experimental and molecular modelling studies of the interaction with DNA and BSA; and in vitro cytotoxicity investigations against five human cancer cell lines, New J. Chem., 2016, 40, 9081 RSC.
- A. M. Mansour and K. Radacki, Spectroscopic and antimicrobial activity of photoactivatable tricarbonyl Mn(I) terpyridine compounds, Inorg. Chim. Acta, 2020, 511, 119806 CrossRef CAS.
- A. M. Mansour, Antifungal activity, DNA and lysozyme binding affinity of Pd(II) and Pt(II) complexes bearing N,N pyridylbenzimidazole ligand, J. Coord. Chem., 2018, 71(20), 3381–3391 CrossRef CAS.
- N. M. Aghatabay, M. Somer, M. Senel, B. Dulger and F. Gucin, Raman, FT-IR, NMR spectroscopic data and antimicrobial activity of bis[μ2-(benzimidazol-2-yl)-2-ethanethiolato-N,S,S-chloro-palladium(II)] dimer, [(μ2-CH2CH2NHNCC6H4)PdCl]2·C2H5OH complex, Eur. J. Med. Chem., 2007, 42, 1069 CrossRef CAS PubMed.
- F. Gümüş, İ. Pamuk, T. Özden, S. Yildiz, N. Diril, E. Öksüzoğlu, S. Gür and A. Özkul, Synthesis, characterization and in vitro cytotoxic, mutagenic and antimicrobial activity of platinum(II) complexes with substituted benzimidazole ligands, J. Inorg. Biochem., 2003, 94, 255–262 CrossRef.
- N. Bandyopadhyay, M. Das, A. Samanta, M. Zhu, L. Lu and J. P. Naskar, Promising Antimicrobial Activity of an Oxime Based Palladium(II) Complex, ChemistrySelect, 2017, 2, 230 CrossRef CAS.
- G. P. Radić, V. V. Glodović, I. D. Radojević, O. D. Stefanović, L. R. Čomić, Z. R. Ratković, A. Valkonen, K. Rissanen and S. R. Trifunović, Synthesis, characterization and antimicrobial activity of palladium(II) complexes with some alkyl derivates of thiosalicylic acids: Crystal structure of the bis(S-benzyl-thiosalicylate)-palladium(II) complex, [Pd(S-bz-thiosal)2, Polyhedron, 2012, 31, 69 CrossRef.
- A. M. Mansour, K. Radacki and O. R. Shehab, Half-sandwich triazolato Rh(III) compound of pyridylbenzimidazole ligand with cell selective toxicity towards Cryptococcus neoformans, J. Organomet. Chem., 2021, 949, 121928 CrossRef CAS.
- M. J. Frisch, G. W. Trucks, H. B. Schlegel, G. E. Scuseria, M. A. Robb, J. R. Cheeseman, V. G. Zakrzewski, J. A. Montgomery, R. E. Stratmann, J. C. Burant, S. Dapprich, J. M. Millam, A. D. Daniels, K. N. Kudin, M. C. Strain, O. Farkas, J. Tomasi, V. Barone, M. Cossi, R. Cammi, B. Mennucci, C. Pomelli, C. Adamo, S. Clifford, J. Ochterski, G. A. Petersson, P. Y. Ayala, Q. Cui, K. Morokuma, D. K. Malick, A. D. Rabuck, K. Raghavachari, J. B. Foresman, J. Cioslowski, J. V. Ortiz, A. G. Baboul, B. B. Stefanov, G. Liu, A. Liashenko, P. Piskorz, I. Komaromi, R. Gomperts, R. L. Martin, D. J. Fox, T. Keith, M. A. Al-Laham, C. Y. Peng, A. Nanayakkara, C. Gonzalez, M. Challacombe, P. M. W. Gill, B. G. Johnson, W. Chen, M. W. Wong, J. L. Andres, M. Head-Gordon, E. S. Replogle and J. A. Pople, Gaussian 03 (Revision A.9), Gaussian, Inc., Pittsburgh, 2003 Search PubMed.
- A. Frisch, A. B. Nielson and A. J. Holder, Gaussview User Manual, Gaussian, Inc., Pittsburgh, PA 2000 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1ra06559a |
| This journal is © The Royal Society of Chemistry 2021 |