DOI:
10.1039/D1RA06338C
(Paper)
RSC Adv., 2021,
11, 37649-37660
Natural deep eutectic solvents as tailored and sustainable media for the extraction of five compounds from compound liquorice tablets and their comparison with conventional organic solvents†
Received
21st August 2021
, Accepted 25th October 2021
First published on 25th November 2021
Abstract
An efficient and environmentally friendly ultrasound-assisted (UAE) natural deep eutectic solvent (NADES) extraction method was applied for the extraction of five bioactive compounds (liquiritin, isoliquiritin, liquiritigenin, glycyrrhizic acid and isoliquiritigenin) from compound liquorice tablets (CPLTs), and the antioxidant activities of these compounds were evaluated. In this study, eighteen different NADES systems based on either two or three components were tested and a 1,4-butanediol–levulinic acid system (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 molar ratio) was selected as a topgallant solvent for maximizing analyte extraction yields. Various extraction parameters, such as water content, liquid/solid ratio, extraction time and temperature, were systematically optimized by single-factor and response surface methodology (RSM) experiments. The results indicated that the optimum extraction conditions for the analytes featured a water content of 17%, a liquid/solid ratio of 42 mL g−1 and an extraction time of 30 min. The extracted amounts of liquiritin, isoliquiritin, liquiritigenin, glycyrrhizic acid and isoliquiritigenin reached 5.60, 3.17, 1.27, 74.62 and 1.34 mg g−1, respectively, under optimized conditions, which were much higher than those extracted using conventional organic solvents. In addition, antioxidant tests revealed that the NADES extracts showed higher DPPH and hydroxyl radical-scavenging capacity than the conventional solvent extracts used for comparison. This study provides a suitable approach for efficiently extracting the bioactive compounds of CPLTs. Meanwhile, NADESs can be extended to other natural products as green extraction media.
3 molar ratio) was selected as a topgallant solvent for maximizing analyte extraction yields. Various extraction parameters, such as water content, liquid/solid ratio, extraction time and temperature, were systematically optimized by single-factor and response surface methodology (RSM) experiments. The results indicated that the optimum extraction conditions for the analytes featured a water content of 17%, a liquid/solid ratio of 42 mL g−1 and an extraction time of 30 min. The extracted amounts of liquiritin, isoliquiritin, liquiritigenin, glycyrrhizic acid and isoliquiritigenin reached 5.60, 3.17, 1.27, 74.62 and 1.34 mg g−1, respectively, under optimized conditions, which were much higher than those extracted using conventional organic solvents. In addition, antioxidant tests revealed that the NADES extracts showed higher DPPH and hydroxyl radical-scavenging capacity than the conventional solvent extracts used for comparison. This study provides a suitable approach for efficiently extracting the bioactive compounds of CPLTs. Meanwhile, NADESs can be extended to other natural products as green extraction media.
1. Introduction
The dry root of Glycyrrhiza uralensis Fisch, or liquorice, of the family Leguminosae, is recognized as “the progenitor of herbs”. This plant is widely distributed across northeast and northwest China.1–2 In addition to liver protection3 and antioxidant and anti-aging effects,1 the main pharmacological effects of Glycyrrhiza uralensis Fisch are anti-tussive,4 anti-inflammatory5,6 and antiviral action in traditional and modern medicine;7 hence, it is widely used in the prevention and treatment of various diseases, as well as in foods and cosmetics.8,9 Studies have shown that the main pharmacological bioactive compounds of Glycyrrhiza uralensis Fisch are saponins and flavonoids, among which flavonoids, including liquiritin and liquiritigenin, mainly have anti-cancer,10 anti-inflammatory,11 and immune regulatory12 functions. Meanwhile, saponins also have numerous pharmacological effects, such as anti-inflammatory and anti-viral activities.13 Glycyrrhizic acid is the major chemical constituent of these saponins.14 Nevertheless, the content of some compounds such as isoliquiritin and liquiritigenin is low, which leads to the inability to play a better pharmacological role. Compound liquorice tablets (CPLTs) are listed as anti-tussive expectorants in the Chinese Pharmacopoeia, consist of liquorice-immersed ointment, opium, camphor, star anise oil and sodium benzoate, and are particularly enriched in the bioactive compounds found in Glycyrrhiza uralensis Fisch.15–17 CPLTs are commonly used as apophlegmatic and anti-inflammatory drugs for the symptomatic treatment of various respiratory diseases.18 In CPLTs, a large amount of bioactive compounds of Glycyrrhiza uralensis Fisch play a pharmacological role exhibiting anti-inflammatory and anti-viral action, and other components assist. For example, camphor and anise oil stimulate the bronchial mucosa and enhance cough-reflex sensitivity.16,17 Therefore, not only can the extraction of CPLTs yield high contents of bioactive compounds, but excipients in CPLTs can also enhance the pharmacological action of the extract. The bioactive compounds of CPLTs are usually extracted with conventional organic solvents. According to the Chinese Pharmacopoeia,19 the conventional extract solvent for liquiritin and glycyrrhizic acid was 70% ethanol. Guo et al.20 reported that ultrasonic-assisted 50% methanol has higher extraction efficiency for the extraction of liquiritin and glycyrrhizic acid from CPLTs. However, organic solvents have the disadvantages of volatility, pollution and low biodegradability, which do not conform to the concept of green chemistry.21–25 Thus, it is crucial to develop a green and efficient extraction solvent for CPLTs in order to minimize environmental pollution and cost, and improve safety and health.
Ionic liquids (ILs) can potentially replace organic solvents due to their chemical stability, negligible volatility, high solvency, and favorable thermal properties.26,27 However, ILs have limited application in the pharmaceutical and food industries due to the high synthesis costs and the toxicity of some components.28 To overcome these issues, new solvents with similar physical properties to ILs, which are known as deep eutectic solvents (DESs), were first described by Abbot and his colleagues in 2003. DESs are naturally occurring, safe and inexpensive mixtures of two or more components including choline chloride (ChCl), polyols, organic acids, and sugars.29–32 In addition, DESs have other advantages such as biodegradability, non-toxicity, low cost of production and simple synthesis.33,34 They are widely used in material chemistry, electrochemistry, organic synthesis and extraction processes.35–37 To extend the range of green solvents, Choi et al. have recently described DES-like media that are primarily composed of natural primary metabolites, and named them NADESs.38–40 NADESs exhibit greater biodegradability and compatibility, and lower toxicity than the DESs, and are therefore increasingly replacing DESs and ILs for the extraction of natural products.41–45 However, little is known regarding the application of NADESs for extracting CPLT compounds.
UAE extraction facilitates the release of extractable compounds and the penetration of solvents to plant matrices by disrupting plant cell walls, thereby providing equivalent or higher extraction yields of targeted compounds than conventional extractions do.46 Therefore, in this study, we evaluate the efficiency of UAE extraction of five CPLT bioactive compounds including liquiritin, isoliquiritin, liquiritigenin, glycyrrhizic acid and isoliquiritigenin, using different NADESs. To the best of our knowledge, studies related to the use of UAE-NADESs for the extraction of CPLT have yet to be reported. The molar ratio and water content of the NADESs, liquid/solid ratio, extraction time and temperature were optimized by single-factor and RSM experiments. The latter is a practical tool for optimizing complex extraction procedures, and is used to evaluate multiple parameters and their interactions through mathematical models that avoid the influence of external factors.47 Thereinto, Box–Behnken design (BBD) can effectively design and interpret experiments.48 In addition, the antioxidant activities of different solvent extracts were explored by DPPH and hydroxyl radical-scavenging activity. Overall, the aim of the current study is to develop an efficient and sustainable extraction method based on UAE-NADESs for the extraction of liquiritin, isoliquiritin, liquiritigenin, glycyrrhizic acid and isoliquiritigenin from CPLT and quantification by high-performance liquid chromatography (HPLC), which can be a technical reference for further research.
2. Materials and methods
2.1 Sample of CPLT
The CPLT (Inner Mongolia Yili Pharmaceutical Co. Ltd.) was ground into powder with a particle size of 355 ± 13 μm, and preserved under dry conditions at room temperature away from light.
2.2 Chemicals
ChCl, 1,3-propylene glycol, 1,3-butanediol, 1,4-butanediol, lactic acid, malonic acid, levulinic acid, L-proline and 1,1-dipheny-2-picrylhydrazyl (DPPH) were provided by Shanghai Roen Reagent Co., Ltd (Shanghai, China). Liquiritin (purity 98%), isoliquiritin (purity 98%), liquiritigenin (purity 98%), glycyrrhizic acid (purity 98%) and isoliquiritigenin (purity 98%) were purchased from Chengdu Alfa Biotechnology Co., Ltd (Chengdu, China). Methanol, ethanol, acetonitrile and phosphoric acid (all HPLC grade) were obtained from MREDA technology (USA). Deionized water was obtained through a Milli-Q water facility (Themo, USA). All samples were filtered through a 0.45 μm membrane (MEMBRANA, Germany) before being injected into the HPLC system.
2.3 HPLC analysis
2.3.1 HPLC conditions. HPLC was performed using a Supersil ODS2 5 μm column (4.6 mm × 250 mm, 5 μm). The mobile phase consisted of acetonitrile (mobile phase A) and 0.05% phosphoric acid aqueous solution (mobile phase B) at a flow rate of 1 mL min−1. The gradient elution is as follows: 0–8 min, 81% B; 8–30 min, 81–57%; 30–33.5 min, 57% B; 33.5–40 min, 57–55% B. The temperature of the column was maintained at 25 °C, the injection volume was 10 μL and samples were detected at 237 nm for liquiritin, liquiritigenin, glycyrrhizic acid and isoliquiritigenin, and 360 nm for isoliquiritin.
2.3.2 Preparation of standard curve. Liquiritin (9.87 mg), isoliquiritin (10.64 mg), liquiritigenin (9 mg), glycyrrhizic acid (25.89 mg) and isoliquiritigenin (9.43 mg) standards were accurately weighed and respectively transferred to a 10 mL volumetric flask and diluted as a stock solution. Then, 1.5, 0.8, 0.4, 6.9 and 0.4 mL of liquiritin, isoliquiritin, liquiritigenin, glycyrrhizic acid and isoliquiritigenin stock solution were transferred to a 10 mL volumetric flask as mixed standard solutions. The mixed standard solution was diluted to get a battery of concentrations for developing the calibration curves.
2.4 Preparation of NADESs
NADESs were synthesized by the heating method according to the previously published procedure.49,50 Different components were mixed in the appropriate molar ratio and heated at 80 °C for 20–60 min with constant stirring until a homogeneous liquid was obtained. For the first screening experiment, 20% (v/v) of water was added to each NADES. Eighteen NADESs were synthesized, and they are summarized in Table 1.
Table 1 Composition of different NADESs
| Abbreviation |
Component 1 |
Component 2 |
Component 3 |
Mole ratio |
| NADES-1 |
ChCl |
Lactic acid |
|
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 2 |
| NADES-2 |
Levulinic acid |
|
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 2 |
| NADES-3 |
Malonic acid |
|
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 2 |
| NADES-4 |
1,3-Propylene glycol |
|
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 2 |
| NADES-5 |
1,3-Butanediol |
|
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 2 |
| NADES-6 |
1,4-Butanediol |
|
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 2 |
| NADES-7 |
L-Proline |
Levulinic acid |
|
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 2 |
| NADES-8 |
Malonic acid |
|
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 2 |
| NADES-9 |
1,3-Butanediol |
|
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 2 |
| NADES-10 |
1,4-Butanediol |
|
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 2 |
| NADES-11 |
1,4-Butanediol |
Malonic acid |
|
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 2 |
| NADES-12 |
Levulinic acid |
|
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 2 |
| NADES-13 |
1,3-Butanediol |
Malonic acid |
|
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 2 |
| NADES-14 |
Levulinic acid |
|
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 2 |
| NADES-15 |
Choline chloride |
1,4-Butanediol |
Levulinic acid |
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
| NADES-16 |
1,4-Butanediol |
Malonic acid |
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
| NADES-17 |
L-Proline |
1,4-Butanediol |
Levulinic acid |
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
| NADES-18 |
1,4-Butanediol |
Malonic acid |
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
2.5 Optimization of the extraction procedure
Different types of NADESs were tested under the same initial screening condition (molar ratio, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2; water content, 20%; liquid/solid ratio, 15 mL g−1; extraction time, 20 min; extraction temperature, 50 °C). First, 0.3 g of CPLT sample was extracted with 4.5 mL solvents in a conical flask. The mixture was vortexed for 30 s, and ultrasonicated for 40 min at room temperature. The extracts obtained were then centrifuged at 5000 rpm for 10 min. The suspension was diluted with water and filtered through a 0.45 μm membrane before testing. Each extraction was performed in triplicate, and the extraction yield was calculated as follows:
2; water content, 20%; liquid/solid ratio, 15 mL g−1; extraction time, 20 min; extraction temperature, 50 °C). First, 0.3 g of CPLT sample was extracted with 4.5 mL solvents in a conical flask. The mixture was vortexed for 30 s, and ultrasonicated for 40 min at room temperature. The extracts obtained were then centrifuged at 5000 rpm for 10 min. The suspension was diluted with water and filtered through a 0.45 μm membrane before testing. Each extraction was performed in triplicate, and the extraction yield was calculated as follows:
where Y is the extraction yield, C is the concentration of analytes, N is the diluted multiples, V is the sample solution volume and M is the mass of the sample.
The optimization of the extraction procedure parameters including optimal NADES molar ratios of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 1
1, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2, 1
2, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3, 1
3, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 or 1
4 or 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5, water contents of 10, 20, 30, 40 or 50%, liquid/solid ratios of 10, 20, 30, 40 or 50 mL g−1, extraction time periods of 10, 20, 30, 40, 50, 60 or 70 min, and extraction temperatures of 30, 40, 50, 60 or 70 °C was carried out based on single-factor experiments. When examining the effect of one factor on the extraction yield, the other factors were maintained immobile.
5, water contents of 10, 20, 30, 40 or 50%, liquid/solid ratios of 10, 20, 30, 40 or 50 mL g−1, extraction time periods of 10, 20, 30, 40, 50, 60 or 70 min, and extraction temperatures of 30, 40, 50, 60 or 70 °C was carried out based on single-factor experiments. When examining the effect of one factor on the extraction yield, the other factors were maintained immobile.
2.6 Experimental design
On the basis of single-factor experiments, three-level and three-factor BBD was performed in order to obtain optimum extraction conditions. The three main independent variables were water content (X1), liquid/solid ratio (X2) and extraction time (X3), and the extraction yield of five bioactive compounds was the dependent variable of the equation. The experimental design of BBD is outlined in Table 2.
Table 2 Independent variables and levels used for BBD
| Variables |
Level |
| −1 |
0 |
1 |
| Water content (%) |
10 |
20 |
30 |
| Liquid/solid ratio (mL g−1) |
30 |
40 |
50 |
| Extraction time (min) |
20 |
30 |
40 |
2.7 Comparison between the optimal NADES and conventional organic solvents
2.7.1 Comparison of extraction efficiency. The extraction capacities of the optimal NADES and conventional organic solvents were compared under optimal conditions in terms of the CPLT yield. According to the Chinese Pharmacopoeia,19 the conventional extract solvent for analytes was 70% ethanol. Hence, 70% ethanol was selected for the comparison.
2.7.2 Comparison of antioxidant activity.
2.7.2.1 DPPH radical-scavenging activity. DPPH radical scavenging capability was investigated to compare the antioxidant activity of CPLT extracted by the optimal NADES and 70% ethanol according to the results previously reported in the literature.51,52 Briefly, the desired sample (0.1 mL) was mixed with 0.1 mL DPPH solution (0.2 mM) dissolved in ethanol, and shaken and incubated at 25 °C for 30 min in the dark. The absorbance of the samples was measured at 517 nm. The DPPH radical-scavenging activity (%) was calculated according to the following equation:
| DPPH radical-scavenging activity (%) = [1 − (A − A0)/A1] × 100% |
where A and A1 are the desired samples replaced with the CPLT extract and extraction solvents respectively, and A0 is the DPPH solution replaced with ethanol.
2.7.2.2 Hydroxyl radical-scavenging activity. The hydroxyl assay was used to determine the radical-scavenging activity of the CPLT extract of the optimal NADES and 70% ethanol. As described previously,53,54 1,10-phenanthroline solution (1 mL, 7.5 mM) was mixed with twice the volume of PBS (0.2 mM, pH 7.4). After adding 1 mL each of the desired sample, FeSO4 (7.5 mM, pH 7.4) and H2O2, the samples were mixed and fully incubated at 37 °C for 30 min. Finally, the absorbance of the samples was measured at 536 nm. The hydroxyl radical-scavenging activity (%) was calculated according to the following equation:
| Hydroxyl radical-scavenging activity (%) = (A − A1)/(A2 − A1) × 100% |
where A and A1 are the desired samples replaced with the CPLT extract and extraction solvents respectively, and A0 is H2O2 replaced with deionized water.
2.8 Statistical analysis
In this study, all experiments were performed in triplicate, and the experimental data were expressed as the mean ± SD of three parallel measurements. The effects of parameters on extraction efficiency were analysed via a one-way analysis of variance (ANOVA) test (p < 0.05 significance level; SPSS software v. 17). All diagrams were drawn using the Origin 2018. The SD is indicated as error bars in the figures. Design Expert (version 12.0) was used for the BBD experimental design and statistical analysis.
3. Results and discussion
3.1 HPLC analysis
The five compounds of CPLT extracts of conventional solvent and NADES were better separated under optimized chromatographic conditions, and the chromatograms are shown in Fig. 1. The calibration curve between the concentration (X) of the standard solution and the corresponding peak area (Y) was established (Table S1†). The calibration curves of the five components showed good linearity with correlation coefficients (R2) larger than 0.9991.
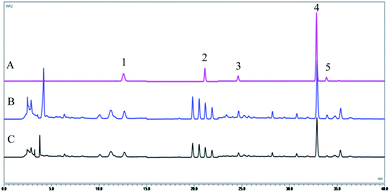 |
| | Fig. 1 HPLC chromatograms of mixed standard solutions (A), CPLT extract![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) NADES (B) and 70% ethanol (C). (1) liquiritin; (2) isoliquiritin; (3) liquiritigenin; (4) glycyrrhizic acid; (5) isoliquiritigenin. The detection wavelength was set at 237 (liquiritin, liquiritigenin, glycyrrhizic acid and isoliquiritigenin) and 360 (isoliquiritin) nm. NADES (B) and 70% ethanol (C). (1) liquiritin; (2) isoliquiritin; (3) liquiritigenin; (4) glycyrrhizic acid; (5) isoliquiritigenin. The detection wavelength was set at 237 (liquiritin, liquiritigenin, glycyrrhizic acid and isoliquiritigenin) and 360 (isoliquiritin) nm. | |
3.2 Optimization of the NADES extraction parameters
3.2.1 Extraction efficiency of different NADESs. Studies show that acid-, alcohol- and amide-based NADESs have superior extraction efficiencies to sugar-based solvents for most compounds.48,55 Therefore, we synthesized eighteen NADESs with ChCl, L-proline, acids and alcohols and extracted compounds (n = 3, p < 0.05). Owing to the high viscosity of NADESs, 20% (w/w) water was added for preferable extraction fields56,57 (Fig. 2). It is obvious that the extraction capacity of the tested NADESs varied greatly depending on the type of hydrogen bond donors (HBDs) and hydrogen bond acceptors (HBAs). For liquiritin, isoliquiritin and liquiritigenin, NADES-1, NADES-2 and NADES-3 that consisted of ChCl and acids exhibited generally better extraction yields compared to the other NADESs. However, compared with NADESs that consisted of ChCl and acids, the yield of the less polar compounds glycyrrhizic acid and isoliquiritigenin were higher with NADESs composed of ChCl and alcohols (NADES-4 and NADES-6). In addition, as HBA for NADESs, the extraction efficiency of most choline chloride is higher than L-proline. Based on the results, two or three components of acid–alcohol based NADESs were synthesized (NADES-11–NADES-18). Comparison of all NADESs showed that NADES-12 and NADES-14 exhibited optimum yields of different polarities. Among the total yields of the five targeted compounds of NADES-12 is 80.41 ± 0.04 mg g−1, which was higher than that amount of NADES-14 (69.16 ± 0.31 mg g−1). The components of NADESs affect not only their physicochemical properties such as polarity, viscosity and solubilization ability, but also hydrogen bond interactions between the solvent and analytes. Based on the above-mentioned results, NADES-12 synthesized with 1,4-butanediol and levulinic acid was selected as the optimal solvent for the subsequent experiments.
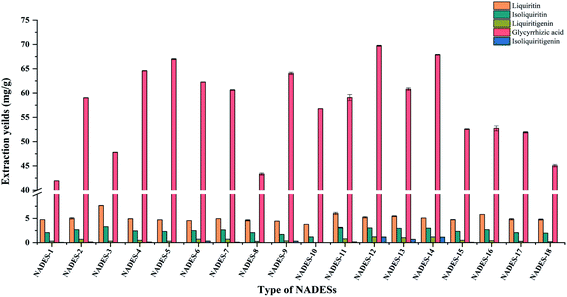 |
| | Fig. 2 The effect of different types of NADESs on the extraction yield of the five targeted compounds. | |
3.2.2 Extraction effects of different molar ratios of NADES. The extraction efficiency of NADES-12 of different molar ratios (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 1
1, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2, 1
2, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3, 1
3, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4, and 1
4, and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5) was evaluated (n = 3, p > 0.05). As shown in Fig. 3A, it is apparent that the yield of all five compounds increased as the molar ratio was raised from 1
5) was evaluated (n = 3, p > 0.05). As shown in Fig. 3A, it is apparent that the yield of all five compounds increased as the molar ratio was raised from 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 to 1
1 to 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3, most likely due to enhanced hydrogen bond formation. However, a higher molar ratio decreased the yield since the increase in HBDs resulted in considerable steric hindrance, which prevented interactions between the targeted compounds and HBAs.48,58,59 Hence, 1,4-butanediol and levulinic acid at a molar ratio of 1
3, most likely due to enhanced hydrogen bond formation. However, a higher molar ratio decreased the yield since the increase in HBDs resulted in considerable steric hindrance, which prevented interactions between the targeted compounds and HBAs.48,58,59 Hence, 1,4-butanediol and levulinic acid at a molar ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 were used for subsequent experiments.
3 were used for subsequent experiments.
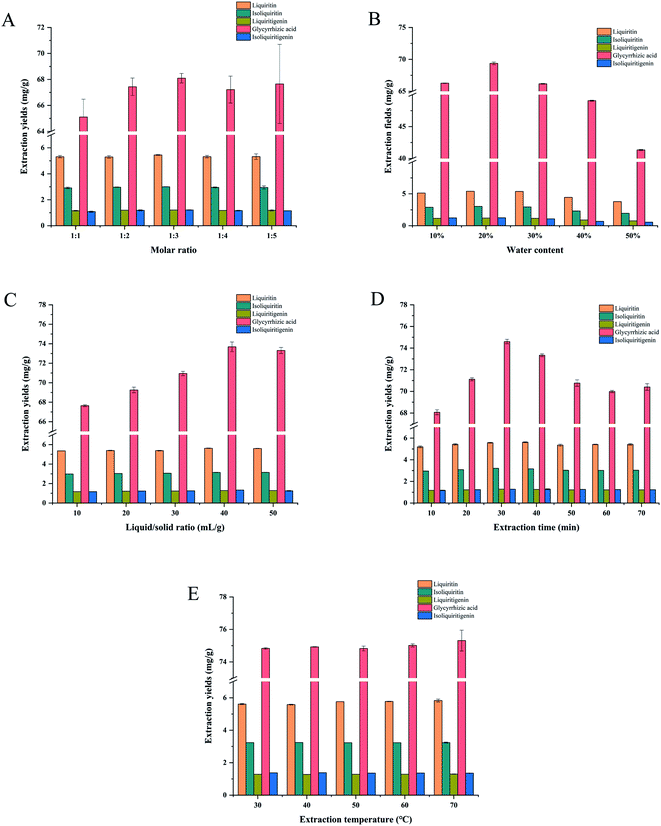 |
| | Fig. 3 The effect of molar ratio (A), water content (B), liquid/solid ratio (C), extraction time (D) and extraction temperature (E) on the extraction yield of the five targeted compounds. | |
3.2.3 Extraction effect of different water content in NADES. We next compared the extraction efficiency of NADES-12 (molar ratio 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3) with 10, 20, 30, 40 and 50% water content (n = 3, p < 0.05). Fig. 3B shows that the extraction efficiency for all compounds increased significantly when the water content was raised from 10 to 20%, which could be attributed to the corresponding decrease in viscosity that promoted the formation of an ordered hydrogen bond network. However, high water contents (30, 40, and 50%) can limit the interactions between the solute and solvents, and thus, lower the extraction efficiency.60–62 Based on these results, NADES-12 with 20% water was used for the subsequent experiments.
3) with 10, 20, 30, 40 and 50% water content (n = 3, p < 0.05). Fig. 3B shows that the extraction efficiency for all compounds increased significantly when the water content was raised from 10 to 20%, which could be attributed to the corresponding decrease in viscosity that promoted the formation of an ordered hydrogen bond network. However, high water contents (30, 40, and 50%) can limit the interactions between the solute and solvents, and thus, lower the extraction efficiency.60–62 Based on these results, NADES-12 with 20% water was used for the subsequent experiments.
3.2.4 Extraction effect of different liquid/solid ratios. The effect of liquid/solid ratio ranging from 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 to 50
1 to 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 on the extraction efficiency of the NADES-12 was examined (n = 3, p < 0.05). As shown in Fig. 3C, the extraction yield of the solute increased significantly with the increase in solvent volume and peaked at the liquid/solid ratio of 40 mL g−1, before plateauing and then decreasing. This is likely due to the fact that excess solvent volume increased dissolution of impurities along with that of targeted analytes, which eventually hindered the dissolution of the latter.47,63 Jing et al.64 similarly reported that the solvent volume above a certain threshold can inhibit analyte dissolution due to the presence of impurities.
1 on the extraction efficiency of the NADES-12 was examined (n = 3, p < 0.05). As shown in Fig. 3C, the extraction yield of the solute increased significantly with the increase in solvent volume and peaked at the liquid/solid ratio of 40 mL g−1, before plateauing and then decreasing. This is likely due to the fact that excess solvent volume increased dissolution of impurities along with that of targeted analytes, which eventually hindered the dissolution of the latter.47,63 Jing et al.64 similarly reported that the solvent volume above a certain threshold can inhibit analyte dissolution due to the presence of impurities.
3.2.5 Extraction effect of different extraction time periods. Extraction durations of 10 to 70 min were tested, and as shown in Fig. 3D, there was a strong correlation between the extraction efficiency and time (n = 3, p < 0.05). There was a significant time-dependent increase in the extraction yield within 30 min, whereas the prolonged extraction process decreased the yield. Acoustic cavitation and mechanical agitation due to ultrasound effectively broke down the plant cytoderm and allowed dissolution of the active compounds. However, prolonged sonication can alter or even decompose the target compounds, resulting in lower yields.60,62,65 Thus, the targeted compounds could be completely extracted within 30 min and the condition was used for subsequent experiments.
3.2.6 Extraction effect of different extraction temperatures. To determine the optimal extraction temperature, the samples were extracted in the range of 30–70 °C (n = 3, p > 0.05). The results are outlined in Fig. 3E, and the extraction yield did not increase noticeably with the increase in temperature.
3.3 Optimization of extraction conditions by RSM
3.3.1 Statistical analysis and model fitting. Based on the results of single-factor experiments, the content of water in NADES-12 (10–30%), the liquid/solid ratio (30–50 mL g−1) and extraction time (20–40 min) were selected as independent variables and extraction yields of five compounds were taken as the response value. BBD was used to optimize the extraction conditions for five compounds in CPLT with three conditions and three levels. The data obtained from the 17-run experiments are summarized in Table 3. On the basis of multivariate regression analysis of experimental data, the second-order polynomial equation of five compounds was obtained as follows:
| Yliquiritin = 5.62 − 0.0503X1 + 0.0688X2 + 0.0197X3 − 0.0411X1X2 + 0.0079X1X3 + 0.0010X2X3 − 0.2411X12 − 0.1239X22 − 0.0660X32 |
| Yisoliquiritin = 3.13 − 0.0555X1 + 0.0115X2 − 0.0088X3 − 0.0194X1X2 + 0.0049X1X3 − 0.0004X2X3 − 0.1089X12 − 0.0551X22 − 0.0477X32 |
| Yliquiritigenin = 1.26 − 0.0300X1 + 0.0063X2 + 0.0008X3 − 0.0080X1X2 + 0.0043X1X3 + 0.0003X2X3 − 0.0387X12 − 0.0298X22 − 0.0204X32 |
| Yglycyrrhizic acid = 73.11 − 2.41X1 + 0.6047X2 − 0.2159X3 − 0.2620X1X2 + 0.1645X1X3 − 0.0622X2X3 − 3.08X12 − 1.49X22 − 1.04X32 |
| Yisoliquiritigenin = 1.30 − 0.0918X1 + 0.0248X2 + 0.0087X3 + 0.0037X1X2 + 0.0023X1X3 − 0.0117X2X3 − 0.0660X12 − 0.0466X22 − 0.0073X32 |
Y is the extraction yield of the different analytes (mg g−1), X1 is the content of water in NADES-12 (%), X2 is the liquid/solid ratio (mL g−1) and X3 is the extraction time (min).
Table 3 Experimental conditions for BBD and the corresponding responses
| Run |
X1 |
X2 |
X3 |
Yliquiritin (mg g−1) |
Yisoliquiritin (mg g−1) |
Yliquiritigenin (mg g−1) |
Yglycyrrhizic acid (mg g−1) |
Yisoliquiritigenin (mg g−1) |
| 1 |
1 |
1 |
0 |
5.26 |
2.92 |
1.16 |
66.80 |
1.14 |
| 2 |
0 |
−1 |
−1 |
5.36 |
3.02 |
1.20 |
70.14 |
1.21 |
| 3 |
0 |
0 |
0 |
5.68 |
3.15 |
1.27 |
73.38 |
1.31 |
| 4 |
0 |
0 |
0 |
5.58 |
3.11 |
1.26 |
72.99 |
1.30 |
| 5 |
1 |
0 |
1 |
5.28 |
2.90 |
1.17 |
66.22 |
1.14 |
| 6 |
0 |
−1 |
1 |
5.39 |
3.02 |
1.20 |
69.96 |
1.26 |
| 7 |
0 |
1 |
1 |
5.50 |
3.03 |
1.21 |
70.91 |
1.27 |
| 8 |
0 |
1 |
−1 |
5.47 |
3.04 |
1.21 |
71.33 |
1.27 |
| 9 |
0 |
0 |
0 |
5.56 |
3.13 |
1.25 |
73.04 |
1.28 |
| 10 |
−1 |
−1 |
0 |
5.16 |
2.97 |
1.20 |
69.77 |
1.25 |
| 11 |
0 |
0 |
0 |
5.66 |
3.13 |
1.26 |
73.31 |
1.33 |
| 12 |
1 |
0 |
−1 |
5.21 |
2.92 |
1.16 |
66.46 |
1.13 |
| 13 |
0 |
0 |
0 |
5.61 |
3.12 |
1.25 |
72.83 |
1.31 |
| 14 |
−1 |
0 |
1 |
5.39 |
3.02 |
1.23 |
71.21 |
1.33 |
| 15 |
−1 |
0 |
−1 |
5.36 |
3.06 |
1.24 |
72.10 |
1.32 |
| 16 |
1 |
−1 |
0 |
5.17 |
2.92 |
1.16 |
65.97 |
1.07 |
| 17 |
−1 |
1 |
0 |
5.41 |
3.05 |
1.23 |
71.64 |
1.31 |
As shown in Table 4, the coefficients of determination (R2) of the quadratic models of five analytes were 0.9674, 0.9822, 0.9699, 0.9928 and 0.9835, suggesting that these models can be used to describe the response of the experiment pertaining to compounds. The experimental data was in agreement with the prediction model (Fig. 4). The high F-value and the small p-value mean significant corresponding variables. The F-values of the five compounds respectively were 23.09, 42.82, 25.06, 107.44 and 46.42 respectively, and all p-values were less than 0.0005, indicating that these models are highly significant. Moreover, the values of “lack of fit” of five quadratic models were not significant, indicating good fitting. Furthermore, X1 and X12 were significant for all models, X2 and X22 were significant for most models, whereas X32 was partly significant. Accordingly, the water content of NADES-12 had the highest influence on the extraction efficiency, followed by the liquid/solid ratio and extraction time.
Table 4 Analysis of the variance of the experimental results of the BBD
| Variables |
Liquiritin |
Isoliquiritin |
Liquiritigenin |
Glycyrrhizic acid |
Isoliquiritigenin |
| Mean square |
F-Value |
p-Value |
Mean square |
F-Value |
p-Value |
Mean square |
F-Value |
p-Value |
Mean square |
F-Value |
p-Value |
Mean square |
F-Value |
p-Value |
| Model |
0.0470 |
23.09 |
0.0002 |
0.0119 |
42.82 |
<0.0001 |
0.0023 |
25.06 |
0.0002 |
12.05 |
107.44 |
<0.0001 |
0.0015 |
46.42 |
<0.0001 |
| X1 |
0.0202 |
9.92 |
0.0162 |
0.0246 |
88.62 |
<0.0001 |
0.0072 |
77.46 |
<0.0001 |
46.36 |
413.51 |
<0.0001 |
0.0674 |
272.75 |
<0.0001 |
| X2 |
0.0378 |
18.57 |
0.0035 |
0.0011 |
3.83 |
0.0912 |
0.0003 |
3.37 |
0.1091 |
2.93 |
26.10 |
0.0014 |
0.0049 |
19.92 |
0.0029 |
| X3 |
0.0031 |
1.53 |
0.2562 |
0.0006 |
2.23 |
0.1786 |
4.805 × 10−6 |
0.0518 |
0.8265 |
0.3728 |
3.33 |
0.1110 |
0.0006 |
2.42 |
0.1637 |
| X1X2 |
0.0068 |
3.33 |
0.1110 |
0.0015 |
5.40 |
0.0531 |
0.0003 |
2.78 |
0.1396 |
0.2745 |
2.45 |
0.1616 |
0.0001 |
0.2185 |
0.6544 |
| X1X3 |
0.0002 |
0.1218 |
0.7374 |
0.0001 |
0.3489 |
0.5733 |
0.0001 |
0.7878 |
0.4042 |
0.1083 |
0.9658 |
0.3585 |
0.0000 |
0.0856 |
0.7783 |
| X2X3 |
3.802 × 10−6 |
0.0019 |
0.9667 |
6.400 × 10−7 |
0.0023 |
0.9631 |
3.025 × 10−7 |
0.0033 |
0.9561 |
0.0155 |
0.1380 |
0.7212 |
0.0005 |
2.21 |
0.1803 |
| X12 |
0.2448 |
120.20 |
<0.0001 |
0.0499 |
179.48 |
<0.0001 |
0.0063 |
67.95 |
<0.0001 |
39.48 |
355.34 |
<0.0001 |
0.0183 |
74.15 |
<0.0001 |
| X22 |
0.0647 |
31.75 |
0.0008 |
0.0128 |
46.05 |
0.0003 |
0.0038 |
40.42 |
0.0004 |
9.31 |
83.06 |
<0.0001 |
0.0092 |
37.04 |
0.0005 |
| X32 |
0.0183 |
9.01 |
0.0199 |
0.0096 |
34.52 |
0.0006 |
0.0018 |
18.97 |
0.0033 |
4.51 |
40.27 |
0.0004 |
0.0002 |
0.8982 |
0.3748 |
| Lack of fit |
0.0012 |
0.4378 |
0.7383 |
0.0004 |
1.72 |
0.3008 |
0.0001 |
1.12 |
0.4413 |
0.1905 |
3.57 |
0.1252 |
0.0001 |
0.3882 |
0.7685 |
| R2 |
0.9674 |
0.9822 |
0.9699 |
0.9928 |
0.9835 |
| Adjusted R2 |
0.9255 |
0.9592 |
0.9312 |
0.9132 |
0.9623 |
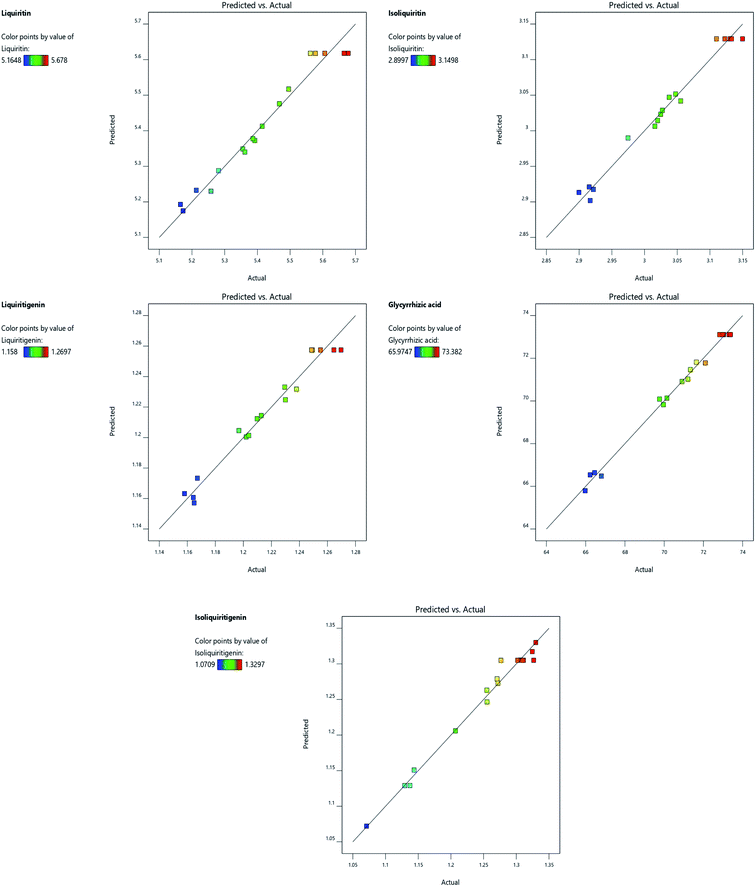 |
| | Fig. 4 Correlation graphs of predicted values and actual yields of the five targeted compounds. | |
3.3.2 Response surface analysis. Three-dimensional (3D) response surface plots were used to determine the interaction between independent variables.35 Higher gradients indicated significant interaction between two factors. For Fig. 5(A)–9(A), the extraction yields of the analytes increased with the increase in the water content, and declined when the latter exceeded 17%. This is likely due to the fact that excessive water can alter the hydrogen bonding network of the NADESs. The extraction time had a lower impact than the water content, and the yield of the compounds did not increase after 30 min (Fig. 5(B)–9(B)). With the optimum extraction time, the yield increased with the increase in liquid/solid ratio and peaked at 42 mL g−1 before declining (Fig. 5(C)–9(C)). It was because that high amounts of the solvent quenched the ultrasonic energy and influenced extraction efficiency. The predicted maximum extraction yields of liquiritin, isoliquiritin, liquiritigenin, glycyrrhizic acid and isoliquiritigenin were 5.62, 3.14, 1.26, 73.66 and 1.33 mg g−1 respectively at 16.625% water content, 42.260 mL g−1 liquid/solid ratio and 29.794 min extraction time. For the convenience of practical operation, we selected 17% water content, 42 mL g−1 liquid/solid ratio and 30 min extraction time for extraction. Under these conditions, the extraction yields of the five compounds were 5.60, 3.17, 1.27, 74.62 and 1.34 mg g−1, respectively, and the relative standard deviation (RSD) values were less than 0.9%.
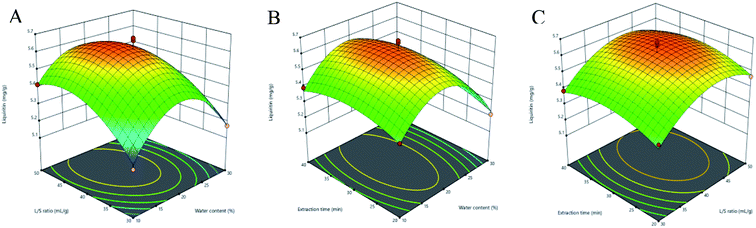 |
| | Fig. 5 Three-dimensional response surface plots of liquiritin. (A) Water content and liquid/solid ratio; (B) water content and extraction time; (C) liquid/solid ratio and extraction time. | |
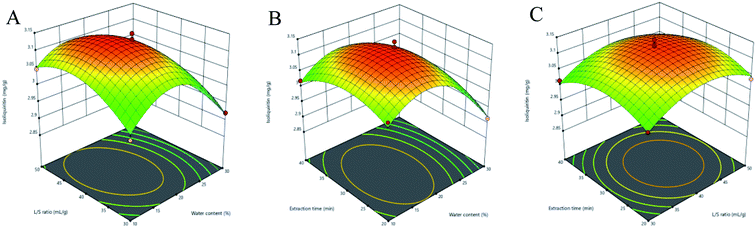 |
| | Fig. 6 Three-dimensional response surface plots of isoliquiritin. (A) Water content and liquid/solid ratio; (B) water content and extraction time; (C) liquid/solid ratio and extraction time. | |
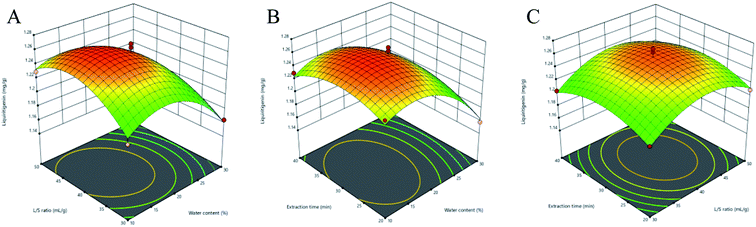 |
| | Fig. 7 Three-dimensional response surface plots of liquiritigenin. (A) Water content and liquid/solid ratio; (B) water content and extraction time; (C) liquid/solid ratio and extraction time. | |
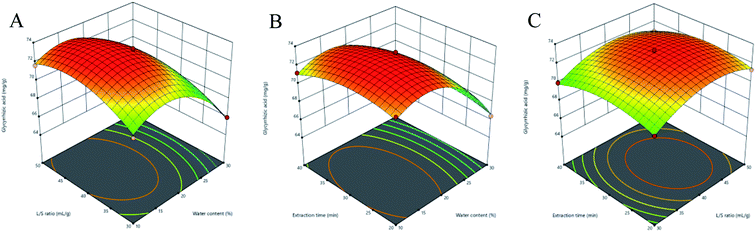 |
| | Fig. 8 Three-dimensional response surface plots of glycyrrhizic acid. (A) Water content and liquid/solid ratio; (B) water content and extraction time; (C) liquid/solid ratio and extraction time. | |
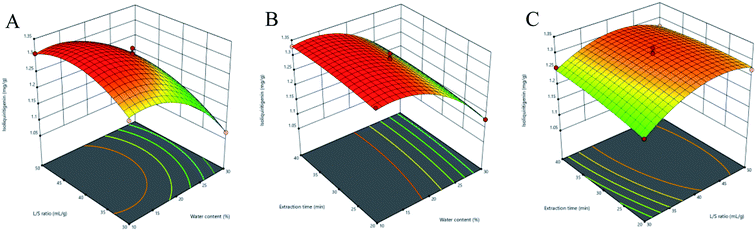 |
| | Fig. 9 Three-dimensional response surface plots of isoliquiritigenin. (A) Water content and liquid/solid ratio; (B) water content and extraction time; (C) liquid/solid ratio and extraction time. | |
3.4 Comparison of NADESs with a conventional extraction solvent
3.4.1 Comparison of extraction efficiencies. In order to evaluate the extraction efficiency of the optimal NADES, the extraction efficiencies of the optimal NADES and 70% ethanol were compared, and the results are outlined in Fig. 10A. It could be found that the extraction efficiency of NADES-12 was significantly higher than that of 70% ethanol (p < 0.05). Therefore, NADESs were a more efficient alternative solvent for extracting bioactive compounds from CPLT.
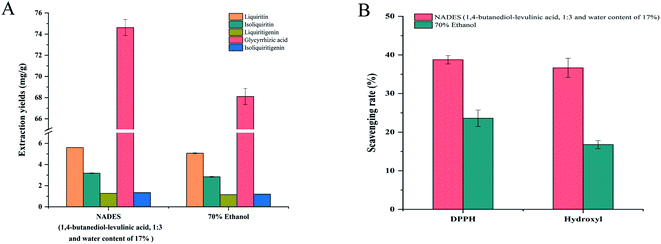 |
| | Fig. 10 Comparison of the extract of NADES-12 and 70% ethanol for extraction efficiency (A) and DPPH and hydroxyl radical-scavenging activity (B). | |
3.4.2 Comparison of antioxidant activities. Besides extraction ability, the antioxidant activity of the NADES-12 extract was another evaluation criterion for the feasibility of NADES-12 as a green solvent. Therefore, we next examined DPPH and hydroxyl radical-scavenging capacity to compare the antioxidant activity of NADES-12 and 70% ethanol extracts. Indeed, the NADES-12 extracts showed superior radical-scavenging capacity to 70% ethanol extracts under the same conditions (p < 0.05) (Fig. 10B), probably because of the higher extraction efficiency of the NADES-12 system.66 Another hypothesis is that NADES-12 components also have certain antioxidant capacity as natural primary metabolites. A study conducted by N. M. Woo et al.67 highlighted that DES (L-proline![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) glycerol) had antioxidant activity, which was an important reason why the antioxidant activity of DES extracts was significantly higher than that of methanol. On this basis, DPPH and hydroxyl radical assays of pure NADES (1,4-butanediol
glycerol) had antioxidant activity, which was an important reason why the antioxidant activity of DES extracts was significantly higher than that of methanol. On this basis, DPPH and hydroxyl radical assays of pure NADES (1,4-butanediol![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) levulinic acid) were performed under the same conditions, which also displayed results consistent with our hypothesis. DPPH and hydroxyl radical-scavenging capacities of pure NADES-12 were 15.69 ± 0.69% and 14.23 ± 0.53% respectively. The results indicated that pure NADES-12 had antioxidant effects, which had an additive effect on the antioxidant activity of the targeted extract. Taken together, NADESs is a more efficient alternative solvent for extracting bioactive compounds from CPLT in terms of both the yield and the antioxidant activity of the extract.
levulinic acid) were performed under the same conditions, which also displayed results consistent with our hypothesis. DPPH and hydroxyl radical-scavenging capacities of pure NADES-12 were 15.69 ± 0.69% and 14.23 ± 0.53% respectively. The results indicated that pure NADES-12 had antioxidant effects, which had an additive effect on the antioxidant activity of the targeted extract. Taken together, NADESs is a more efficient alternative solvent for extracting bioactive compounds from CPLT in terms of both the yield and the antioxidant activity of the extract.
3.5 Application and prospects
NADESs have attracted great attention from the scientific community due to their advantages of simple and easy synthesis, insignificant volatility, chemical and thermal stability, non-flammability, highly biodegradable and low toxicity. Moreover, the components of NADESs are abundantly present in nature, have a low cost and are biorenewable.44,50,68 At present, NADESs have been widely used for the extraction of bioactive compounds from traditional Chinese medicine. Compared with traditional organic solvents, NADESs have higher extraction efficiency and lower cost. Another major advantage is that since NADESs are generally composed of natural primary metabolites, they may be directly incorporated into drugs, food formulations and so on without additional purification steps.39,45 In addition, NADESs are also widely used in the extraction and separation of residual drugs and metal ions and increase of bioavailability of compounds.69 Therefore, NADESs can be applied for large-scale extraction of bioactive compounds from traditional Chinese medicine as extraction solvents without economic issues, which meet the principles of green chemistry.
4. Conclusions
In this study, a green, efficient and simple UAE-NADES method has been developed for the extraction of five bioactive compounds from CPLTs. NADESs were synthesized and applied as tailored solvents for the CPLT. Single-factor and RSM-BBD experiments were applied for optimizing extraction parameters to maximize the extraction efficiency of the five compounds (liquiritin, isoliquiritin, liquiritigenin, glycyrrhizic acid and isoliquiritigenin). The optimal experimental conditions are as follows: NADES system, 1,4-butanediol–levulinic acid (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 molar ratio); NADES-12 water content, 17%; liquid/solid ratio, 42 mL g−1 and extraction time, 30 min. Under the optimal extraction conditions, the NADES-12 extract exhibited higher extraction efficiency and in vitro antioxidant activity than the conventional solvent extract. The developed method is a promising strategy for the extraction of bioactive compounds, which provides high efficiency for the natural product. The proposed method is proved to be efficient, simple, environmentally friendly and sustainable for obtaining five bioactive compounds from the CPLT, which contributes to the urgent need of research on the green extraction of compounds in Chinese herbal medicines.
3 molar ratio); NADES-12 water content, 17%; liquid/solid ratio, 42 mL g−1 and extraction time, 30 min. Under the optimal extraction conditions, the NADES-12 extract exhibited higher extraction efficiency and in vitro antioxidant activity than the conventional solvent extract. The developed method is a promising strategy for the extraction of bioactive compounds, which provides high efficiency for the natural product. The proposed method is proved to be efficient, simple, environmentally friendly and sustainable for obtaining five bioactive compounds from the CPLT, which contributes to the urgent need of research on the green extraction of compounds in Chinese herbal medicines.
Author contributions
Jia-ni Dong and Guo-dong Wu: responsible for establishing the HPLC method, the synthesis of the NADESs and wrote the manuscript and revised the manuscript. Zhi-qiang Dong, Dan Yang and Yu-kun Bo: responsible for data collection and statistical analysis. Ming An and Long-shan Zhao: designed the experiments, supervised, and participated in the entire work. All the authors revised and approved the final manuscript.
Conflicts of interest
The authors declare no competing financial interests.
Acknowledgements
This study was funded by grants from the Science and Technology Innovation Guidance Project of Inner Mongolia Autonomous Region (No. 00120209), the Natural Science Foundation of Inner Mongolia Autonomous Region, China (2021MS08011).
References
- G. D. Maria Pia, F. Sar, F. Mario and S. Lorenza, Mini-Rev. Med. Chem., 2019, 19(8), 647–656 CrossRef CAS PubMed.
- T.-T. Li, S.-Y. Hua, J.-H. Ma, L. Dong, F. Xu and X.-Y. Fu, J. Anal. Methods Chem., 2020, 2020, 8838290 Search PubMed.
- S.-H. Gou, M. He, B.-B. Li, N.-Y. Zhu and J.-M. Ni, Nat. Prod. Res., 2020, 1–5 CrossRef PubMed.
- M.-Y. Jiang, S.-J. Zhao, S.-S. Yang, X. Lin, X.-G. He, X.-Y. Wei, Q. Song, R. Li, C.-M. Fu, J.-M. Zhang and Z. Zhang, J. Ethnopharmacol., 2020, 249, 112439 CrossRef CAS PubMed.
- R. Yang, B.-C. Yuan, Y.-S. Ma, S. Zhou and Y. Liu, Pharm. Biol., 2017, 55(1), 5–18 CrossRef CAS PubMed.
- L. Yin, E.-S. Guan, Y.-B. Zhang, Z.-H. Shu, B. Wang, X.-L. Wu, J. Chen, J.-X. Liu, X.-Y. Fu, W.-H. Sun and M.-F. Liu, Iran. J. Pharm. Res., 2018, 17(2), 726–734 CAS.
- J. Chen, X.-Q. Zhu, L. Yang, Y. Luo, M.-Y. Wang, X.-T. Liu, K.-X. Liang and X.-L. Gu, Asian Pac. J. Trop. Med., 2016 Nov, 9(11), 1078–1083 Search PubMed.
- C.-C. Wang, L.-H. Chen, C.-Q. Xu, J.-J. Shi, S.-Y. Chen, M.-X. Tan, J.-L. Chen, L.-S. Zou, C.-H. Chen, Z.-X. Liu and X.-H. Liu, Am. J. Chin. Med., 2020, 48(1), 17–45 CrossRef CAS PubMed.
- X.-Y. Wang, H. Zhang, L.-L. Chen, L.-H. Shan, G.-W. Fan and X.-M. Gao, J. Ethnopharmacol., 2013, 150(3), 781–790 CrossRef CAS PubMed.
- Z.-Z. Dong, Z.-H. Zhu and H.-Q. Huang, et al., J. China Univ., 2020, 34(06), 58–63 Search PubMed.
- M.-L. Shen, Y.-F. Liang and G.-X. Liu, et al., Chin. Tradit. Pat. Med., 2021, 43(01), 154–159 Search PubMed.
- X. Jiang, S.-F. Sun and Y. Wang, et al., Chem. Ind. Times, 2017, 31(07), 25–28 Search PubMed.
- P. Mlh, X.-F. Guo and X. Yin, et al., J. Agric. Sci. Technol., 2019,(03), 211–212 Search PubMed.
- A. Karkanis, N. Martins, S. A. Petropoulos and I. C. F. R. Ferreira, Food Rev. Int., 2016, 1261300 Search PubMed.
- L.-L. Lan, Y.-J. Zhang, M.-T. Zhang and G.-X. Sun, J. Pharm. Biomed. Anal., 2019, 175, 112715 CrossRef CAS PubMed.
- Z.-S. Wu, O. Tao, W. Cheng, L. Yu, X.-Y. Shi and Y.-J. Qiao, Spectrochim. Acta, Part A, 2012, 86, 631–636 CrossRef CAS PubMed.
- Z.-F. Hou and G.-X. Sun, Phytochem. Anal., 2021, 32(6), 1118–1130 CrossRef CAS PubMed.
- X.-W. He, F.-C. Zhang and Y. Jiang, J. Chromatogr. Sci., 2012 Jul, 50(6), 457–463 Search PubMed.
- Chinese Pharmacopoeia Commission, Chinese Pharmacopoeia, China Medical Science Press, Beijing, China, 2020 Search PubMed.
- W.-B. Guo, S.-F. Zheng and M.-S. Hu, et al., Chin. J. Pharm. Anal., 2013, 33(01), 141–145 Search PubMed.
- F. Chemat, M. A. Vian and G. Cravotto, Int. J. Mol. Sci., 2012, 13(7), 8615–8627 CrossRef CAS PubMed.
- L.-N. Li, Y.-M. Liu, Z.-T. Wang, L. Yang and H. Liu, J. Sep. Sci., 2021, 44(6), 1098–1121 CrossRef CAS PubMed.
- Y. Dai, J. van Spronsen, G. J. Witkamp, R. Verpoorte and Y. H. Choi, Anal. Chim. Acta, 2013, 766, 61–68 CrossRef CAS PubMed.
- G. Wang, Q. Cui, L.-J. Yin, X. Zheng, M.-Z. Gao, Y. Meng and W. Wang, J. Pharm. Biomed. Anal., 2019, 170, 285–294 CrossRef CAS PubMed.
- M. H. Zainal-Abidin, M. Hayyan, A. Hayyan and N. S. Jayakumar, Anal. Chim. Acta, 2017, 979, 1–23 CrossRef CAS PubMed.
- J. Tang, H. Song, X.-T. Feng, A. Yohannes and S. Yao, Curr. Med. Chem., 2019, 26(32), 5947–5967 CrossRef CAS PubMed.
- T.-D. Ho, C. Zhang, L. W. Hantao and J. L. Anderson, Anal. Chem., 2014, 86(1), 262–285 CrossRef CAS PubMed.
- J. Huang, X.-Y. Guo, T.-Y. Xu, L.-Y. Fan, X.-P. Zhou and S.-H. Wu, J. Chromatogr. A, 2019, 1598, 1–19 CrossRef CAS PubMed.
- A. P. Abbott, G. Capper, D. L. Davies, R. K. Rasheed and V. Tambyrajah, Chem. Commun., 2003,(1), 70–71 RSC.
- W. W. Oomen, P. Begines, N. R. Mustafa, E. G. Wilson, R. Verpoorte and Y. H. Choi, Molecules, 2020, 25(3), 617 CrossRef CAS PubMed.
- M. Wang, J.-Q. Wang, Y. Zhang, Q. Xia, W.-T. Bi, X.-D. Yang and D. Y. Chen, J. Chromatogr. A, 2016, 1443, 262–266 CrossRef CAS PubMed.
- J. Chen, M.-L. Liu, Q. Wang, H.-Z. Du and L.-W. Zhang, Molecules, 2016, 21(10), 1383 CrossRef PubMed.
- W.-T. Bi, M.-L. Tian and K. H. Row, J. Chromatogr. A, 2013, 1285, 22–30 CrossRef CAS PubMed.
- K. M. Jeong, M. S. Lee, M. W. Nam, J. Zhao, Y. Jin, D. K. Lee, S. W. Kwon, J. H. Jeong and J. Lee, J. Chromatogr. A, 2015, 1424, 10–17 CrossRef CAS PubMed.
- M. Zdanowicz, K. Wilpiszewska and T. Spychaj, Carbohydr. Polym., 2018, 200, 361–380 CrossRef CAS PubMed.
- X. Li and K. H. Row, J. Sep. Sci., 2016, 39(18), 3505–3520 CrossRef CAS PubMed.
- Y. P. Mbous, M. Hayyan, A. Hayyan, W. F. Wong, M. A. Hashim and C. Y. Looi, Biotechnol. Adv., 2017, 35(2), 105–134 CrossRef CAS PubMed.
- Y. H. Choi, J. van Spronsen, Y. Dai, M. Verberne, F. Hollmann, I. W. Arends, G. J. Witkamp and R. Verpoorte, Plant Physiol., 2011, 156(4), 1701–1705 CrossRef CAS PubMed.
- A. Mišan, J. Nađpal, A. Stupar, M. Pojić, A. Mandić, R. Verpoorte and Y. H. Choi, Crit. Rev. Food Sci. Nutr., 2020, 60(15), 2564–2592 CrossRef PubMed.
- E. L. P. de Faria, R. S. do Carmo, A. F. M. Cláudio, C. S. R. Freire, M. G. Freire and A. J. D. Silvestre, Int. J. Mol. Sci., 2017, 18(11), 2276 CrossRef PubMed.
- S. Sut, M. Faggian, V. Baldan, G. Poloniato, I. Castagliuolo, I. Grabnar, B. Perissutti, P. Brun, F. Maggi, D. Voinovich, G. Peron and S. Dall' Acqua, Molecules, 2017, 22(11), 1921 CrossRef CAS PubMed.
- Y. Dai, R. Verpoorte and Y. H. Choi, Food Chem., 2014, 159, 116–121 CrossRef CAS PubMed.
- T. Jeliński, M. Przybyłek and P. Cysewski, Pharm. Res., 2019, 36(8), 116 CrossRef PubMed.
- M. Espino, M. d. l. Á. Fernández, F. J. V. Gomez and M. Fernanda Silva, Trends Anal. Chem., 2016, 76, 126–136 CrossRef CAS.
- Z. Yang, Adv. Biochem. Eng.Biotechnol., 2019, 168, 31–59 CrossRef CAS PubMed.
- Y. C. Wu, P. Wu, Y. B. Li, T. C. Liu, L. Zhang and Y. H. Zhou, RSC Adv., 2018, 8(27), 15069–15077 RSC.
- W. T. Bi, M. L. Tian and K. H. Row, J. Chromatogr. A, 2013, 1285, 22–30 CrossRef CAS PubMed.
- Y.-Q. Ma, M.-H. Liu, T. Tan, A.-P. Yan, L. Guo, K. Jiang, C.-H. Tan and Y.-Q. Wan, Phytochem. Anal., 2018, 29(6), 639–648 CrossRef CAS PubMed.
- P. Yu, Q. Li, Y.-M. Feng, S.-N. Ma, Y.-Y. Chen and G.-C. Li, Molecules, 2021, 26(5), 1310 CrossRef CAS PubMed.
- Y. Dai, J. van Spronsen, G. J. Witkamp, R. Verpoorte and Y. H. Choi, Anal. Chim. Acta, 2013, 766, 61–68 CrossRef CAS PubMed.
- S. Pinteus, J. Silva, C. Alves, A. Horta, N. Fino, A. I. Rodrigues, S. Mendes and R. Pedrosa, Food Chem., 2017, 218, 591–599 CrossRef CAS PubMed.
- Y. Wang, Y. Gao, H. Ding, S. Liu, X. Han, J. Gui and D. Liu, Food Chem., 2017, 218, 152–158 CrossRef CAS PubMed.
- H. Cheng and G. Huang, Int. J. Biol. Macromol., 2018, 114, 415–419 CrossRef CAS PubMed.
- Z. Sroka and W. Cisowski, Food Chem. Toxicol., 2003, 41(6), 753–758 CrossRef CAS PubMed.
- B. Zhuang, L. L. Dou, P. Li and E. H. Liu, J. Pharm. Biomed. Anal., 2017, 134, 214–219 CrossRef CAS PubMed.
- J. Cao, H. Wang, W. Zhang, F. Cao, G. Ma and E. Su, Molecules, 2018, 23(12), 3214 CrossRef PubMed.
- Y. Dai, G. J. Witkamp, R. Verpoorte and Y. H. Choi, Anal. Chem., 2013, 85(13), 6272–6278 CrossRef CAS PubMed.
- Z.-F. Wei, X.-Q. Wang, P. Xiao, W. Wang, C.-J. Zhao, Y.-G. Zu and Y.-J. Fu, Ind. Crops Prod., 2015, 63, 175–181 CrossRef CAS.
- J. Cao, L. Chen, M. Li, F. Cao, L. Zhao and E. Su, J. Pharm. Biomed. Anal., 2018, 158, 317–326 CrossRef CAS PubMed.
- X. Shang, J. N. Tan, Y. Du, X. Liu and Z. Zhang, Molecules, 2018, 23(9), 2110 CrossRef PubMed.
- L. Zhang and M. Wang, Int. J. Biol. Macromol., 2017, 95, 675–681 CrossRef CAS PubMed.
- N. D. Oktaviyanti, K. Kartini, M. A. Hadiyat, E. Rachmawati, A. C. Wijaya, H. Hayun and A. Mun'im, R. Soc. Open Sci., 2020, 7(10), 201116 CrossRef CAS PubMed.
- S. Bajkacz and J. Adamek, Talanta, 2017, 168, 329–335 CrossRef CAS PubMed.
- C. L. Jing, X. F. Dong and J. M. Tong, Molecules, 2015, 20(9), 15550–15571 CrossRef CAS PubMed.
- F. C. Chong and X. F. Gwee, Nat. Prod. Res., 2015, 29(15), 1485–1487 CrossRef CAS PubMed.
- C. Bakirtzi, K. Triantafyllidou and D. P. Makris, J. Appl. Res. Med. Aromat. Plants, 2016, 3(3), 120–127 Search PubMed.
- N. M. Woo, J. Zhao and M.-S. Lee, et al., Green Chem., 2015, 17(3), 1718–1727 RSC.
- M. L. Á. Fernández, J. Boiteux, M. Espino, F. J. V. Gomez and M. F. Silva, Anal. Chim. Acta, 2018, 1038, 1–10 CrossRef PubMed.
- Z. Zhao, Y. Ji, X. Liu and L. Zhao, Chin. J. Chromatogr., 2021, 39(2), 152–161 CrossRef PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1ra06338c |
| ‡ These authors contributed equally. |
|
| This journal is © The Royal Society of Chemistry 2021 |
Click here to see how this site uses Cookies. View our privacy policy here.  Open Access Article
Open Access Article a,
Guo-dong Wu‡a,
Zhi-qiang Dongc,
Dan Yanga,
Yu-kun Boa,
Ming An*a and
Long-shan Zhao
a,
Guo-dong Wu‡a,
Zhi-qiang Dongc,
Dan Yanga,
Yu-kun Boa,
Ming An*a and
Long-shan Zhao *b
*b
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 molar ratio) was selected as a topgallant solvent for maximizing analyte extraction yields. Various extraction parameters, such as water content, liquid/solid ratio, extraction time and temperature, were systematically optimized by single-factor and response surface methodology (RSM) experiments. The results indicated that the optimum extraction conditions for the analytes featured a water content of 17%, a liquid/solid ratio of 42 mL g−1 and an extraction time of 30 min. The extracted amounts of liquiritin, isoliquiritin, liquiritigenin, glycyrrhizic acid and isoliquiritigenin reached 5.60, 3.17, 1.27, 74.62 and 1.34 mg g−1, respectively, under optimized conditions, which were much higher than those extracted using conventional organic solvents. In addition, antioxidant tests revealed that the NADES extracts showed higher DPPH and hydroxyl radical-scavenging capacity than the conventional solvent extracts used for comparison. This study provides a suitable approach for efficiently extracting the bioactive compounds of CPLTs. Meanwhile, NADESs can be extended to other natural products as green extraction media.
3 molar ratio) was selected as a topgallant solvent for maximizing analyte extraction yields. Various extraction parameters, such as water content, liquid/solid ratio, extraction time and temperature, were systematically optimized by single-factor and response surface methodology (RSM) experiments. The results indicated that the optimum extraction conditions for the analytes featured a water content of 17%, a liquid/solid ratio of 42 mL g−1 and an extraction time of 30 min. The extracted amounts of liquiritin, isoliquiritin, liquiritigenin, glycyrrhizic acid and isoliquiritigenin reached 5.60, 3.17, 1.27, 74.62 and 1.34 mg g−1, respectively, under optimized conditions, which were much higher than those extracted using conventional organic solvents. In addition, antioxidant tests revealed that the NADES extracts showed higher DPPH and hydroxyl radical-scavenging capacity than the conventional solvent extracts used for comparison. This study provides a suitable approach for efficiently extracting the bioactive compounds of CPLTs. Meanwhile, NADESs can be extended to other natural products as green extraction media.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2
2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2
2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2
2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2
2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2
2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2
2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2
2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2
2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2
2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2
2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2
2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2
2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2
2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2
2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2; water content, 20%; liquid/solid ratio, 15 mL g−1; extraction time, 20 min; extraction temperature, 50 °C). First, 0.3 g of CPLT sample was extracted with 4.5 mL solvents in a conical flask. The mixture was vortexed for 30 s, and ultrasonicated for 40 min at room temperature. The extracts obtained were then centrifuged at 5000 rpm for 10 min. The suspension was diluted with water and filtered through a 0.45 μm membrane before testing. Each extraction was performed in triplicate, and the extraction yield was calculated as follows:
2; water content, 20%; liquid/solid ratio, 15 mL g−1; extraction time, 20 min; extraction temperature, 50 °C). First, 0.3 g of CPLT sample was extracted with 4.5 mL solvents in a conical flask. The mixture was vortexed for 30 s, and ultrasonicated for 40 min at room temperature. The extracts obtained were then centrifuged at 5000 rpm for 10 min. The suspension was diluted with water and filtered through a 0.45 μm membrane before testing. Each extraction was performed in triplicate, and the extraction yield was calculated as follows:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 1
1, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2, 1
2, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3, 1
3, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 or 1
4 or 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5, water contents of 10, 20, 30, 40 or 50%, liquid/solid ratios of 10, 20, 30, 40 or 50 mL g−1, extraction time periods of 10, 20, 30, 40, 50, 60 or 70 min, and extraction temperatures of 30, 40, 50, 60 or 70 °C was carried out based on single-factor experiments. When examining the effect of one factor on the extraction yield, the other factors were maintained immobile.
5, water contents of 10, 20, 30, 40 or 50%, liquid/solid ratios of 10, 20, 30, 40 or 50 mL g−1, extraction time periods of 10, 20, 30, 40, 50, 60 or 70 min, and extraction temperatures of 30, 40, 50, 60 or 70 °C was carried out based on single-factor experiments. When examining the effect of one factor on the extraction yield, the other factors were maintained immobile.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 1
1, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2, 1
2, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3, 1
3, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4, and 1
4, and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5) was evaluated (n = 3, p > 0.05). As shown in Fig. 3A, it is apparent that the yield of all five compounds increased as the molar ratio was raised from 1
5) was evaluated (n = 3, p > 0.05). As shown in Fig. 3A, it is apparent that the yield of all five compounds increased as the molar ratio was raised from 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 to 1
1 to 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3, most likely due to enhanced hydrogen bond formation. However, a higher molar ratio decreased the yield since the increase in HBDs resulted in considerable steric hindrance, which prevented interactions between the targeted compounds and HBAs.48,58,59 Hence, 1,4-butanediol and levulinic acid at a molar ratio of 1
3, most likely due to enhanced hydrogen bond formation. However, a higher molar ratio decreased the yield since the increase in HBDs resulted in considerable steric hindrance, which prevented interactions between the targeted compounds and HBAs.48,58,59 Hence, 1,4-butanediol and levulinic acid at a molar ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 were used for subsequent experiments.
3 were used for subsequent experiments.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3) with 10, 20, 30, 40 and 50% water content (n = 3, p < 0.05). Fig. 3B shows that the extraction efficiency for all compounds increased significantly when the water content was raised from 10 to 20%, which could be attributed to the corresponding decrease in viscosity that promoted the formation of an ordered hydrogen bond network. However, high water contents (30, 40, and 50%) can limit the interactions between the solute and solvents, and thus, lower the extraction efficiency.60–62 Based on these results, NADES-12 with 20% water was used for the subsequent experiments.
3) with 10, 20, 30, 40 and 50% water content (n = 3, p < 0.05). Fig. 3B shows that the extraction efficiency for all compounds increased significantly when the water content was raised from 10 to 20%, which could be attributed to the corresponding decrease in viscosity that promoted the formation of an ordered hydrogen bond network. However, high water contents (30, 40, and 50%) can limit the interactions between the solute and solvents, and thus, lower the extraction efficiency.60–62 Based on these results, NADES-12 with 20% water was used for the subsequent experiments.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 to 50
1 to 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 on the extraction efficiency of the NADES-12 was examined (n = 3, p < 0.05). As shown in Fig. 3C, the extraction yield of the solute increased significantly with the increase in solvent volume and peaked at the liquid/solid ratio of 40 mL g−1, before plateauing and then decreasing. This is likely due to the fact that excess solvent volume increased dissolution of impurities along with that of targeted analytes, which eventually hindered the dissolution of the latter.47,63 Jing et al.64 similarly reported that the solvent volume above a certain threshold can inhibit analyte dissolution due to the presence of impurities.
1 on the extraction efficiency of the NADES-12 was examined (n = 3, p < 0.05). As shown in Fig. 3C, the extraction yield of the solute increased significantly with the increase in solvent volume and peaked at the liquid/solid ratio of 40 mL g−1, before plateauing and then decreasing. This is likely due to the fact that excess solvent volume increased dissolution of impurities along with that of targeted analytes, which eventually hindered the dissolution of the latter.47,63 Jing et al.64 similarly reported that the solvent volume above a certain threshold can inhibit analyte dissolution due to the presence of impurities.



![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) glycerol) had antioxidant activity, which was an important reason why the antioxidant activity of DES extracts was significantly higher than that of methanol. On this basis, DPPH and hydroxyl radical assays of pure NADES (1,4-butanediol
glycerol) had antioxidant activity, which was an important reason why the antioxidant activity of DES extracts was significantly higher than that of methanol. On this basis, DPPH and hydroxyl radical assays of pure NADES (1,4-butanediol![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) levulinic acid) were performed under the same conditions, which also displayed results consistent with our hypothesis. DPPH and hydroxyl radical-scavenging capacities of pure NADES-12 were 15.69 ± 0.69% and 14.23 ± 0.53% respectively. The results indicated that pure NADES-12 had antioxidant effects, which had an additive effect on the antioxidant activity of the targeted extract. Taken together, NADESs is a more efficient alternative solvent for extracting bioactive compounds from CPLT in terms of both the yield and the antioxidant activity of the extract.
levulinic acid) were performed under the same conditions, which also displayed results consistent with our hypothesis. DPPH and hydroxyl radical-scavenging capacities of pure NADES-12 were 15.69 ± 0.69% and 14.23 ± 0.53% respectively. The results indicated that pure NADES-12 had antioxidant effects, which had an additive effect on the antioxidant activity of the targeted extract. Taken together, NADESs is a more efficient alternative solvent for extracting bioactive compounds from CPLT in terms of both the yield and the antioxidant activity of the extract.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 molar ratio); NADES-12 water content, 17%; liquid/solid ratio, 42 mL g−1 and extraction time, 30 min. Under the optimal extraction conditions, the NADES-12 extract exhibited higher extraction efficiency and in vitro antioxidant activity than the conventional solvent extract. The developed method is a promising strategy for the extraction of bioactive compounds, which provides high efficiency for the natural product. The proposed method is proved to be efficient, simple, environmentally friendly and sustainable for obtaining five bioactive compounds from the CPLT, which contributes to the urgent need of research on the green extraction of compounds in Chinese herbal medicines.
3 molar ratio); NADES-12 water content, 17%; liquid/solid ratio, 42 mL g−1 and extraction time, 30 min. Under the optimal extraction conditions, the NADES-12 extract exhibited higher extraction efficiency and in vitro antioxidant activity than the conventional solvent extract. The developed method is a promising strategy for the extraction of bioactive compounds, which provides high efficiency for the natural product. The proposed method is proved to be efficient, simple, environmentally friendly and sustainable for obtaining five bioactive compounds from the CPLT, which contributes to the urgent need of research on the green extraction of compounds in Chinese herbal medicines.






