Open Access Article
Open Access ArticleDevelopment of novel, biocompatible, polyester amines for microglia-targeting gene delivery†
Boomin Choi‡
 a,
Min-Hye Ahn‡b,
Seojin Hongb,
Ellane Eda Barcelona,
Jaiprakash Sangshetti
a,
Min-Hye Ahn‡b,
Seojin Hongb,
Ellane Eda Barcelona,
Jaiprakash Sangshetti c,
Rohidas B. Arote
c,
Rohidas B. Arote *b and
Sung Joong Lee
*b and
Sung Joong Lee *a
*a
aDepartment of Neuroscience and Physiology, Dental Research Institute, School of Dentistry, Seoul National University, Seoul 08826, Republic of Korea. E-mail: sjlee87@snu.ac.kr
bDepartment of Molecular Genetics and Dental Research Institute, School of Dentistry, Seoul National University, Seoul 08826, Republic of Korea. E-mail: rohi06@snu.ac.kr
cDepartment of Medicinal Chemistry, Y. B. Chavan College of Pharmacy, Dr Rafiq Zakaria Campus, Rauza Baugh, Aurangabad, MS, India
First published on 16th November 2021
Abstract
Recent progress in personalized medicine and gene delivery has created exciting opportunities in therapeutics for central nervous system (CNS) disorders. Despite the interest in gene-based therapies, successful delivery of nucleic acids for treatment of CNS disorders faces major challenges. Here we report the facile synthesis of a novel, biodegradable, microglia-targeting polyester amine (PEA) carrier based on hydrophilic triethylene glycol dimethacrylate (TG) and low-molecular weight polyethylenimine (LMW-PEI). This nanocarrier, TG-branched PEI (TGP), successfully condensed double-stranded DNA into a size smaller than 200 nm. TGP nanoplexes were nontoxic in primary mixed glial cells and showed elevated transfection efficiency compared with PEI-25K and lipofector-EZ. After intrathecal and intracranial administration, PEA nanoplexes delivered genes specifically to microglia in the spinal cord and brain, respectively, proposing TGP as a novel microglia-specific gene delivery nanocarrier. The microglia-specific targeting of the TGP nanocarrier offers a new therapeutic strategy to modulate CNS disorders involving aberrant microglia activation while minimizing off-target side effects.
Introduction
Microglia are innate immune cells of the central nervous system (CNS) and play pivotal roles in CNS physiology and pathophysiology.1 During neurodegeneration and natural brain aging, microglia lose their homeostatic cellular properties, show aberrant activation such as increased production of pro-inflammatory cytokines and reactive oxygen species (ROS), and develop dysfunctional lysosomal deposits showing impaired phagocytosis.2 Moreover, microglia are involved in the regulation of CNS homeostasis while pruning unnecessary synapses and managing cell debris.3 Microglia play a significant role in the clearance of amyloid plaques and aggregated α-synuclein in Alzheimer's disease (AD)4 and Parkinson's disease (PD),5 respectively. Increased cell-to-cell propagation of α-synuclein in grafted dopaminergic neurons and PD was observed due to pharmacological ablation of microglia using PLX5622.6 In contrast, uncontrolled over-activation of microglia and subsequent neurotoxic mediators are suggested to contribute to these neurodegenerative diseases.7 In neuropathic pain animal models, microglial activation and subsequent pro-inflammatory gene expression in the spinal cord elicit central pain sensitization to induce chronic pain.8–10 All these studies point to the critical role of microglia in CNS disease and propose microglia as a prime therapeutic target for these CNS disorders.Given such a role of microglia, efforts have been made to specifically deliver drugs to microglia in the CNS using nanoparticle-mediated delivery. In neuropathic pain studies, specific targeting of microglia using the CD11b antibody-conjugated nanozyme significantly reduced microglial reactive oxygen species production and thereby alleviated neuropathic pain.11 In addition, poly-ε-caprolactone-based nanoparticles12 and polyamidoamine dendrimers13 were utilized to deliver drugs specifically to microglia in a neuropathic pain animal model. Although these nanoparticles have successfully delivered drug molecules to microglia and show a certain level of therapeutic efficacy, the development of microglia-targeting nanocarriers that efficiently deliver nucleic acids for gene therapy is lacking.
Despite recent technical advances in gene delivery systems, several issues such as cytotoxicity, low transfection efficiency, and cell specificity have remained the main challenges for researchers. For the past few decades, different vectors have been tested as gene carriers. Though gene therapy using viral vectors has shown clinical progress, restrictions associated with use, such as adverse immune reactions, smaller cargo size, and repeated administration, persist.14,15 Non-viral vectors have emerged as promising alternatives for nucleic acid delivery, with advantages such as high target specificity and low immunogenicity compared with their viral counterparts. Nevertheless, polymeric nanocarriers offer great potential for target-specific delivery of nucleic acids with not only high transfection efficiency, but also strong biocompatibility and stability.16,17 Among non-viral vectors, polyethylenimine (PEI) is one of the most widely used gene carriers because of its proton sponge effect, high transfection efficiency, and ability to condense DNA/RNA into nanoplexes.18,19 However, cell toxicity is one of the major limitations of PEIs. Moreover, the transfection efficiency and toxicity of PEI are molecular weight-dependent.16
Various biodegradable cationic nanocarriers such as polyesteramine (PEA) have been reported and investigated as successful non-viral gene carriers owing to their low cytotoxicity and high transfection efficiency.20–22 To overcome the toxicity-related limitations of PEI, we induced alkylation via the Michael addition reaction to convert primary and secondary amines to respective secondary and tertiary amines without changing the buffering capacity of the backbone. Various PEA-based carriers synthesized using this method with low-molecular weight PEI (LMW-PEI) and acrylate monomers have been reported.20–22 Apart from high transfection efficiency, the biodegradability of PEA offers less toxicity and thereby rapid clearance, an important feature of PEAs that distinguishes them from PEI, a highly used cationic gene carrier.23,24 PEAs employ excellent gene delivery by utilizing their biodegradable ester backbone for low cytotoxicity and the proton sponge effect, thereby resulting in remarkable transfection efficiency using a PEI fraction.15,25–27
Here we report the design, synthesis, and evaluation of a PEA based on triethylene glycol dimethacrylate (TG) and LMW-PEI via the Michael addition reaction. Immunofluorescence studies with labeling antibodies were carried out in cultured glial cells as well as in vivo to determine the carrier's specificity for microglia. Subsequent cell toxicity studies and transfection efficiency studies were conducted in vitro and in vivo utilizing immunohistochemistry to determine successful microglial transfection. Here, we present data demonstrating that our newly synthesized novel polymer, TG-branched PEI (TGP), has high transfection efficiency and targeting specificity for microglia without showing any signs of cytotoxicity, proposing TGP as an ideal gene carrier for microglia-targeting gene delivery (Scheme 1).
Experimental
Materials
Branched PEI (BPEI) (Mn: 1200 Da and 25 kDa), anhydrous dimethylsulfoxide (DMSO), and bafilomycin A1 were purchased from Sigma-Aldrich (St. Louis, Mo, USA). TG (C14H22O6, 286.32 g mol−1) was purchased from Monomer-polymer and DAJAC LABs (Ambler, PA, USA). The MTS and Luciferase reporter assay kits for in vitro use were purchased from Promega (Madison, WI, USA). Plasmid mini- and midi-prep systems for the pGL3 vector with SV-40 promoter purification were purchased from QIAGEN (Hilden, Germany). Lipofector-EZ was purchased from AptaBio (Yongin, Korea).Synthesis of TGP
TG with BPEI (1.2 kDa) was prepared at a molar ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 following a method similar to that reported by Arote et al.26 Briefly, both TG and BPEI solutions were dissolved in anhydrous DMSO and continuously purged with nitrogen. The TG mixture was added dropwise into BPEI solution and stirred overnight at 60 °C under nitrogen. Afterward, the modified TG-PEI solution was purified by dialysis using a membrane with a molecular weight cut-off of 3500 Da for 24 h at 4 °C against distilled water. The purified TGP was obtained by lyophilization for 3 days.
2 following a method similar to that reported by Arote et al.26 Briefly, both TG and BPEI solutions were dissolved in anhydrous DMSO and continuously purged with nitrogen. The TG mixture was added dropwise into BPEI solution and stirred overnight at 60 °C under nitrogen. Afterward, the modified TG-PEI solution was purified by dialysis using a membrane with a molecular weight cut-off of 3500 Da for 24 h at 4 °C against distilled water. The purified TGP was obtained by lyophilization for 3 days.
Characterization of TGP
The composition of TGP was assessed by 1H high-resolution nuclear magnetic resonance (NMR) spectroscopy using a Bruker AVANCE 600 MHz spectrometer (Bruker, Billerica, MA, USA). The TG, PEI, and TGP were dissolved in deuterium oxide (D2O) at a concentration of 5 mg ml−1 and measured. The molecular weight of TGP was analyzed using gel permeation chromatography with a Dionex UltiMate 3000 instrument (Dionex, Sunnyvale, CA, USA). Briefly, the sample (50 μl) was injected and measured in triplicate. The flow rate of the column was 1.0 ml min−1, and 0.1 M sodium nitrate solution was used as the mobile phase.Observation of transmission electron microscopy
The morphology of TGP/DNA nanoplexes (N/P 20) was observed by energy-filtering transmission electron microscopy (EF-TEM) (LIBRA 120, Carl Zeiss, Oberkochen, Germany). The particle solution was placed on the copper grid, stained with 2% aqueous uranyl acetate for 5 s, washed twice with distilled water, and the resulting liquid was removed with filter paper.Measurement of zeta-potential and particle sizes
The zeta-potential and sizes of TGP/DNA nanoplexes at N/P ratios of 5, 10, 20, and 30 were measured using a dynamic light scattering spectrophotometer (DLS, ELS 8000, Otsuka Electronics, Osaka, Japan) with 90° and 20° scattering angles. Average and standard deviation were calculated from three measurements of each sample.Gel retardation assay
The nanoplex formation ability of TGP with DNA at various N/P ratios was evaluated by the agarose gel shift assay. Different N/P ratios of TGP were complexed with DNA (0.1 μg pGL3) and incubated for 30 min at room temperature (RT). Incubated samples consisting of 6 X loading dye were loaded onto 1% agarose gel in Tris-acetate-EDTA buffer. DNA bands were visualized under ultraviolet illumination.Primary glial cells and microglia culture
One-day-old C57BL/6 mice pups were used to culture primary glia using a procedure previously described.28 After removing the meninges from the cerebral hemisphere, tissue was dissociated into a single-cell suspension through gentle repetitive pipetting. Cells were cultured in Dulbecco's Modified Eagle's medium supplemented with 10 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid, 10% fetal bovine serum (FBS), 2 mM L-glutamine, 1 × NEAA, and 1 × antibiotic/antimycotic in 75 cm2 flasks at 37 °C in a 5% CO2 incubator; the medium was changed every five days. To isolate microglia from the mixed glial cells after 15–20 days of culturing, we kept the flasks in a rotating shaker at 250 rpm for 3 h. The floating cells were collected and plated on poly-D-lysine (PDL)-coated glass coverslips in a four-well plate with 2 × 104 cells per well. After 15 min, the media was replaced with fresh media in order to eliminate unbound non-microglial cells and debris.In vitro transfection efficiency
The efficiency of transfection in primary mixed glia was evaluated using the luciferase assay. Cells were seeded at 3.0 × 104 cells per well in a 24-well plate and treated with TGP/pGL3 nanoplexes at 70–80% confluency. TGP and PEI were complexed with purified pGL3 plasmids (0.5 μg) contained in serum-free media at various N/P ratios (5, 10, 20, 30). After 5 h of incubation, the prior media was replaced with serum-containing media (5%). Luciferase assay was conducted according to the manufacturer's protocol. Relative light units (RLU) were standardized by total protein, which was measured by a bicinchoninic acid assay (BCA) and performed in triplicate with a chemiluminometer (SPARK 10M, Tecan, Männedorf, Switzerland).Bafilomycin A1 treatment
Since hyper-branched PEI contains a high concentration of amine groups, we assumed that TGP has the proton sponge effect and facilitates the escape of nanoplexes from the endosome. In order to characterize the pH buffering capacity of TGP, transfection experiments with bafilomycin A1 treatment were carried out. Primary mixed glial cells were seeded in a 24-well plate at 3 × 104 cells per well and treated with 200 nM bafilomycin A1 for 15 min before transfection with TGP/pGL3 (N/P ratio of 20) for an additional 5 h. Afterward, the medium was replaced with serum-containing media (5% FBS), and the cells were incubated for 18–24 h. Luciferase activity was measured in triplicate with a chemiluminometer (SPARK 10M).Flow cytometry analysis
The whole hippocampus and spinal cord tissue from the cervical, thoracic, lumbar, and sacral spine was removed separately and homogenized mechanically to a single-cell suspension. For in vitro experiments, the mixed glial cells were detached using 0.25% trypsin with 3 min incubation at 37 °C and collected. Cells were washed with ice-cold PBS containing 2% FBS and incubated with Fc BlockerTM (BD Bioscience, San Jose, CA, USA) for 10 min at 4 °C prior to staining. Then, cells stained with CD11b-APC (Biolegend Inc., San Diego, CA, USA) were analyzed with a BD FACSVERSE flow cytometer (BD Bioscience) to measure the CD11b-positive microglia population. Likewise, cells were stained with ACSA A-2-PE (Miltenyi Biotec, Bergisch Gladbach, Germany) and Thy-1.2 violet (Biolegend Inc.) to identify astrocytes and neurons, respectively. Data were acquired and analyzed with BD FACSuite v1.2 (BD Biosciences).Immunofluorescence
Immunostaining was carried out using previously established protocols.29 The spinal cord sections were incubated in a blocking solution (5% normal goat serum, 2% bovine serum albumin (BSA), and 0.1% Triton X-100) for 1 h at RT. Sections were incubated overnight at 4 °C with the primary antibody for rabbit-anti-Iba1 (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000; Wako, Osaka, Japan) or rabbit-anti-GFAP (1
1000; Wako, Osaka, Japan) or rabbit-anti-GFAP (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000; Dako, Agilent, CA, USA). After rinsing in 0.1 M PBS, the sections were incubated for 1 h at RT with a mixture of Cy3-conjugated secondary antibodies (1
1000; Dako, Agilent, CA, USA). After rinsing in 0.1 M PBS, the sections were incubated for 1 h at RT with a mixture of Cy3-conjugated secondary antibodies (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200; Jackson ImmunoResearch, PA, USA) and mounted with VectaShield medium (Vector Labs, Burlingame, CA, USA). Fluorescent images were obtained using a confocal microscope (LSM800; Carl Zeiss). For immunocytochemical analysis, mixed glial cells or pure microglial cells were seeded onto a PDL-coated cover glass (2 × 104 cells per 12 mm2). After 2 days of proper attachment and growth, the cells were treated with Alexa488-conjugated nanoparticles. Cells were fixed with 2% PFA in 0.1 M PBS (pH 7.4) for 15 min. The following steps were the same as described in the histochemistry procedure. For quantification of immunoreactivity, stained sections from mice spinal cords were collected and quantified using LSM 800 software.
200; Jackson ImmunoResearch, PA, USA) and mounted with VectaShield medium (Vector Labs, Burlingame, CA, USA). Fluorescent images were obtained using a confocal microscope (LSM800; Carl Zeiss). For immunocytochemical analysis, mixed glial cells or pure microglial cells were seeded onto a PDL-coated cover glass (2 × 104 cells per 12 mm2). After 2 days of proper attachment and growth, the cells were treated with Alexa488-conjugated nanoparticles. Cells were fixed with 2% PFA in 0.1 M PBS (pH 7.4) for 15 min. The following steps were the same as described in the histochemistry procedure. For quantification of immunoreactivity, stained sections from mice spinal cords were collected and quantified using LSM 800 software.
MTS assay
Mixed glial cells were grown on a 96-well plate with 3 × 104 cells per well and treated with a cytotoxic reagent for 24 h. Then, 20 μl of MTS reagent (Promega) was added directly into the cell culture media and incubated at 37 °C for 1 h. Reduction of the MTS tetrazolium compound by viable cells to generate the formazan product, soluble in the serum-free media, was quantified by measuring the absorbance at 492 nm using an EMax® Plus Microplate Reader (Molecular Devices, San Jose, CA, USA). Absorbance obtained from samples treated with cytotoxic reagents was compared to absorbance in control samples to obtain the percentage of cell viability.Animals
Male C57BL/6 mice aged 7–10 weeks were purchased from DBL (Eumsung, Korea) for experimental use. Four to five mice were housed in a plastic cage with standard bedding. They had access to food and water ad libitum. They were accommodated at a constant room temperature of 23 °C ± 2 °C and a 12 h dark/light cycle. All animal procedures were performed in accordance with the Guidelines for Care and Use of Laboratory Animals of Seoul National University and approved by the Animal Ethics Committee of the Seoul National University Institutional Animal Care and Use Committee (SNU IACUC).Intracranial injection
For administration of Alexa488-conjugated TGP, mice were anesthetized and placed on a stereotaxic apparatus (myNeuroLab, St. Louis, MO, USA). The animals were injected with saline or Alexa488-TGP/pGL3 nanoplexes at a rate of 0.5 μl min−1. After 5 min, the needle was removed in three intermediate steps over 3 min to minimize backflow. The incision was cleaned with saline and sutured, after which the animals were kept on a warm pad during recovery.Intrathecal injection
For administration of Alexa488-conjugated TGP, mice were injected with avertin (2%) for anesthesia. After shaving the back of the mice, Alexa488-TGP/pGL3 nanoplexes (5 μl) were injected using a 10 μl Hamilton syringe (Hamilton Company, Reno, NV, USA) with a 30-gauge one-half-inch needle into the subarachnoid space; a slight tail-flick denoted proper administration of the test compounds.Statistical analysis
All data are presented as the mean value with standard error of the mean (SEM). Differences between groups were determined by one-way ANOVA with a Newman–Keuls multiple comparison test. Our threshold for statistical significance was p <0.05.Results and discussion
Synthesis scheme & NMR analysis
The novel PEA was synthesized from the nucleophilic addition reaction of amines and acrylate monomers under specific experimental conditions. The preparation of TGP proceeds through formation of an ester bond via the Michael addition reaction between the amine groups of PEI and the dimethacrylate groups of TG. The schematic illustration of the TGP synthesis reaction is shown in Fig. 1. The synthesis was confirmed using 1H NMR spectroscopy. For comparison, the 1H NMR spectra of LMW PEI (1.2 kDa) and TG were analyzed. The intense peaks at 2.5–2.8 ppm (–NHCH2) represent protons in the amine groups of PEI, and 5.8–6.5 ppm peaks (–C(CH3)R![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH2) are associated with the dimethacrylate proton signals of TG.30,31 When PEI was crosslinked with TG, 1H NMR showed absence of the dimethacrylate proton peaks of TG, indicating successful copolymerization between the two compounds. Additionally, the 1H NMR spectrum of the ester bond (–OCH2) and 1′-alkyl (–CH3) and 3′-alkyl (-CH) groups showed signals at 3.6–3.7 ppm, 1.1–1.2 ppm, and 1.5 ppm, respectively (Fig. S1†). The composition and proportions of TG and PEI were calculated using 1H NMR data as 32 and 68 mol%, respectively. Moreover, the final molecular weight of TGP was measured by gel permeation chromatography (GPC) to reveal an Mn of 1777 Da and a polydispersity index (Mw/Mn) of 1.02 relative to polyethylene glycol (PEG) standards (Table S1†).
CH2) are associated with the dimethacrylate proton signals of TG.30,31 When PEI was crosslinked with TG, 1H NMR showed absence of the dimethacrylate proton peaks of TG, indicating successful copolymerization between the two compounds. Additionally, the 1H NMR spectrum of the ester bond (–OCH2) and 1′-alkyl (–CH3) and 3′-alkyl (-CH) groups showed signals at 3.6–3.7 ppm, 1.1–1.2 ppm, and 1.5 ppm, respectively (Fig. S1†). The composition and proportions of TG and PEI were calculated using 1H NMR data as 32 and 68 mol%, respectively. Moreover, the final molecular weight of TGP was measured by gel permeation chromatography (GPC) to reveal an Mn of 1777 Da and a polydispersity index (Mw/Mn) of 1.02 relative to polyethylene glycol (PEG) standards (Table S1†).
Physiochemical properties of TGP nanoplexes
Controlling the morphology, size, and zeta potential of the nanoparticle with the ability to condense DNA into compact particles is an essential factor in transfection efficiency, cellular uptake, and toxicity.32 Since hyper-branched PEI contain a high concentration of amine groups, we assumed that TGP could easily interact with DNA and condense it into compact particles. Representative TEM pictures of the self-assembled TGP copolymers with DNA at N/P 20 are shown in Fig. 2C and S2.† In DLS analysis of size distribution, the average size of TGP/DNA complexes was about 150 nm. This result was consistent with that measured by DLS, as shown in Fig. 2A. Interestingly, the average surface charge of the TGP/pGL3 nanoplexes also depends on the N/P ratio. As depicted in Fig. 2B, the surface charge was decreased from +16 to −0.2 mV when the N/P ratio was increased from 5 to 30. As the PEG part of TGP masks the positive charge of PEI (shielding effect),33 it is assumed that the surface charge decreases as the N/P ratio increases. These results suggest that TGP/DNA nanoplexes show a promising delivery system even at a high N/P ratio, which results in an optimal size and surface charge ideal for cellular uptake in primary mixed glia.34,35 Nanoplex formation and subsequent release of DNA from the nanoplex were studied using the gel retardation assay. To evaluate the DNA compression ability of the polymer, we performed a gel retardation assay. TGP efficiently and stably condensed DNA (pGL3) at all N/P ratios from 1 to 30. As shown in Fig. 2D, the mobility of nanoplexes was blocked in the agarose gel at an N/P ratio 1 or greater by perfect condensation with DNA.Transfection efficiency and cytotoxicity of TGP/pGL3
The cytotoxicity of TGP/pGL3 was evaluated using the MTS assay in primary mixed glia. High-molecular weight PEIs are the “gold standard” in gene delivery. However, they have high toxicity due to damage to the cell membrane via aggregation on the cell surface.36,37 To minimize the cytotoxicity of the polymer, we used LMW BPEI (1.2 kDa) instead of the high-MW counterpart. Since cross-linked branched PEI with TG have degradable ester linkages, they undergo hydrolysis at intracellular pH, leading to formation of nontoxic building blocks.20–22,38 Fig. 3A shows that TGP nanoparticles did not show apparent cytotoxicity (average of cell viability of 100%) at N/P ratios of 5 and 10 and exhibited remarkably lower cytotoxicity than PEI (25 kDa, PEI-25K)/pGL3 even at a high dose. TGP/pGL3 nanoplexes showed approximately 70–100% cell viability, which decreased by 25–68% when PEI-25K was used. To compare the in vitro transfection ability of TGP and PEI-25K at various N/P ratios, we transfected primary mixed glia with 0.5 μg of pGL3 complexed with TGP, PEI-25K, and lipofector-EZ as controls. Compared to PEI-25K/pGL3, TGP/pGL3 nanoplexes exhibited significantly higher transfection efficacy at N/P ratios of 20 and 30 by 188- and 252-fold, respectively. Comparable transfection efficiency was observed in the presence of serum (Fig. S3†). Interestingly, the DNA delivery capacity of PEI-25K/pGL3 decreased with increasing N/P ratio because of their non-biodegradability, leading to cytotoxicity. At an N/P ratio of 30, the transfection efficiency of TGP/pGL3 was approximately 7-fold higher than that of lipofector-EZ, a widely used commercial transfecting agent (Fig. 3B). These data imply that the hydrophobic parts [(–CH2CH2O–)n] in TGP improved cellular uptake through hydrophobic interactions with the cell membrane.20,39We attributed the high transfection efficiency of TGP to the proton sponge effect of the PEI backbone in TGP. To evaluate the proton sponge effect of TGP,20–22,40 we pre-treated primary mixed glia with bafilomycin A1, a specific vacuolar-type H+ ATPase inhibitor, and then transfected TGP/pGL3 nanoplexes at an N/P ratio of 20. The result indicated that the transfection efficiency of TGP/pGL3 and PEI-25K/pGL3 complexes was significantly decreased by 365-fold and 3-fold, respectively, compared to controls, as shown in Fig. 3C. Interestingly, endosomal release of TGP was significantly affected by H+ ATPase inhibitor, more so than by PEI. This result indicates that TGP produces the proton sponge effect and can serve as an efficient DNA transfer vector by preventing lysosomal degradation.20,41
In vitro microglia uptake of TGP in primary microglia
To visualize microglial targeting of TGP by fluorescent microscopy, we prepared Alexa488-conjugated TGP (Alexa488-TGP) and Alexa546-conjugated pGL3 DNA (Alexa546-pGL3). Using these fluorescent molecules, we synthesized two fluorescent nanoplexes, Alexa488-TGP/pGL3 and TGP/Alexa546-pGL3. Primary pure microglia were transfected with 0.5 μg of the fluorescent-conjugated TGP/pGL3 nanoplexes at an N/P ratio of 20, and live-cell images were produced for 5 and 24 h to determine the uptake kinetics. Hoechst staining was performed to assess the nuclei of live cells. The fluorescence signal of Alexa546-conjugated pGL3 (red dot) was detected around the nucleus at 5 h (Fig. 4, left panel). A denser signal was detected near the nucleus at 24 h and even inside the nucleus (Fig. 4, arrows) indicating localization of pGL3 DNA in the nucleus. Also, green dots representing Alexa488-conjugated TGP were observed near the nucleus both at 5 and 24 h and are compartmented near a Hoechst signal at 24 h (Fig. 4, right panel). These data indicate that TGP/pGL3 nanoplexes were taken up by microglia in 5 h. Then, the DNA were released from the nanoplexes, which can localize at the nucleus in 24 h. These results imply that TGP/pGL3 nanoplexes can deliver to the DNA microglial nucleus within 24 h.Cell type-specificity of TGP in primary mixed glial cells in vitro
To investigate the cell type-specific targeting of TGP, we treated primary mixed glia consisting of around 80% astrocytes and 20% microglia, which is similar to an in vivo environment,42 with Alexa488-TGP/pGL3 nanoplexes. Immunocytochemistry using ionized calcium-binding adaptor molecule 1 (Iba-1) and glial fibrillary acidic protein (GFAP) antibody as markers for microglia and astrocytes, respectively, showed Alexa488 signal in both cell types at 5 and 24 h (Fig. 5A), indicating that both microglia and astrocytes can uptake TGP/pGL3 in vitro. We also measured the intensity of Alexa488-TGP fluorescent signal in each cell by flow cytometry using cluster of differentiation molecule 11b (CD11b) and astrocyte cell surface antigen-2 (ACSA-2) antibody as markers for microglia and astrocytes, respectively (Fig. 5B). At four N/P ratios, CD11b-positive microglia showed higher uptake efficiency than astrocytes at each N/P ratio at both 1 and 5 h time points. The mean fluorescence intensity (MFI) of CD11b-positive microglia was 3387, 4196, 2888, and 3264 at N/P 5, 10, 20, and 30, respectively, at 1 h and 7106, 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 186, 6601, and 7656 at 5 h. Meanwhile, the MFI of ACSA-2-positive astrocytes was 1112, 1871, 1766, and 1653 in each N/P ratio at 1 h and 2681, 4443, 3911, and 3916 at 5 h, which was much lower than that of CD11b-positive cells. These data suggest that, even though Alexa488-TGP/pGL3 nanoplexes can be taken up by both microglia and astrocytes, it has better targeting-specificity to microglia in vitro.
186, 6601, and 7656 at 5 h. Meanwhile, the MFI of ACSA-2-positive astrocytes was 1112, 1871, 1766, and 1653 in each N/P ratio at 1 h and 2681, 4443, 3911, and 3916 at 5 h, which was much lower than that of CD11b-positive cells. These data suggest that, even though Alexa488-TGP/pGL3 nanoplexes can be taken up by both microglia and astrocytes, it has better targeting-specificity to microglia in vitro.
Microglia-specific targeting of Alexa488-TGP/pGL3 nanoplexes in vivo
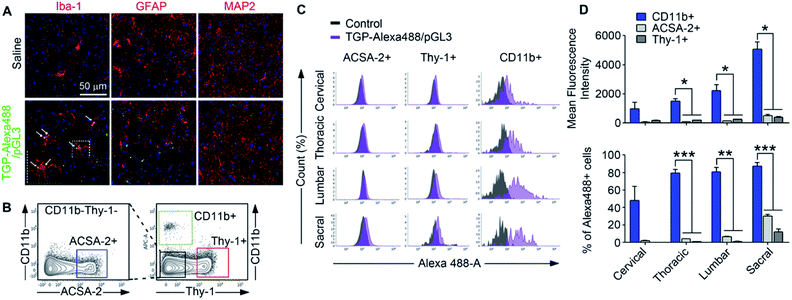
We tested the targeting specificity of TGP nanoplexes in vivo. It has been long sought after to deliver drugs or genes to spinal cord microglia to modulate neurological diseases such as multiple sclerosis43,44 and neuropathic pain.11,45 Therefore, using Alexa488-TGP/pGL3, we first investigated whether the nanoplexes have cell-type specificity for spinal microglia in vivo. Alexa488-TGP/pGL3 (0.1 μg) nanoplexes were administered intrathecally into the mouse spinal canal, which was analyzed via immunohistochemistry at 24 h after injection. The lumbar segment 4–6 (L4–L6) tissue samples were immunostained with cell type-specific antibodies, and localization of the Alexa488 signal was assessed. Unlike in vitro data, the Alexa488 signal mainly co-localized with Iba-1-positive microglia but not with GFAP-positive astrocytes or microtubule-associated protein 2 (MAP-2)-positive neurons (Fig. 6A). For further conformation, Alexa488-positive cell types in the spinal cord tissue were analyzed by flow cytometry 24 h after nanoplex administration. Alexa488-positive signal was detected in 48.1, 79.2, 80.8, and 87.4% of CD11b-positive microglia in cervical, thoracic, lumbar, and sacral regions of the spine, respectively. Meanwhile, they were detected in only 1.8, 4.0, 6.4, and 30.0% of ACSA-2-positive astrocytes (Fig. 6D). A much lower proportion was detected in Thy-1 positive neurons (Fig. 6D). Taken together, these data demonstrate that Alexa488-TGP/pGL3 nanoplexes are mainly targeted to microglia in the spinal cord in vivo, especially in the cervical, thoracic, and lumbar regions. Although Alexa488 signal was present in 30% and 13% of astrocytes and neurons, respectively, in the sacral region near which the intrathecal injection was introduced, the percentage of Alexa488-positive population was much lower than that of microglia (80%). In addition, the mean fluorescent intensities of Alexa488 signal in astrocytes or neurons were less than 10% that of microglia, indicating that the amount of Alexa488-TGP/pGL3 nanoplex uptake by each astrocyte or neuron was much lower than that of microglia (Fig. 6D). In this regard, our data suggest that TGP nanoparticles can be utilized as a microglia-specific gene carrier for neurological diseases involving aberrant microglia function in the spinal cord.We also tested the cell type specificity of TGP in the brain. For this, 0.1 μg of nanoplexes was injected intracranially into the mouse hippocampus, and, after 24 h, the brain slices (bregma −2.18 mm) were analyzed via IHC. Alexa488-positive signal (green dot) was co-localized mostly with Iba-1-positive microglia but not with GFAP-positive astrocytes or NeuN-positive neurons (Fig. 7A). In flow cytometry analysis, Alexa488 fluorescent signal was detected in 12.5% of the CD11b-positive microglia population but in only 0.4% of ACSA-2-positive astrocytes and 0.2% of Thy-1-positive neurons (Fig. 7B–D). These data show that Alexa488-TGP/pGL3 nanoplexes have far greater microglia-targeting specificity in the central nervous system in vivo than in vitro. It is well known that astrocytes in primary culture manifest more highly activated phenotypes compared to cells in vivo,46 which might be underlying the difference in targeting efficacy of the TGP nanoplexes to astrocytes in vitro vs. in vivo. Despite such a slight difference of specificity between in vitro and in vivo experimentation, our data show that TGP specifically and efficiently delivers genes to microglia in vivo and can be utilized as a nanocarrier for gene delivery targeting microglia in vivo.
Conclusions
An effective cell-type-specific non-viral vector system is of critical importance for gene therapy for CNS diseases. For microglia-specific gene delivery, we synthesized TGP nano-molecules by conjugating TG and LMW-PEI. The nanoscale TGP/DNA nanoplexes (<200 nm) were quickly internalized by microglia and avoid lysosomal degradation by the proton sponge effect. In comparison to PEI, TGP showed better transfection efficiency with low cytotoxicity. We showed that TGP could localize inside primary microglia within 5 h, and DNA can enter the nucleus within 24 h. In mixed glia culture, TGP/pGL3 are taken up by microglia rather than by astrocytes. In vivo, TGP/pGL3 are taken up mostly by microglia in the spinal cord and hippocampus, demonstrating that TGP can be a novel microglia-specific gene delivery system. Given the pivotal role of microglia in regulating neurological disorders, TGP can be of instrumental importance to regulate microglia-specific in vivo function, which might have a clinical implication for neurodegenerative diseases and neuropathic pain, in which aberrant microglia dysfunction is involved.Author contributions
Sung Joong Lee and Rohidas B. Arote conceived and designed the experiments. Boomin Choi and Min-Hye Ahn performed the experiments and analyzed the data with equal contribution. Sung Joong Lee, Rohidas B. Arote, Boomin Choi, and Min-Hye Ahn wrote and refined the article. Jaiprakash Sangshetti helped in designing and synthesizing the multifunctional nanocarrier used for delivery of genes. Seojin Hong performed the experiments of nanoparticle synthesis and analysis. Ellane Eda Barcelon performed the biological experiments.Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
This work was supported by the National Research Foundation of Korea (NRF-2016M3C7A1905074).Notes and references
- D. P. Schafer, E. K. Lehrman, A. G. Kautzman, R. Koyama, A. R. Mardinly, R. Yamasaki, R. M. Ransohoff, M. E. Greenberg, B. A. Barres and B. Stevens, Neuron, 2012, 74, 691–705 CrossRef CAS PubMed.
- K. I. Mosher and T. Wyss-Coray, Biochem. Pharmacol., 2014, 88, 594–604 CrossRef CAS PubMed.
- U. K. Hanisch and H. Kettenmann, Nat. Neurosci., 2007, 10, 1387–1394 CrossRef CAS.
- S. Wang and M. Colonna, J. Leukoc. Biol., 2019, 106, 219–227 CrossRef CAS PubMed.
- L. Qin, X. Wu, M. L. Block, Y. Liu, G. R. Breese, J. S. Hong, D. J. Knapp and F. T. Crews, Glia, 2007, 55, 453–462 CrossRef PubMed.
- S. George, N. L. Rey, T. Tyson, C. Esquibel, L. Meyerdirk, E. Schulz, S. Pierce, A. R. Burmeister, Z. Madaj, J. A. Steiner, M. L. Escobar Galvis, L. Brundin and P. Brundin, Mol. Neurodegener., 2019, 14, 34 CrossRef PubMed.
- S. Hickman, S. Izzy, P. Sen, L. Morsett and J. El Khoury, Nat. Neurosci., 2018, 21, 1359–1369 CrossRef CAS PubMed.
- Z. H. Guan, J. A. Kuhn, X. D. Wang, B. Colquitt, C. Solorzano, S. Vaman, A. K. Guan, Z. Evans-Reinsch, J. Braz, M. Devor, S. L. Abboud-Werner, L. L. Lanier, S. Lomvardas and A. I. Basbaum, Nat. Neurosci., 2016, 19, 94–101 CrossRef CAS PubMed.
- H. Lim, J. Lee, B. You, J. H. Oh, H. J. Mok, Y. S. Kim, B. E. Yoon, B. G. Kim, S. K. Back, J. S. Park, K. P. Kim, R. L. Schnaar and S. J. Lee, EMBO J., 2020, 39, 14 CrossRef PubMed.
- J. A. Coull, S. Beggs, D. Boudreau, D. Boivin, M. Tsuda, K. Inoue, C. Gravel, M. W. Salter and Y. De Koninck, Nature, 2005, 438, 1017–1021 CrossRef CAS PubMed.
- B. Choi, M. Soh, Y. Manandhar, D. Kim, S. I. Han, S. Baik, K. Shin, S. Koo, H. J. Kwon, G. Ko, J. Oh, H. Hwang, T. Hyeon and S. J. Lee, Nanoscale, 2019, 11, 19437–19447 RSC.
- S. Papa, I. Caron, E. Erba, N. Panini, M. De Paola, A. Mariani, C. Colombo, R. Ferrari, D. Pozzer, E. R. Zanier, F. Pischiutta, J. Lucchetti, A. Bassi, G. Valentini, G. Simonutti, F. Rossi, D. Moscatelli, G. Forloni and P. Veglianese, Biomaterials, 2016, 75, 13–24 CrossRef CAS PubMed.
- H. Kim, B. Choi, H. Lim, H. Min, J. H. Oh, S. Choi, J. G. Cho, J. S. Park and S. J. Lee, Mol. Pain, 2017, 13, 1744806917697006 CrossRef CAS PubMed.
- I. M. Verma and N. Somia, Nature, 1997, 389, 239–242 CrossRef CAS PubMed.
- S. J. Hong, M. H. Ahn, J. Sangshetti, P. H. Choung and R. B. Arote, Carbohydr. Polym., 2018, 181, 1180–1193 CrossRef CAS PubMed.
- A. P. Pandey and K. K. Sawant, Mater. Sci. Eng. C, 2016, 68, 904–918 CrossRef CAS PubMed.
- J. K. Raty, H. P. Lesch, T. Wirth and S. Yla-Herttuala, Curr. Drug Saf., 2008, 3, 46–53 CrossRef CAS PubMed.
- O. Boussif, F. Lezoualc'h, M. A. Zanta, M. D. Mergny, D. Scherman, B. Demeneix and J. P. Behr, Proc. Natl. Acad. Sci. U. S. A., 1995, 92, 7297–7301 CrossRef CAS PubMed.
- R. B. Arote, S. K. Hwang, H. T. Lim, T. H. Kim, D. Jere, H. L. Jiang, Y. K. Kim, M. H. Cho and C. S. Cho, Biomaterials, 2010, 31, 2435–2445 CrossRef CAS PubMed.
- R. Arote, T. H. Kim, Y. K. Kim, S. K. Hwang, H. L. Jiang, H. H. Song, J. W. Nah, M. H. Cho and C. S. Cho, Biomaterials, 2007, 28, 735–744 CrossRef CAS PubMed.
- R. B. Arote, S. K. Hwang, M. K. Yoo, D. Jere, H. L. Jiang, Y. K. Kim, Y. J. Choi, J. W. Nah, M. H. Cho and C. S. Cho, J. Gene Med., 2008, 10, 1223–1235 CrossRef CAS PubMed.
- Y. D. Kim, T. E. Park, B. Singh, K. S. Cho, J. N. Sangshetti, Y. J. Choi, R. B. Arote and C. S. Cho, J. Mater. Chem. B, 2016, 4, 2208–2218 RSC.
- D. M. Lynn and R. Langer, J. Am. Chem. Soc., 2000, 122, 10761–10768 CrossRef CAS.
- R. B. Arote, D. Jere, H. L. Jiang, Y. K. Kim, Y. J. Choi and C. S. Cho, Biomed. Mater., 2009, 4, 044102 CrossRef PubMed.
- D. Jere, C. X. Xu, R. Arote, C. H. Yun, M. H. Cho and C. S. Cho, Biomaterials, 2008, 29, 2535–2547 CrossRef CAS PubMed.
- R. B. Arote, E. S. Lee, H. L. Jiang, Y. K. Kim, Y. J. Choi, M. H. Cho and C. S. Cho, Bioconjugate Chem., 2009, 20, 2231–2241 CrossRef CAS PubMed.
- R. B. Arote, M. K. Yoon, T. H. Kim, D. Jere, H. L. Jiang, Y. K. Kim, I. K. Park and C. S. Cho, J. Nanosci. Nanotechnol., 2010, 10, 3294–3298 CrossRef CAS PubMed.
- S. J. Lee, T. Zhou, C. Choi, Z. Wang and E. N. Benveniste, J. Immunol., 2000, 164, 1277–1285 CrossRef CAS PubMed.
- I. H. Cho, J. Hong, E. C. Suh, J. H. Kim, H. Lee, J. E. Lee, S. Lee, C. H. Kim, D. W. Kim, E. K. Jo, K. E. Lee, M. Karin and S. J. Lee, Brain, 2008, 131, 3019–3033 CrossRef PubMed.
- K. H. Son and J. W. Lee, Materials, 2016, 9, 854 CrossRef PubMed.
- J. H. Yu, J. S. Quan, J. Huang, J. W. Nah and C. S. Cho, J. Mater. Sci. Mater. Med., 2009, 20, 2501–2510 CrossRef CAS PubMed.
- S. Behzadi, V. Serpooshan, W. Tao, M. A. Hamaly, M. Y. Alkawareek, E. C. Dreaden, D. Brown, A. M. Alkilany, O. C. Farokhzad and M. Mahmoudi, Chem. Soc. Rev., 2017, 46, 4218–4244 RSC.
- V. Kumar, J. Qin, Y. Jiang, R. G. Duncan, B. Brigham, S. Fishman, J. K. Nair, A. Akinc, S. A. Barros and P. V. Kasperkovitz, Mol. Ther.—Nucleic Acids, 2014, 3, e210 CrossRef PubMed.
- M. Zhu, G. Nie, H. Meng, T. Xia, A. Nel and Y. Zhao, Acc. Chem. Res., 2013, 46, 622–631 CrossRef CAS PubMed.
- P. R. Lockman, R. J. Mumper, M. A. Khan and D. D. Allen, Drug Dev. Ind. Pharm., 2002, 28, 1–13 CrossRef CAS PubMed.
- M. Thomas and A. M. Klibanov, Appl. Microbiol. Biotechnol., 2003, 62, 27–34 CrossRef CAS PubMed.
- D. Fischer, T. Bieber, Y. X. Li, H. P. Elsasser and T. Kissel, Pharmaceut. Res., 1999, 16, 1273–1279 CrossRef CAS PubMed.
- Y. B. Lim, S. M. Kim, H. Suh and J. S. Park, Bioconjugate Chem., 2002, 13, 952–957 CrossRef CAS PubMed.
- L. W. C. Ho, W. Y. Yung, K. H. S. Sy, H. Y. Li, C. K. K. Choi, K. C. Leung, T. W. Y. Lee and C. H. J. Choi, ACS Nano, 2017, 11, 6085–6101 CrossRef CAS PubMed.
- Q. F. Guo, T. T. Liu, X. Yan, X. H. Wang, S. A. Shi, F. Luo and Z. Y. Qian, Int. J. Nanomed., 2011, 6, 1641–1649 CAS.
- T. Bus, A. Traeger and U. S. Schubert, J. Mater. Chem. B, 2018, 6, 6904–6918 RSC.
- S. H. Chen, E. A. Oyarzabal and J. S. Hong, Methods Mol. Biol., 2013, 1041, 231–240 CrossRef CAS PubMed.
- J. C. Nissen, K. K. Thompson, B. L. West and S. E. Tsirka, Exp. Neurol., 2018, 307, 24–36 CrossRef CAS PubMed.
- F. L. Heppner, M. Greter, D. Marino, J. Falsig, G. Raivich, N. Hovelmeyer, A. Waisman, T. Rulicke, M. Prinz, J. Priller, B. Becher and A. Aguzzi, Nat. Med., 2005, 11, 146–152 CrossRef CAS PubMed.
- D. Kim, B. You, E. K. Jo, S. K. Han, M. I. Simon and S. J. Lee, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 14851–14856 CrossRef CAS PubMed.
- J. D. Cahoy, B. Emery, A. Kaushal, L. C. Foo, J. L. Zamanian, K. S. Christopherson, Y. Xing, J. L. Lubischer, P. A. Krieg, S. A. Krupenko, W. J. Thompson and B. A. Barres, J. Neurosci., 2008, 28, 264–278 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1ra06277h |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2021 |