Open Access Article
Open Access ArticleNew carvacrol and thymol derivatives as potential insecticides: synthesis, biological activity, computational studies and nanoencapsulation†
Carolina M. Natala,
Maria José G. Fernandesa,
Nuno F. S. Pintoa,
Renato B. Pereira b,
Tatiana F. Vieira
b,
Tatiana F. Vieira cd,
Ana Rita O. Rodrigues
cd,
Ana Rita O. Rodrigues e,
David M. Pereira
e,
David M. Pereira b,
Sérgio F. Sousa
b,
Sérgio F. Sousa cd,
A. Gil Fortesa,
Elisabete M. S. Castanheira
cd,
A. Gil Fortesa,
Elisabete M. S. Castanheira e and
M. Sameiro T. Gonçalves
e and
M. Sameiro T. Gonçalves *a
*a
aCentre of Chemistry, Department of Chemistry, University of Minho, Campus of Gualtar, 4710-057 Braga, Portugal. E-mail: msameiro@quimica.uminho.pt
bREQUIMTE/LAQV, Laboratory of Pharmacognosy, Department of Chemistry, Faculty of Pharmacy, University of Porto, R. Jorge Viterbo Ferreira, 228, 4050-313 Porto, Portugal
cAssociate Laboratory i4HB – Institute for Health and Bioeconomy, Faculty of Medicine, University of Porto, 4200-319 Porto, Portugal
dUCIBIO – Applied Molecular Biosciences Unit, BioSIM – Department of Biomedicine, Faculty of Medicine, University of Porto, 4200-319 Porto, Portugal
eCentre of Physics of Minho and Porto Universities (CF-UM-UP), University of Minho, Campus of Gualtar, 4710-057 Braga, Portugal
First published on 21st October 2021
Abstract
New compounds with potential insecticide activity were synthesized by structural modifications performed in the monoterpenoid phenolic moieties of carvacrol and thymol, resulting in a set of derivatives with the ether function containing the propyl, chloropropyl or hydroxypropyl chains, as well as a bicyclic ether with an unsaturated chain containing a carboxylic acid terminal. In addition, an analogue of carvacrol and thymol isomers bearing methoxyl, 1-hydroxyethyl and (3-chlorobenzoyl)oxy, instead of the three original methyl groups, was also synthesized. Several structural changes that resulted in diminished insecticide activity have been identified, but two significantly active molecules have been synthesized, one of them being less toxic to human cells than the naturally-derived starting materials. Structure-based inverted virtual screening and molecular dynamics simulations demonstrate that these active molecules likely target the insect odorant binding proteins and/or acetylcholinesterase and are able to form stable complexes. For the most promising compounds, nanoencapsulation assays were carried out in liposomes of egg phosphatidylcholine/cholesterol (7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3) prepared by both thin film hydration and ethanolic injection methods. The compound-loaded liposomes were generally monodisperse and with sizes smaller than or around 200 nm. The thin film hydration method allowed high encapsulation efficiencies (above 85%) for both compounds and a delayed release, while for the systems prepared by ethanolic injection the encapsulation efficiency is lower than 50%, but the release is almost complete in two days.
3) prepared by both thin film hydration and ethanolic injection methods. The compound-loaded liposomes were generally monodisperse and with sizes smaller than or around 200 nm. The thin film hydration method allowed high encapsulation efficiencies (above 85%) for both compounds and a delayed release, while for the systems prepared by ethanolic injection the encapsulation efficiency is lower than 50%, but the release is almost complete in two days.
Introduction
Crop destruction by pests, mainly by insects, is one of the main problems responsible for losses in agricultural production.1–3 The strategy that has shown the best results in controlling insect pests is the use of synthetic pesticides, but they are associated with adverse impacts on the natural environment. Synthetic insecticides possess a broad spectrum of activity against various groups of insects, leading to almost complete elimination of the pests from the crops; nonetheless, the action of these compounds is not just limited to crop areas. Substantial amounts of insecticides applied in fields exert a toll in aquatic and land ecosystems, negatively impacting invertebrates and also vertebrates. Synthetic insecticides display significant environmental persistence, due to their large half-life, and have also propensity to accumulate in diverse trophic levels of the food net.4–6The urgent need to adopt Integrated Pest Management to protect arable crop production, with the goal of decreasing the current overuse of synthetic insecticides, gives to plant products an important role.7,8 For sustainable agriculture and public health, the future of pest management programs stresses on phytochemicals, either extracts or isolated molecules, as well as other biopesticides.9,10 Bioinsecticides are associated with a number of advantages, namely lower toxicity to non-target organisms than synthetic, efficiency at low concentrations and ready biodegradability, which circumvents environmental pollution problems.7,8,11,12
Monoterpenes, secondary metabolites of plants are one of the most abundant and active groups possessing biological activity against various pests.13,14 Carvacrol (5-isopropyl-2-methylphenol) and thymol (2-isopropyl-5-methylphenol) are important phenolic monoterpenes obtained from the essential oil of some Laminaceae members, such as oregano, thyme, and savory, functioning as a chemical defence mechanism against phytopathogenic microorganisms.15–21
In addition to other monoterpenes, carvacrol and thymol have been structurally modified to enhance their biological activities.22,23 For example, carvacrol, thymol and their derivatives have demonstrated larvicidal activity against Aedes aegypti.24 The insecticidal action of T. vulgaris oil against P. shantungensis could be due to the presence of carvacrol and thymol as reported by Park et al. The authors also showed that thymol analogues have promising potential as first-choice insecticides against P. shantungensis adults and nymphs.25 Chitosan nanoparticles functionalized with β-cyclodextrin containing carvacrol and linalool revealed insecticidal activity against the species Helicoverpa armigera (corn earworm) and Tetranychus urticae (spider mite), as well as repellent activity and reduction in oviposition for the mites.26
However, more studies are necessary to obtain other derivatives/analogues that can help to understand the structure–activity relationship, thus helping to plan structural changes and design of novel insecticides.
Considering all these facts and meeting the current interest in alternative botanical-based pesticides,27,28 the present work is focused on the synthesis of a new set of carvacrol and thymol derivatives, including ether derivatives possessing the propyl chain without and with hydroxyl group and a chlorine atom as terminals, as well as the corresponding bicyclic ethers with an unsaturated chain containing a carboxylic acid terminal. The analogue bearing methoxyl, 1-hydroxyethyl and (3-chlorobenzoyl)oxy instead of the three original methyl groups present in carvacrol or thymol skeletons was also synthesized.
The insecticide activity of all compounds against the insect cell line Sf9 (Spodoptera frugiperda) was evaluated. Moreover, computational studies were carried out to identity the most likely protein targets responsible for the observed insecticide activity of the new carvacrol and thymol derivatives. The two most active compounds were encapsulated in liposomes, considering the advantage in protecting them from early degradation and to surpass the high volatility generally observed in monoterpenes. Encapsulation efficiencies and release assays were carried out. Pursuing full biocompatibility, a natural phospholipid extract, phosphatidylcholine from egg yolk (egg-PC) was chosen as main component for the formulation of liposomes, considering that phosphatidylcholines are major constituents of biological membranes. Liposomes composed of phosphatidylcholine (PC) and cholesterol (Ch) mixture, in a 7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 ratio, are widely used as biological membrane models in studies of membrane permeation by antimicrobial peptides29,30 and antitumor drug release.31,32 Moreover, cholesterol is also known for its essential role as modulator of membrane fluidity. A promising utility for the encapsulated novel compounds is anticipated in future development of pesticide nanoformulations.
3 ratio, are widely used as biological membrane models in studies of membrane permeation by antimicrobial peptides29,30 and antitumor drug release.31,32 Moreover, cholesterol is also known for its essential role as modulator of membrane fluidity. A promising utility for the encapsulated novel compounds is anticipated in future development of pesticide nanoformulations.
Results and discussion
Synthesis
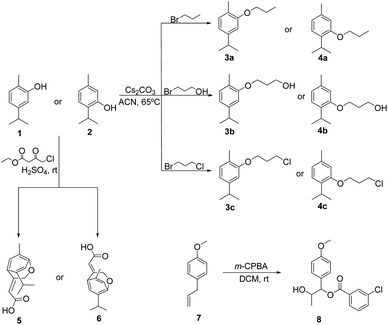
Essential oils components, such as carvacrol and thymol, can be sensitive to environmental conditions, such as temperature, light and oxygen, resulting in phenoxide radicals, which further proceed to oxidative, and polymerization reactions, that may lead to loss of insecticidal efficacy. They are safe and good starting materials allowing structural changes, namely through the hydroxyl group, and the obtention of derivatives can contribute to circumvent their stability limitations, improve physicochemical properties and result in compounds with improved biological activity. In this sense, carvacrol, 5-isopropyl-2-methylphenol 1 was reacted with 1-bromopropane, 3-bromopropan-1-ol and 1-bromo-3-chloropropane, using cesium carbonate as a base, by heating at 65 °C in acetonitrile, followed by column chromatography on silica gel using dichloromethane/light petroleum as the eluent, to give the corresponding O-alkylated derivatives, namely 4-isopropyl-1-methyl-2-propoxybenzene 3a, 3-(5-isopropyl-2-methylphenoxy)propan-1-ol 3b, and 2-(3-chloropropoxy)-4-isopropyl-1-methylbenzene 3c (Scheme 1). Although chemical structures of carvacrol and thymol isomers are very similar, only differing in the insertion of the OH-group placed in ortho (carvacrol) or meta (thymol) positions their activity may be different. Thus, starting from thymol, 2-isopropyl-5-methylphenol 2 and using again 1-bromopropane, 3-bromopropan-1-ol and 1-bromo-3-chloropropane in similar reaction conditions as mentioned above, 1-isopropyl-4-methyl-2-propoxybenzene 4a, 3-(2-isopropyl-5-methylphenoxy)propan-1-ol 4b and 2-(3-chloropropoxy)-1-isopropyl-4-methylbenzene 4c were also obtained.Compounds 3a–c and 4a–c were obtained as oils in 8 to 73% yields and were fully characterized by 1H and 13C NMR spectroscopy and HRMS. The 1H NMR spectra of all compounds showed the methylenic protons of the ether-linked aliphatic chains introduced by alkylation reaction, namely OCH2CH2CH2 as triplet (δ 3.99–4.38 ppm), OCH2CH2CH2 as multiplet, triplet (3b) or quintet (4c) (δ 1.83–4.38 ppm) and OCH2CH2CH2 as doublet (3b) or triplet (δ 3.80–4.09 ppm), as well as the methyl groups (δ 1.12–1.14 ppm, in 3a and 4a), in addition to the aromatic protons (δ 6.67–7.18 ppm) related to the carvacrol and thymol structures. 13C NMR spectra showed the signals of methylenic carbons of the ether-linked aliphatic chains, namely OCH2CH2CH2 (δ 64.11–69.36 ppm), OCH2CH2CH2 (δ 22.65–32.56 ppm) and OCH2CH2CH2 (δ 1.03–4.30 ppm), as well as the methyl carbons (δ ∼10.60 ppm, in 3a and 4a), in addition to the remaining aromatic carbons (δ 109.32–157.15 ppm) related to carvacrol and thymol structures.
In order to obtain bicyclic ether derivatives possessing a conjugated system with simultaneously an aliphatic chain with a double linkage and an electron withdrawing group, carvacrol 1 and thymol 2 were reacted with ethylchloroacetoacetate in acid medium at room temperature,33 followed by column chromatography purification in silica gel using dichloromethane/light petroleum as the eluent. Compounds 5 and 6 were obtained as solid materials in moderate yields and their structures were confirmed by the usual analytical techniques. 1H NMR spectra showed the methylenic protons as doublets (δ 5.11 and 5.34 ppm), the double bond protons as triplets (δ 6.1 and 6.21 ppm), in addition to the remaining aliphatic protons for the dimethyl group (δ 1.24 and 1.15 ppm) and the isopropyl group protons (δ 3.10–3.22 ppm); the aromatic protons show up as singlets (δ 6.86–7.19 ppm). 13C NMR spectra main features confirmed the presence of methylenic carbons (δ 73.45 and 72.42 ppm), and the double bond carbons (δ 116.08 and 112.32 ppm), in addition to the carbonyl group carbon (δ 174.39 and 174.29 ppm). Considering that a carboxylic ester as well as a chlorine atom in lateral chains of natural derivatives may increase their insecticidal activity, it was decided to prepare carvacrol/thymol analogues possessing (3-chlorobenzoyl)oxy, in addition to 1-hydroxyethyl and methoxyl groups instead of the three methyl original groups present in the considered isomers. Thus, reaction of anethole, 1-allyl-4-methoxybenzene 7 with m-chloroperbenzoic acid in dichloromethane gave 2-hydroxy-1-(4-methoxyphenyl)propyl 3-chlorobenzoate 8 as an oil in 89%. In the 1H NMR spectrum stands out the protons of the 1-hydroxyethyl group as a multiplet (δ 4.19–4.26 ppm) and methoxyl group as a singlet (δ 3.81 ppm), as well as the aromatic protons of both rings as a series of six signals (δ 6.91 and 8.05 ppm). 13C NMR spectrum showed the carbons of the 1-hydroxyethyl group (δ 70.20 ppm), the methoxyl group (δ 55.25 ppm), and carbonyl group (δ 164.71 ppm), as well as the aromatic carbons of both rings as 10 signals (δ 114.06 and 159.74 ppm).
Screening of toxicity towards insect cells
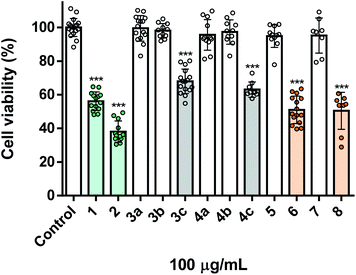
Aiming the evaluation of the insecticidal activity of the synthesized carvacrol and thymol derivatives/analogues, Spodoptera frugiperda cells, a common pest, were used. For comparison purposes of their potency, all the molecules under study were screened at the same concentration (100 μg mL−1). As can be seen in Fig. 1, the naturally-derived starting materials carvacrol 1 and thymol 2, displayed a marked toxicity towards Sf9 insect cells, decreasing cell viability to ca. 55% and 40%, respectively. In a general way, the alkylation of the phenolic hydroxyl group of both carvacrol 1 and thymol 2 resulted in derivatives with lower toxicity (compounds 3a–c and 4a–c), pointing out the importance of this group to the activity displayed (Fig. 1). Particularly, the O-alkylated derivatives containing propane and propan-1-ol (3a, b and 4a, b) were completely devoid of toxicity. Noteworthy, when compared with 3a, b and 4a, b, the presence of a chlorine atom in the lateral chains of the natural derivatives significantly increased their toxicity (3c and 4c). On the other hand, the syntheses of the bicyclic ether derivatives (compounds 5 and 6) seem to lead to a reduction in toxicity, with a complete loss of activity in the case of compound 5. However, compound 6 is still significantly active, with 50% of viability loss of Sf9 cells (Fig. 1). Among all derivatives/analogues synthesized, compounds 6 and 8 were the most potent, both eliciting ca. 50% of decrease in viability of insect cells (Fig. 1).Screening of toxicity towards human lung fibroblasts
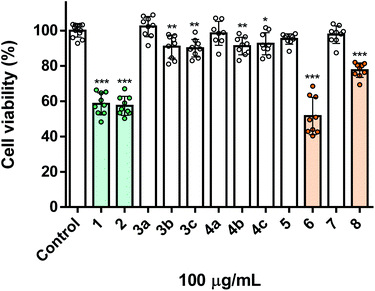
Apart from their toxicity towards insect cells, we were also interested in evaluating the toxicity of these molecules in human cells, thus assessing their potential selectivity. Depending on how insecticides are used, they may affect human and environmental health in a variety of settings. Exposure to insecticides may occur through multiple routes, such as dermal contact, ingestion, and inhalation, the latter being associated to the occupational or residential exposure during or after pesticides application, particularly on the form of aerosols. Thereby, as a model we used human lung fibroblasts (MRC-5 cell line), as they represent a cell population mimicking the major organ (lung) involved in pesticide exposure, namely via inhalation. As shown in Fig. 2, the naturally derived starting materials carvacrol 1 and thymol 2, displayed marked and equivalent toxicity towards MRC-5 cells, both decreasing cell viability to ca. 60%. This result indicates that the position of the hydroxyl group in the two molecules is irrelevant to the toxicity towards fibroblasts, contrary to what was verified against insect cells. On the other hand, the O-alkylated derivatives (3a–c and 4a–c) were completely or nearly devoid of toxicity (Fig. 2), derivatives holding a chlorine atom in the lateral chain (3c and 4c) displaying a selective effect towards insect cells. It is also important to mention that compounds 6 and 8 were of equal or lower toxicity to MRC-5, when compared to the natural counterparts (Fig. 2), respectively. Notably, compound 8 had a marginal effect in human cells (ca. 20% viability loss) (Fig. 2) at the same concentration in which it caused ca. 50% of cell viability loss in insect cells (Fig. 1), pointing out to its potential use as a semisynthetic insecticide.Nanoencapsulation and release assays
The most active compounds against Sf9 cells, compounds 6 and 8, were encapsulated in egg-PC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Ch (7
Ch (7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3) liposomes, prepared by two methods, thin film hydration (TFH)34 and ethanolic injection (EI),35 and hydrodynamic sizes and polydispersity were measured by dynamic light scattering (Table 1). Encapsulation efficiencies of both compounds were determined and are also presented in Table 1.
3) liposomes, prepared by two methods, thin film hydration (TFH)34 and ethanolic injection (EI),35 and hydrodynamic sizes and polydispersity were measured by dynamic light scattering (Table 1). Encapsulation efficiencies of both compounds were determined and are also presented in Table 1.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Ch (7
Ch (7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3) containing compounds 6 and 8
3) containing compounds 6 and 8
| Cpd | Method | Size ± SD (nm) | PDI ± SD | EE (%) ± SD (%) |
|---|---|---|---|---|
| 6 | EI | 101.3 ± 22 | 0.24 ± 0.02 | 48.9 ± 0.5 |
| TFH | 206.9 ± 28 | 0.26 ± 0.04 | 85.4 ± 0.8 | |
| 8 | EI | 86.4 ± 19 | 0.21 ± 0.02 | 41 ± 6 |
| TFH | 165.4 ± 15 | 0.15 ± 0.02 | 94.4 ± 2 |
It can be observed that the hydrodynamic sizes of compound-loaded nanosystems are around or below 200 nm and with a low polydispersity. Considering encapsulation efficiencies, the thin film hydration is the most suitable method for both compounds, exhibiting encapsulation efficiencies higher than 85% (attaining more than 94% for compound 8). In the case of EI preparation method, the encapsulation efficiencies are lower than 50%.
The release of the encapsulated compounds 6 and 8 was followed for 48 h at room temperature towards a buffer of neutral pH (Fig. 3).
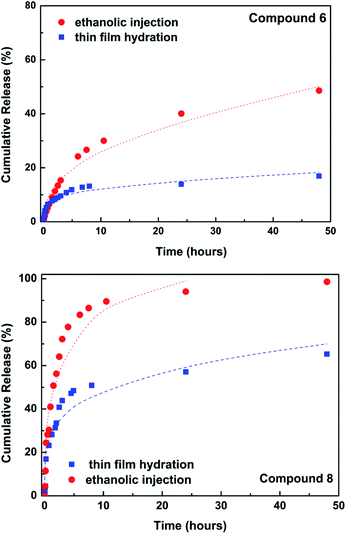 | ||
| Fig. 3 Release profiles and fitting to Korsmeyer–Peppas model. The lines are the fittings to the model. | ||
The release profiles were fitted to the Korsmeyer–Peppas model36 (fitted curves also in Fig. 3), the results being presented in Table 2.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Ch liposomal nanosystems. R is the coefficient of determination
Ch liposomal nanosystems. R is the coefficient of determination
| Cpd | Method | K (min−1) | n | R2 |
|---|---|---|---|---|
| 6 | EI | 1.82 × 10−2 | 0.416 | 0.98 |
| TFH | 2.49 × 10−2 | 0.250 | 0.97 | |
| 8 | EI | 1.23 × 10−1 | 0.305 | 0.97 |
| TFH | 1.01 × 10−1 | 0.252 | 0.95 |
The release is more effective from liposomes prepared with EI method, but the amount of compounds encapsulated is also significantly lower (considering EE% values). The liposomal systems obtained by TFH method exhibit a delayed release, with a much lower amount of compounds released in 48 h. In the case of liposomes loaded with compound 8 and prepared by EI method, the release was fitted only until 24 h (almost complete release at that time). Release of compound 8 is much faster than that of compound 6, which seems to be retained in the liposomes. In all cases, the release mechanism is diffusion-controlled (n < 0.45).
Inverted virtual screening results
Table S1† summarizes the average scores obtained for each protein-target with compounds 6 and 8. GOLD scores are dimensionless, and a higher score means higher affinity, whereas Vina is the opposite. It uses a metric that is a more precise approximation of binding free energy, so a more negative value means better affinity. For each group of targets, the molecular structure with the highest score was selected and ranked from highest to lowest affinity according to the prediction of each scoring function. The scoring values obtained with the 5 independents scoring functions employed demonstrate that the most likely protein targets for the compounds 6 and 8 are Odorant Binding proteins (OBP), acetylcholinesterase (AChE), and chitinase. This tendency is consistent across the different methods evaluated.Molecular dynamics simulations and free energy calculations results
Molecular dynamic simulations were performed considering the most likely proteins targets predicted in the inverted virtual screening protocol: AChE and OBPs. This was done to validate the results and evaluate the interactions between protein and ligand. The structures chosen were the ones presented the overall higher score: 1QON for AChE and 3K1E for OBPs.The binding of compounds 6 and 8 towards these targets was evaluated, starting from the target–ligand complexes obtained from the docking calculations. The overall stability and strength of interaction was calculated through root mean square deviation (RMSD) for both the Cα atom of protein and ligand. The solvent accessible surface area (SASA) of the molecules was analysed, as well as the percentage of potential SASA of the ligands that was buried by the target upon binding, and the number of hydrogen bonds formed throughout time. A summary of all these results is presented in Table 3.
| Compound | Average RMSD of the ligand (Å) | SASA (Å2) | Percentage of ligand SASA buried (%) | Average H-bonds | ΔGbind (kcal mol−1) | Main contributors | |
|---|---|---|---|---|---|---|---|
| AChE | 6 | 0.7 ± 0.2 | 45.5 ± 14.8 | 89 | 1.5 ± 0.8 | −19.4 ± 0.2 | Arg70 (−6.4 ± 3.2) |
| Tyr374 (−2.5 ± 2.0) | |||||||
| Trp83 (−2.2 ± 0.5) | |||||||
| 8 | 1.1 ± 0.2 | 68.7 ± 18.2 | 87 | 0.5 ± 0.6 | −32.2 ± 0.2 | Trp83 (−2.8 ± 0.5) | |
| Tyr374 (−1.9 ± 0.7) | |||||||
| Tyr71 (−1.7 ± 0.9) | |||||||
| OBP | 6 | 0.6 ± 0.3 | 52.1 ± 19.2 | 87 | 0.7 ± 0.9 | −21.3 ± 0.5 | Arg85 (−3.1 ± 4.4) |
| Ser108 (−2.8 ± 2.0) | |||||||
| Trp105 (−2.5 ± 0.7) | |||||||
| 8 | 1.5 ± 0.4 | 19.2 ± 8.6 | 96 | 0.3 ± 0.5 | −32.5 ± 0.2 | Phe114 (−1.6 ± 1.1) | |
| Leu71 (−1.5 ± 0.5) | |||||||
| Trp105 (−1.2 ± 0.5) |
Analysis of the RMSD values presented in Fig. S1† shows that compounds 6 and 8 retain the overall positions predicted from docking along the 100 ns of MD simulation performed (average RMSD values between 0.6 and 1.5 Å). In addition, an analysis of the percentage of the potential SASA of the ligand buried by the target (Table 3 and Fig. S2†) demonstrates that the protein-ligand complexes remain quite stable throughout the simulations, remaining well-shielded from the solvent by AChE and OBP. Compound 8 remains strongly associated to OBP, maintaining on average 96% of its surface non-solvent accessible. With AChE, compound 8 has only 87% of its surface protected by the protein. Compound 6 remains also strongly associated to OBP and AChE with average values of non-solvent accessible surface of 87 and 89%, respectively.
Analysing the number of hydrogen bonds formed between each molecule and their putative target throughout the simulation is helpful to understand the strength and type of interactions formed (Table 3 and Fig. S3†). Compound 6 establishes on average more hydrogen bonds with both targets than compound 8. This effect is more evident for AchE (2 hydrogen bonds on average for compound 6 vs. 0.5 for compound 8).
Table 3 also presents the values for the overall Gibbs energy of association calculated using MM-GBSA. The results show that compound 8 has stronger affinity towards both targets than compound 6, with binding free energies ranging from −32.5 and −32.2 kcal mol−1 for OBP and AChE, against −21.3/−19.4 kcal mol−1. As exhibited by the MM-GBSA values, compounds 6 and 8 display very similar preferences for AChE and OBP.
OBP are a class of proteins that are also present in many different organisms, from mammals to invertebrates. Their function is to bind and transport small hydrophobic molecules into the olfactory receptors. In insects they are present in high number, but they present common features such as their small size, six alpha-helix domains and six cysteine residues bound by three disulfide bonds.37–39
There is a virtual screening study regarding the effect of several essential oils as pesticide agents, with promising results with thymol, carvacrol and OBP, when compared with DEET, a compound commonly used as mosquito repellent.40 There is also deposited in the Protein Data Bank, a structure of a mammal OBP bound to thymol (PDB: 1E02). While the structure is different from insect OBP, it maintains a similar structural function of binding to small odorant molecules. Both compounds 6 and 8 are lipophilic molecules but compound 8 seems to have a higher binding affinity toward OBP.
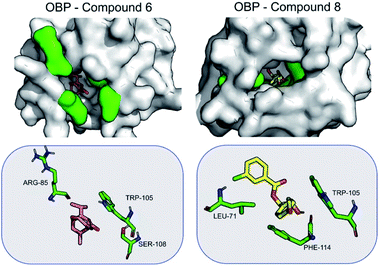
When bound to OBP1, compound 6 is stabilized primarily by electrostatic interactions with Arg85 (−3.1 ± 4.4), Ser108 (−2.8 ± 2.0) and Trp105 (−2.5 ± 0.7). Compound 8 is stabilized mainly by non-polar interactions with Trp105 (−1.2 ± 0.5), Phe114 (−1.6 ± 1.1) and Leu71 (−1.5 ± 0.5) (Fig. 4). The results seem to suggest that compound 8 can be a good antagonist for OBP.
AChE is a common target for several pesticides, due to its crucial role in the hydrolysis of acetylcholine. This serine hydrolase is a common neurotransmitter regulator in many species, from mammals to insects, hence, the lack of specificity of pesticides directed to this target lead to a series of health and environmental problems.41,42 Structural studies with AChE led to the discovery that the pesticides worked by phosphorylating a conserved serine residue in the active site. However, targeting specific structural differences between AChE of different species can be a powerful tool in the discovery of new and more specific pesticides.41
To assess the specificity of these two molecules towards insect AChE in comparison with human AChE, docking studies were performed to the human form of AChE (PDB: 5HFA) with all the scoring functions tested in this study (data shown in ESI, Table S2†). Relevantly, the results consistently showed that compounds 6 and 8 presented higher docking scores against insect AChE than towards human AChE. This tendency was observed with the five independent scoring functions evaluated, suggesting that these molecules show strong preference to insect AChE binding in comparison to human AChE.
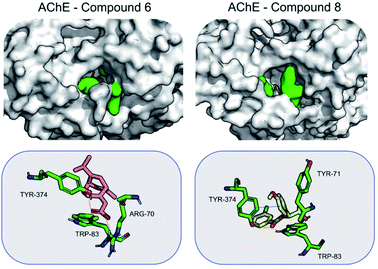
When bound to insect AChE (1QON), compound 6 is stabilized mainly by Arg70 (−6.4 ± 3.2), Tyr374 (−2.5 ± 2.0) and Trp83 (−2.2 ± 0.5) by hydrogen bonds and electrostatic interactions. For compound 8, the stabilization mainly comes from π–π interactions, as well as hydrogen bond formed with Tyr374. Hydrogen bonds are formed also with Trp83 (−1.9 ± 0.4) (Fig. 5).
Experimental
Chemistry
TLC analyses were carried out on 0.25 mm thick precoated silica plates (Merck Fertigplatten Kieselgel 60F254) and spots were visualized under UV light. Chromatography on silica gel was carried out on Merck Kieselgel (230–240 mesh). NMR spectra were obtained on a Bruker Avance III at an operating frequency of 400 MHz for 1H NMR and 100.6 MHz for 13C NMR using the solvent peak as internal reference at 25 °C. All chemical shifts are given in ppm using δ Me4Si = 0 ppm as reference and J values are given in hertz. Assignments were made by comparison of chemical shifts, peak multiplicities and J values and were supported by spin decoupling-double resonance and bidimensional heteronuclear correlation techniques. High resolution mass spectrometry analyses were performed at the “CACTI – Centro de Apoio Científico-Tecnolóxico á Investigación”, at University of Vigo, Spain.General procedure for the synthesis of compounds 3a–c and 4a–c
To a solution of carvacrol, 5-isopropyl-2-methylphenol 1 (1 equiv.), thymol, 2-isopropyl-5-methylphenol 2 (1 equiv.) in acetonitrile (4 mL), the corresponding alkyl halide (1.1 equiv.) and cesium carbonate (5 equiv.) were added, and the resulting mixture was heated at 65 °C for 2 h 30 min. The progress of the reaction was monitored by TLC (light petroleum). The excess of base was filtered, the solvent was evaporated and the crude mixture was purified by column chromatography on silica gel using dichloromethane/light petroleum (mixtures of increasing polarity) as the eluent.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1). 1H NMR (CDCl3, 400 MHz): δH 1.24 (6H, d, J 6.8 Hz CH(CH3)2), 2.15–2.22 (5H, m, CH3Ph and OCH2CH2CH2OH), 2.81–2.91 (1H, m, CH(CH3)2), 4.09 (2H, t, J 6.0 Hz, OCH2CH2CH2OH), 4.38 (2H, t, J 6.0 Hz, OCH2CH2CH2OH), 6.69 (1H, d, J 1.6 Hz, H-6), 6.74 (1H, dd, J 7.6 and 1.6 Hz, H-4), 7.06 (1H, d, J 7.6 Hz, H-3) ppm. 13C NMR (CDCl3, 100.6 MHz): δC 15.76 (CH3Ph), 24.11 (CH(CH3)2), 28.85 (OCH2CH2CH2Cl), 34.12 (CH(CH3)2), 63.77 (OCH2CH2CH2OH), 64.93 (OCH2CH2CH2OH), 109.32 (C-6), 118.18 (C-4), 124.08 (C-2), 130.43 (C-3), 147.89 (C-5), 156.68 (C-1) ppm. HRMS: m/z (ESI-TOF): calcd for C13H21O2 [M + 1]+ 209.1536; found 209.1539.
1). 1H NMR (CDCl3, 400 MHz): δH 1.24 (6H, d, J 6.8 Hz CH(CH3)2), 2.15–2.22 (5H, m, CH3Ph and OCH2CH2CH2OH), 2.81–2.91 (1H, m, CH(CH3)2), 4.09 (2H, t, J 6.0 Hz, OCH2CH2CH2OH), 4.38 (2H, t, J 6.0 Hz, OCH2CH2CH2OH), 6.69 (1H, d, J 1.6 Hz, H-6), 6.74 (1H, dd, J 7.6 and 1.6 Hz, H-4), 7.06 (1H, d, J 7.6 Hz, H-3) ppm. 13C NMR (CDCl3, 100.6 MHz): δC 15.76 (CH3Ph), 24.11 (CH(CH3)2), 28.85 (OCH2CH2CH2Cl), 34.12 (CH(CH3)2), 63.77 (OCH2CH2CH2OH), 64.93 (OCH2CH2CH2OH), 109.32 (C-6), 118.18 (C-4), 124.08 (C-2), 130.43 (C-3), 147.89 (C-5), 156.68 (C-1) ppm. HRMS: m/z (ESI-TOF): calcd for C13H21O2 [M + 1]+ 209.1536; found 209.1539.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1). 1H NMR (CDCl3, 400 MHz): δH 1.21 (6H, d, J 7.2 Hz, CH(CH3)2), 2.16–2.23 (2H, m, OCH2CH2CH2OH), 2.33 (3H, s, CH3Ph), 3.23–3.33 (1H, m, CH(CH3)2), 4.07 (2H, t, J 6.0 Hz, OCH2CH2CH2OH), 4.38 (2H, t, J 6.4 Hz, OCH2CH2CH2OH), 6.67 (1H, d, J 1.6 Hz, H-6), 6.76 (1H, dd, J 8.0 and 1.6 Hz, H-4), 7.10 (1H, d, J 8.0 Hz, H-3) ppm. 13C NMR (CDCl3, 100.6 MHz): δC 21.30 (CH3Ph), 22.73 (CH(CH3)2), 26.54 (CH(CH3)2), 28.87 (OCH2CH2CH2Cl), 63.80 (OCH2CH2CH2OH), 64.94 (OCH2CH2CH2OH), 112.11 (C-6), 121.24 (C-4), 125.87 (C-3), 134.00 (C-2), 136.28 (C-5), 155.67 (C-1) ppm. HRMS: m/z (ESI-TOF): calcd for C13H21O2 [M + H]+ 209.1536; found 209.1530.
1). 1H NMR (CDCl3, 400 MHz): δH 1.21 (6H, d, J 7.2 Hz, CH(CH3)2), 2.16–2.23 (2H, m, OCH2CH2CH2OH), 2.33 (3H, s, CH3Ph), 3.23–3.33 (1H, m, CH(CH3)2), 4.07 (2H, t, J 6.0 Hz, OCH2CH2CH2OH), 4.38 (2H, t, J 6.4 Hz, OCH2CH2CH2OH), 6.67 (1H, d, J 1.6 Hz, H-6), 6.76 (1H, dd, J 8.0 and 1.6 Hz, H-4), 7.10 (1H, d, J 8.0 Hz, H-3) ppm. 13C NMR (CDCl3, 100.6 MHz): δC 21.30 (CH3Ph), 22.73 (CH(CH3)2), 26.54 (CH(CH3)2), 28.87 (OCH2CH2CH2Cl), 63.80 (OCH2CH2CH2OH), 64.94 (OCH2CH2CH2OH), 112.11 (C-6), 121.24 (C-4), 125.87 (C-3), 134.00 (C-2), 136.28 (C-5), 155.67 (C-1) ppm. HRMS: m/z (ESI-TOF): calcd for C13H21O2 [M + H]+ 209.1536; found 209.1530.General procedure for the synthesis of compounds 5 and 6
To a solution of carvacrol, 5-isopropyl-2-methylphenol 1 (1 equiv.) or thymol, 2-isopropyl-5-methylphenol 2 (1 equiv.) in 70% aqueous sulfuric acid (5 mL), ethylchloroacetoacetate (1.5 equiv.) was added and kept under stirring at room temperature for 5 or 3 days, respectively. The reaction mixture was poured into ice water and stirred for 2 h to give a precipitate. The solid was collected by filtration, washed with cold water, dried in a vacuum oven, and the crude mixture was purified by column chromatography on silica gel using dichloromethane/light petroleum 90![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10, as the eluent.
10, as the eluent.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5). 1H NMR (DMSO-d6, 400 MHz): δH 1.15 (6H, d, J 6.8 Hz, CH(CH3)2), 2.32 (3H, s, PhCH3), 3.10–3.18 (1H, m, CH(CH3)2), 5.34 (2H, d, J 1.6 Hz, CCH2O), 6.21 (1H, t, J 1.6 Hz, CHCO2H), 6.73 (1H, s, Ar-H), 7.19 (1H, s, Ar-H), 9.93 (1H, s, OH) ppm. 13C NMR (DMSO-d6, 100.6 MHz): δC 21.68 (CH3), 22.26 (CH(CH3)2), 26.33 (CH(CH3)2), 72.42 (CCH2O), 112.32 (CHCO2H), 118.11 (Ar-C), 120.37 (Ar-C), 126.40 (Ar-C), 132.36 (Ar-C), 136.83 (Ar-C), 156.74 (Ar-C), 164.55 (C(CH2)) 174.29 (CO2H) ppm. HRMS: m/z (ESI-TOF): calcd for C14H17O3 [M + H]+ 233.1172; found 233.1177.
5). 1H NMR (DMSO-d6, 400 MHz): δH 1.15 (6H, d, J 6.8 Hz, CH(CH3)2), 2.32 (3H, s, PhCH3), 3.10–3.18 (1H, m, CH(CH3)2), 5.34 (2H, d, J 1.6 Hz, CCH2O), 6.21 (1H, t, J 1.6 Hz, CHCO2H), 6.73 (1H, s, Ar-H), 7.19 (1H, s, Ar-H), 9.93 (1H, s, OH) ppm. 13C NMR (DMSO-d6, 100.6 MHz): δC 21.68 (CH3), 22.26 (CH(CH3)2), 26.33 (CH(CH3)2), 72.42 (CCH2O), 112.32 (CHCO2H), 118.11 (Ar-C), 120.37 (Ar-C), 126.40 (Ar-C), 132.36 (Ar-C), 136.83 (Ar-C), 156.74 (Ar-C), 164.55 (C(CH2)) 174.29 (CO2H) ppm. HRMS: m/z (ESI-TOF): calcd for C14H17O3 [M + H]+ 233.1172; found 233.1177.Synthesis of (2-hydroxy-1-(4-methoxyphenyl)propyl 3-chlorobenzoate 8
To a suspension of m-CPBA (55%; 1.261 g, 7.31 mmol, 3.0 equiv.) and sodium hydrogen carbonate (0.343 g, 4.08 mmol, 3.0 equiv.) in dichloromethane (2.8 mL) at 0 °C (ice bath), with stirring, a solution of anethole, 1-allyl-4-methoxybenzene 7 (0.2 mL, 1.35 mmol) in dichloromethane (2 mL) was added. The reaction mixture was allowed to stir at room temperature for 23 h. Then, it was washed with aqueous solution of sodium sulfite 10% (2 × 5 mL) and a saturated aqueous solution of sodium hydrogen carbonate (2 × 5 mL). The organic phase was dried with magnesium sulfate and the solvent was evaporated on a rotary evaporator, yielding compound 8 as a yellow oil (0.384 g, 89%). Rf = 0.40 (dichloromethane/methanol, 99![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1). 1H NMR (CDCl3, 400 MHz): δH 1.15 (3H, d, J 6.4 Hz, CH3), 3.81 (3H, s, OCH3), 4.19–4.26 (1H, m, CH(CH3)OH), 5.75 (1H, d, J 7.6 Hz, CHCO2PhCl), 6.78 (1H, s, OH), 6.91 (2H, d, J 8.8 Hz, H-3 and H-5 PhOCH3), 7.35 (2H, d, J 8.8 Hz, H-2 and H-6 PhOCH3), 7.40 (1H, t, J 8.0 Hz, H-5 PhCl), 7.55–7.57 (1H, m, H-6 PhCl), 7.94–7.98 (1H, m, H-4 PhCl), (8.05 (1H, d, J 1.6 Hz, H-2 PhCl) ppm. 13C NMR CDCl3, 100.6 MHz): δC 18.85 (CHCHCH3), 55.25 (OCH3), 70.20 (CHCHCH3), 81.42 (CHCHCH3), 114.06 (Ar-C-2 and Ar-C-6 PhOCH3), 127.83 (Ar-C-3 or Ar-C-5 PhOCH3), 128.52 (Ar-C-5 or Ar-C-3 PhOCH3), 129.32 (Ar-C-4 PhOCH3), 129.64 (Ar-C-6 PhCl), 129.75 (Ar-C-2 and Ar-C-5 PhCl), 131.86 (Ar-C-1 PhCl), 133.14 (Ar-C-4 PhCl), 134.56 (Ar-C-3 PhCl), 159.74 (Ar-C-1 PhOCH3), 164.71 (C
1). 1H NMR (CDCl3, 400 MHz): δH 1.15 (3H, d, J 6.4 Hz, CH3), 3.81 (3H, s, OCH3), 4.19–4.26 (1H, m, CH(CH3)OH), 5.75 (1H, d, J 7.6 Hz, CHCO2PhCl), 6.78 (1H, s, OH), 6.91 (2H, d, J 8.8 Hz, H-3 and H-5 PhOCH3), 7.35 (2H, d, J 8.8 Hz, H-2 and H-6 PhOCH3), 7.40 (1H, t, J 8.0 Hz, H-5 PhCl), 7.55–7.57 (1H, m, H-6 PhCl), 7.94–7.98 (1H, m, H-4 PhCl), (8.05 (1H, d, J 1.6 Hz, H-2 PhCl) ppm. 13C NMR CDCl3, 100.6 MHz): δC 18.85 (CHCHCH3), 55.25 (OCH3), 70.20 (CHCHCH3), 81.42 (CHCHCH3), 114.06 (Ar-C-2 and Ar-C-6 PhOCH3), 127.83 (Ar-C-3 or Ar-C-5 PhOCH3), 128.52 (Ar-C-5 or Ar-C-3 PhOCH3), 129.32 (Ar-C-4 PhOCH3), 129.64 (Ar-C-6 PhCl), 129.75 (Ar-C-2 and Ar-C-5 PhCl), 131.86 (Ar-C-1 PhCl), 133.14 (Ar-C-4 PhCl), 134.56 (Ar-C-3 PhCl), 159.74 (Ar-C-1 PhOCH3), 164.71 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) ppm. HRMS: m/z (ESI-TOF): calcd for C17H1735ClO4 [M + Na]+ 343.0711; found 343.0708; calcd for C17H1737ClO4 [M + Na]+ 345.0684; found 345.0679.
O) ppm. HRMS: m/z (ESI-TOF): calcd for C17H1735ClO4 [M + Na]+ 343.0711; found 343.0708; calcd for C17H1737ClO4 [M + Na]+ 345.0684; found 345.0679.
Cell culture
Sf9 (Spodoptera frugiperda) cells were acquired from ATCC (Manassas, USA), maintained as a suspension culture and cultivated in Grace's medium with 10% FBS and 1% penicillin/streptomycin, at 28 °C. Cells were used in experiments while in the exponential phase of growth. On the other hand, MRC-5 cells (human lung fibroblasts) were also acquired from ATCC and were cultured in MEM supplemented with 10% FBS and 1% penicillin/streptomycin at 37 °C, in a humidified atmosphere of 5% CO2.Viability assessment
For the assessment of viability, a resazurin-based method was used, similarly to what we described before.27 Sf9 and MRC-5 cells were plated at a density of 3.0 × 104 and 2.0 × 104 cells per well, respectively, incubated for 24 h and then exposed to the molecules under study for 24 h. After this period, a commercial solution of resazurin was added (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10) and the kinetic reaction of fluorescence increase monitored at 560/590 nm. For both cell lines, 60 min of incubation were used.
10) and the kinetic reaction of fluorescence increase monitored at 560/590 nm. For both cell lines, 60 min of incubation were used.
Nanoencapsulation studies
Egg yolk phosphatidylcholine (egg-PC) and cholesterol (Ch) were obtained from Sigma-Aldrich. In the ethanolic injection method (EI),35 nanoliposomes were prepared by injection of an ethanolic solution of lipids/compounds mixture (in the ratio egg-PC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Ch 7
Ch 7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3) in an aqueous buffer solution under vigorous stirring. In the thin film hydration (TFH) method,34 a thin film of the egg-PC/Ch mixture was obtained evaporating a lipid solution in chloroform under an ultrapure nitrogen stream. The compound solution was added and, after evaporation, the film was hydrated with the aqueous solution, followed by sonication and ten extrusion cycles through polycarbonate membranes (5 × 400 nm and 5 × 200 nm) in a Lipex™ extruder (from Northern Lipids). In both methods, the final lipid concentration was 1 mM.
3) in an aqueous buffer solution under vigorous stirring. In the thin film hydration (TFH) method,34 a thin film of the egg-PC/Ch mixture was obtained evaporating a lipid solution in chloroform under an ultrapure nitrogen stream. The compound solution was added and, after evaporation, the film was hydrated with the aqueous solution, followed by sonication and ten extrusion cycles through polycarbonate membranes (5 × 400 nm and 5 × 200 nm) in a Lipex™ extruder (from Northern Lipids). In both methods, the final lipid concentration was 1 mM.
The encapsulation efficiency (percent), EE%, was determined through absorbance measurements. After preparation, liposomes were subjected to centrifugation in Amicon® Ultra centrifugal filter units 100 kDa at 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 60 min. Then, the supernatant was removed and its absorption spectrum was measured in a Shimadzu UV-3600 Plus UV/vis/NIR spectrophotometer. Using a previously measured calibration curve of absorbance versus concentration, the encapsulation efficiencies of both compounds were determined through eqn (1), and three independent assays were performed.
000 rpm for 60 min. Then, the supernatant was removed and its absorption spectrum was measured in a Shimadzu UV-3600 Plus UV/vis/NIR spectrophotometer. Using a previously measured calibration curve of absorbance versus concentration, the encapsulation efficiencies of both compounds were determined through eqn (1), and three independent assays were performed.
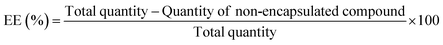 | (1) |
The compound-loaded liposomes' mean hydrodynamic diameter and size distribution (polydispersity) were measured with a Dynamic Light Scattering (DLS) equipment Litesizer 500 from Anton Paar, at 25 °C, using a solid-state laser of 648 nm and 40 mW. Five independent measurements were carried out for each of the samples.
Release assays to phosphate buffer (pH = 7) were performed during 48 h in triplicate, using dialysis membranes. Calibration curves of absorbance versus concentration, previously determined for each compound, were used to calculate the concentration of released compounds. The loaded liposomes were kept under stirring at 25 °C, the solutions being covered to prevent evaporation. The Korsmeyer–Peppas model32 was used to describe the compound release kinetics from the liposomes (eqn (2)):
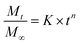 | (2) |
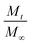 represents the fraction of released drug, K is the release constant, n the transport exponent (dimensionless) and t is the time. The transport exponent is directly related to the release mechanism of the compound: if n > 1, the release is controlled by swelling and material relaxation; 0.89 < n < 1 indicates a relaxation-controlled mechanism, 0.45 < n < 0.89 indicates a combination of diffusion and erosion in drug release (non-Fickian release) and when n < 0.45 the release mechanism is diffusion-controlled (Fickian release).
represents the fraction of released drug, K is the release constant, n the transport exponent (dimensionless) and t is the time. The transport exponent is directly related to the release mechanism of the compound: if n > 1, the release is controlled by swelling and material relaxation; 0.89 < n < 1 indicates a relaxation-controlled mechanism, 0.45 < n < 0.89 indicates a combination of diffusion and erosion in drug release (non-Fickian release) and when n < 0.45 the release mechanism is diffusion-controlled (Fickian release).
Molecular docking and inverted virtual screening optimization
A selection of putative molecular targets associated to insecticide activity was made and was used as a basis for the design of an inverted virtual screening protocol to identity the most likely protein targets responsible for the observed insecticide activity of the carvacrol and thymol derivatives evaluated. The selection of putative molecular targets was made by scanning Scopus using the keywords “Virtual Screening” and “biopesticides”. Some targets were selected from the Protein Data Bank (PDB) based upon the relevance of the target. The relevance and year of publication were considered for the final selection. Of the seventeen studies analyzed, 23 PDB structures were selected to proceed to the optimization of the inverted virtual screening protocol. The molecular targets under consideration are detailed in Table S3.†The 23 molecular targets extracted from the PDB were prepared using the Pymol Autodock Vina plugin43 where crystallographic waters and ligands (when present) were removed. The crystallographic ligands were then saved in separate files and used as reference for active site coordinates, as well as validation for the re-docking steps. In the absence of ligands, the active site coordinates were based on the most important residues described in the literature. Re-docking was used as a validation tool, measuring the ability of the docking software in reproducing the orientation of the crystallographic pose.
Five docking scoring functions were used, and the protocol was optimized for each one and customized to each molecular target with the goal of minimizing the RMSD between the crystallographic and docking poses at the re-docking stage. GOLD44 (PLP, ASP, ChemScore, and GoldScore scoring function) and Autodock Vina45 were the selected docking software as they are well established and are widely used in research.46,47 While it may be difficult to compare different scoring functions, as they handle the target and ligand in different manners, hence different metrics and scales,48 testing multiple alternatives eliminates bias and enables the evaluation of clear interaction tendencies.
Molecular dynamics simulations and free energy calculations
Two of the compounds that presented higher insecticide activity when tested experimentally (compounds 6 and 8) were simulated in complex with the two most promising targets identified from the inverted virtual screening study (acetylcholinesterase – 1QON and odorant binding protein 1 – 3KIE). Molecular dynamics simulations were performed using the Amber18 software.49For MD simulations, the target structures cannot present any gaps. Therefore, a homology model was created for the structure 1QON using SWISS-MODEL50 (detail in ESI, Fig. S4†).
The complexes selected for the MD simulations were the ones that were predicted during the inverted virtual screening experiments using GOLD/PLP and were subsequently treated with the Leap module of AMBER.51 The protein targets were treated with the ff14SB force field,52 while the compounds 6 and 8 were parameterized using ANTECHAMBER, with RESP HF/6-31G(d) charges calculated with Gaussian16 (ref. 53) and the General Amber Force Field (GAFF).54 Counter-ions (Na+) were added to neutralize the overall charge and the complete systems were placed in with TIP3P water boxes with a minimum distance of 12 Å between the protein-surface and the side of the box.
Four consecutive minimizations stages were performed to remove clashes prior to the MD simulation. In these four stages the minimization procedure was applied to the following atoms of the system: 1-water molecules (2500 steps); 2-hydrogens atoms (2500 steps); 3-side chains of all the amino acid residues 2500 steps); 4-full system 10.000 steps). The minimized systems were then subject to a molecular dynamics equilibration procedure, which was divided into two stages: in the first stage (50 ps), the systems were gradually heated to 298 K using a Langevin thermostat at constant volume (NVT ensemble); in the second stage (50 ps) the density of the systems was further equilibrated at 298 K. Finally, molecular dynamic production runs were performed for 100 ns. These were performed with an NPT ensemble at constant temperature (298 K, Langevin thermostat) and pressure (1 bar, Berendsen barostat), with periodic boundary conditions, with an integration time of 2.0 fs using the SHAKE algorithm to constrain all covalent bonds involving hydrogen atoms. A 10 Å cutoff for nonbonded interactions was used during the entire molecular simulation procedure. Final trajectories were analyzed in terms of Root-Mean-Square Deviation (RMSD), confirming that both systems were well equilibrated after the initial 10 ns. The last 90 ns of the simulation were considered for hydrogen bonding analysis, and cluster analysis of the conformations generated. This overall procedure has been previously used with success in the treatment of several biomolecular systems.55–61
In order to estimate the binding free energies of the ligands 6 and 8 toward acetylcholinesterase and odorant binding protein 1, the Molecular Mechanics-Generalized Born Surface Area (MM-GBSA) method62 was applied, taking into account a salt concentration of 0.100 mol dm−3. Also, the contribution of the amino acid residues was estimated using the energy decomposition method. From each MD trajectory, a total of 1400 conformations taken from the last 70 ns of simulation were considered for each MM-GBSA calculation.
Statistical analysis
For biological assays, the Shapiro–Wilk's normality test was performed in the data to ensure that it followed a normal distribution. Comparison between the means of controls and each experimental condition was performed using ANOVA. Outliers were identified by the Grubbs' test. Data was expressed as the mean ± standard deviation (SD) of at least 3 independent experiments. GraphPad Prism 7.0 software was used, and values were considered statistically significant with a p < 0.05.Conclusions
Various derivatives and analogues of carvacrol and thymol isomers were synthesized in which the hydroxyl group was transformed into ethers with different aliphatic chains, such as a propyl without or with a hydroxyl group or chlorine atom as terminals, as well as bicyclic ethers with an unsaturated chain having a carboxylic acid group. In addition, an analogue of the natural isomers was also prepared possessing all the methyl groups changed to methoxyl, 1-hydroxyethyl and (3-chlorobenzoyl)oxy groups.All the synthesized compounds were subjected to tests of biological activity in Sf9, in comparison with the corresponding precursors, in order to evaluate their application as potential natural based insecticides. We identified structural changes that result in diminished insecticide activity and also highlight two molecules, compounds 6 and 8, that are mildly active, the latter being less toxic to human cells than the naturally-derived starting materials.
Structural-based inverted virtual screening studies with five independent methods suggest that the two most potent molecules display their insecticide activity most likely by targeting the insect odorant binding proteins and/or acetylcholinesterase. Molecular dynamics simulations and free energy calculations confirm that these two molecules bind strongly to both targets forming very stable complexes, with well defined-molecular interactions that are maintained through time.
Liposomes of egg phosphatidylcholine/cholesterol (7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3) prepared by the thin film hydration method allow high encapsulation efficiencies and a delayed release of the most active compounds, while the ones prepared by ethanolic injection provide a release higher than 60% in two days.
3) prepared by the thin film hydration method allow high encapsulation efficiencies and a delayed release of the most active compounds, while the ones prepared by ethanolic injection provide a release higher than 60% in two days.
Conflicts of interest
There are no conflicts to declare.Author contributions
Conceptualization, M. S. T. G., D. M. P., S. F. S. and E. M. S. C.; methodology, M. S. T. G., D. M. P., S. F. S. and E. M. S. C.; formal analysis, M. S. T. G., D. M. P., S. F. S., E. M. S. C., R. B. P. and M. J. G. F.; investigation, C. M. N., N. F. S. P., R. B. P., T. F. V. and A. R. O. R.; supervision: M. S. T. G., D. M. P., S. F. S., E. M. S. C. and M. J. G. F.; writing-original draft preparation, M. S. T. G., D. M. P., S. F. S., E. M. S. C. and R. B. P.; writing-review and editing, M. S. T. G., D. M. P., S. F. S., E. M. S. C. R. B. P., A. G. F. and M. J. G. F.; project administration, M. S. T. G. All authors have read and agreed to the published version of the manuscript.Acknowledgements
This study was funded by project PTDC/ASP-AGR/30154/2017 (POCI-01-0145-FEDER-030154) of the COMPETE 2020 program, co-financed by the FEDER and the European Union. The authors acknowledge also to Foundation for Science and Technology (FCT, Portugal), and FEDER-COMPETE-QREN-EU for financial support to the research centres CQ-UM (UID/QUI/00686/2020), CF-UM-UP (UIDB/04650/2020) and REQUIMTE (UIDB/50006/2020). The NMR spectrometer Bruker Avance III 400 is part of the National NMR Network and was purchased within the framework of the National Program for Scientific Re-equipment, contract REDE/1517/RMN/2005 with funds from POCI 2010 (FEDER) and FCT. PRIMA Foundation, H2020-PRIMA 2018-Section 2, MILKQUA project is also acknowledged.Notes and references
- A. Tijjani, K. Bashir, I. Mohammed, A. Muhammad and A. Gambo, J. Biopestic. Agric., 2016, 3(1), 6–13 Search PubMed.
- R. Teixeira, P. Gazolla, A. Silva, M. Borsodi, B. Bergmann, R. Ferreira, B. Vaz, G. Vasconcelos and W. Lima, Eur. J. Med. Chem., 2018, 146, 274–286 CrossRef CAS PubMed.
- B. Gerwick and T. Sparks, Pest Manage. Sci., 2014, 70(8), 1–17 Search PubMed.
- D. Chandler, A. Bailey, G. Mark Tatchell, G. Davidson, J. Greaves and W. Grant, Philos. Trans. R. Soc., B, 2011, 366, 1987–1998 CrossRef PubMed.
- G. Cornelius, G. Lohiya and R. Sharma, Int. J. Eng. Res. Technol., 2019, 8(11), 578–580 Search PubMed.
- J. Oliveira, E. Campos, M. Camara, J. Della Vechia, S. Matos, D. Andrade, K. Gonçalves, J. Nascimento, R. Polanczyk, D. Araújo and L. Fraceto, ACS Appl. Nano Mater., 2020, 3, 207–217 CrossRef.
- J. Oliveira, E. Campos and L. Fraceto, Agric. Food Chem., 2018, 66, 8898–8913 CrossRef PubMed.
- M. Paulraj, S. Ignacimuthu, M. Gandhi, A. Shajahan, P. Ganesan, S. Packiam and N. Al- Dhabi, Int. J. Biol. Macromol., 2017, 104, 1813–1819 CrossRef CAS PubMed.
- E. Fountain and S. Wratten, in Encyclopedia of Ecology, ed. Fath, B., Elsevier, Nova Zelândia, 2013, pp. 377–381 Search PubMed.
- A. Mossa, J. Environ. Sci. Technol., 2016, 9(5), 354–378 CrossRef CAS.
- X. Pang, Y. Feng, X. Qi, Y. Wang, B. Almaz, C. Xi and S. Du, Environ. Sci. Pollut. Res., 2020, 27, 7618–7627 CrossRef CAS PubMed.
- R. Pavela, in Advances in Plant Biopesticides, ed. Singh, D., Springer, Nova Deli, 2014, pp. 347–359 Search PubMed.
- E. Ahmed, M. Arshad, M. Khan, M. Amjad, H. Sadaf, I. Riaz, S. Sabir and N. Sabaoon, J. Pharmacogn. Phytochem., 2017, 6(2), 205–214 CAS.
- I. N. Monteiro, L. O. G. Ferreira, A. K. M. Oliveira, S. Favero, P. L. B. Figueiredo, J. G. S. Maia, O. S. Monteiro and R. Matias, Int. J. Trop. Insect Sci., 2020, 41, 181–187 CrossRef.
- M. El-Miligy, A. Hazzaa, S. El-Zemity and A. Kubeisi, Curr. Bioact. Compd., 2018, 14, 1–13 CrossRef.
- S. Bagul and J. Rajput, Nat. Prod. Chem. Res., 2018, 6(6), 1–3 Search PubMed.
- C. Conceição, L. Morais, D. Campos, J. Chaves, G. Santos, Y. Cid, M. Sousa, F. Scott, D. Chaves and K. Coumendouros, Rev. Bras. Farmacogn., 2020, 30, 774–779 CrossRef.
- A. Escobar, M. Pérez, G. Romanelli and G. Blustein, Arabian J. Chem., 2020, 13, 9243–9269 CrossRef CAS.
- M. Friedman, J. Agric. Food Chem., 2014, 62, 7652–7670 CrossRef CAS PubMed.
- K. Magierowicz, E. Górska-Drabik and C. Sempruch, Pestic. Biochem. Physiol., 2019, 153, 122–128 CrossRef CAS PubMed.
- L. Marinelli, A. Stefano and I. Cacciatore, Phytochem. Rev., 2018, 17, 903–921 CrossRef.
- F. Silva, F. Monte, T. Lemos, P. Nascimento, A. Costa and L. Paiva, Chem. Cent. J., 2018, 12(34), 1–9 Search PubMed.
- O. Hüter, Phytochem. Rev., 2011, 10, 185–194 CrossRef.
- V. B. Silva, D. L. Travassos, A. Nepel, A. Barison, E. V. Costa, L. Scotti, M. T. Scotti, F. J. B. Mendonça-Junior, R. C. Santos and S. C. H. Cavalcanti, J. Arthropod Borne Dis., 2017, 11, 315–330 Search PubMed.
- J.-H. Park, Y.-J. Jeon, C.-H. Lee, N. Chung and H.-S. Lee, Sci. Rep., 2017, 7, 40902 CrossRef CAS PubMed.
- E. V. R. Campos, P. L. F. Proença, J. L. Oliveira, A. E. S. Pereira, L. N. M. Ribeiro, F. O. Fernandes, K. C. Gonçalves, R. A. Polanczyk, T. Pasquoto-Stigliani, R. Lima, C. C. Melville, J. F. D. Vechia, D. J. Andrade and L. F. Fraceto, Sci. Rep., 2018, 8, 7623 CrossRef PubMed.
- M. Fernandes, R. Pereira, D. Pereira, A. Fortes, E. Castanheira and M. Gonçalves, Int. J. Mol. Sci., 2020, 21(9257), 1–14 Search PubMed.
- A. Lopes, M. Monteiro, A. Araújo, A. Rodrigues, E. Castanheira, D. Pereira, P. Olim, A. Fortes and M. Gonçalves, Molecules, 2020, 25, 5855 CrossRef CAS PubMed.
- C. Toniolo, M. Crisma, F. Formaggio, C. Peggion, V. Monaco, C. Goulard, S. Rebuffat and B. Bodo, J. Am. Chem. Soc., 1996, 118(21), 4952–4958 CrossRef CAS.
- M. Lucaciu, S. Rebuffat, C. Goulard, H. Duclohier, G. Molle and B. Bodo, Biochim. Biophys. Acta, Biomembr., 1997, 1323, 85–96 CrossRef CAS.
- A. Abreu, E. Castanheira, M. Queiroz, P. Ferreira, L. Vale-Silva and E. Pinto, Nanoscale Res. Lett., 2011, 6, 482 CrossRef PubMed.
- A. Rodrigues, B. Almeida, J. Rodrigues, M. Queiroz, R. Calhelha, I. Ferreira, A. Pires, A. Pereira, J. Araújo, P. Coutinho and E. Castanheira, RSC Adv., 2017, 7, 15352–15361 RSC.
- Z.-F. Tao, X. Qian and M. Fan, Tetrahedron, 1997, 53, 13329–13338 CrossRef CAS.
- H. Zhang, Methods Mol. Biol., 2017, 1522, 17–22 CrossRef CAS PubMed.
- C. Jaafar-Maalej, R. Diab, V. Andrieu, A. Elaissari and H. Fessi, J. Liposome Res., 2010, 20, 228–243 CrossRef CAS PubMed.
- I. Wu, S. Bala, N. Škalko-Basnet and M. Cagno, Eur. J. Pharm. Sci., 2019, 138, 105026 CrossRef CAS PubMed.
- J. Sun, S. Xiao and J. Carlson, Open Biol., 2018, 8(12), 1–7 Search PubMed.
- P. Pelosi, I. Iovinella, J. Zhu, G. Wang and F. Dani, Biol. Rev., 2018, 93(1), 184–200 CrossRef PubMed.
- P. Pelosi, R. Mastrogiacomo, I. Iovinella, E. Tuccori and K. Persaud, Appl. Microbiol. Biotechnol., 2014, 98(1), 61–70 CrossRef CAS PubMed.
- K. Costa, J. Galúcio, C. Costa, A. Santana, V. Carvalho, L. Nascimento, A. Lima, J. Cruz, C. Alves and J. Lameira, ACS Omega, 2019, 4(27), 22475–22486 CrossRef PubMed.
- Y. Pang, S. Brimijoin, D. Ragsdale, K. Yan Zhu and R. Suranyi, Curr. Drug Targets, 2012, 13(4), 471–482 CrossRef CAS PubMed.
- M. Harel, G. Kryger, T. Rosenberry, W. Mallender, T. Lewis, R. Fletcher, J. Guss, I. Silman and J. Sussman, Protein Sci., 2000, 9(6), 1063–1072 CrossRef CAS PubMed.
- D. Seeliger and B. Groot, J. Comput.-Aided Mol. Des., 2010, 24(5), 417–422 CrossRef CAS PubMed.
- G. Jones, P. Willett, R. Glen, A. Leach and R. Taylor, J. Mol. Biol., 1997, 267, 727–748 CrossRef CAS PubMed.
- O. Trott and A. Olson, J. Comput. Chem., 2010, 31, 455–461 CAS.
- S. Sousa, P. Fernandes and M. Ramos, Proteins: Struct., Funct., Bioinf., 2006, 26, 15–26 CrossRef PubMed.
- S. Sousa, N. Cerqueira, P. Fernandes and M. Ramos, Comb. Chem. High Throughput Screening, 2010, 13, 442–453 CrossRef CAS PubMed.
- T. Vieira and S. Sousa, Appl. Sci., 2019, 9(21), 4538 CrossRef CAS.
- D. Case, T. Cheatham, T. Darden, H. Gohlke, R. Luo, K. Merz Jr, A. Onufriev, C. Simmerling, B. Wang and R. Woods, J. Comput. Chem., 2005, 26, 1668–1688 CrossRef CAS PubMed.
- A. Waterhouse, M. Bertoni, S. Bienert, G. Studer, G. Tauriello, R. Gumienny, F. Heer, T. Beer, C. Rempfer, L. Bordoli, R. Lepore and T. Schwede, Nucleic Acids Res., 2018, 46, W296–W303 CrossRef CAS PubMed.
- J. Wang, R. Wolf, J. Caldwell, P. Kollman and D. Case, J. Comput. Chem., 2004, 25, 1157–1174 CrossRef CAS PubMed.
- J. Maier, C. Martinez, K. Kasavajhala, L. Wickstrom, K. Hauser and C. Simmerling, J. Chem. Theory Comput., 2015, 11, 3696–3713 CrossRef CAS PubMed.
- M. Frisch, G. Trucks and H. Schlegel, et al., Gaussian 09, Revision A.02. Published online 2016 Search PubMed.
- J. Wang, W. Wang, P. Kollman and D. Case, J. Mol. Graphics Modell., 2006, 25, 247–260 CrossRef CAS PubMed.
- S. Sousa, J. Coimbra, D. Paramos, R. Pinto, R. Guimarães, V. Teixeira, P. Fernandes and M. Ramos, J. Mol. Model., 2013, 19, 673–688 CrossRef CAS PubMed.
- S. Sousa, P. Fernandes and M. Ramos, Bioorg. Med. Chem., 2009, 17, 3369–3378 CrossRef CAS PubMed.
- S. Sousa, P. Fernandes and M. Ramos, J. Phys. Chem. B, 2008, 112, 8681–8691 CrossRef CAS PubMed.
- N. Moorthy, S. Sousa, M. Ramos and P. Fernandes, J. Enzyme Inhib. Med. Chem., 2016, 31, 1428–1442 CrossRef CAS PubMed.
- B. Marcial, S. Sousa, I. Barbosa, H. Santos and M. Ramos, J. Phys. Chem. B, 2012, 116, 13644–13654 CrossRef CAS PubMed.
- D. Lapaillerie, C. Charlier, H. Fernandes, S. Sousa, P. Lesbats, P. Weigel, A. Favereaux, V. Guyonnet-Duperat and V. Parissi, Viruses, 2021, 13(3), 365 CrossRef CAS PubMed.
- F. Martins, A. Melo and S. Sousa, Molecules, 2021, 26, 2600 CrossRef CAS PubMed.
- E. Wang, H. Sun, J. Wang, Z. Wang, H. Liu, J. Zhang and T. Hou, Chem. Rev., 2019, 119, 9478–9508 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: 1H and 13C NMR spectra of all compounds and computational data. See DOI: 10.1039/d1ra05616f |
| This journal is © The Royal Society of Chemistry 2021 |