Open Access Article
Open Access ArticlePolyethylene glycol-derived polyelectrolyte–protein nanoclusters for protein drug delivery†
Yuanxiang Yu *ab,
Yi Shaoa,
Mingzhen Zhoua and
Wenjing Li*c
*ab,
Yi Shaoa,
Mingzhen Zhoua and
Wenjing Li*c
aDepartment of Radiation Oncology, Cancer Hospital of Shantou University Medical College, Shantou, 515000, P. R. China. E-mail: sdyxy@163.com
bSchool of Pharmaceutical Sciences, Southern Medical University, Guangzhou, 510515, P. R. China
cDepartment of Radiology, First Affiliated Hospital of Shantou University Medical College, Shantou, 515000, P. R. China. E-mail: Emmaliwj0905@163.com
First published on 25th August 2021
Abstract
Polyelectrolyte–protein nanocomplexes prepared under mild and simple conditions which could have biological activity arising from protein have emerged as fascinating protein delivery systems. However, common polyelectrolytes have problems of biocompatibility and metabolism in vivo, which may limit their further applications. Herein, a novel polyethylene glycol polyelectrolyte was synthesized and used for carrying protein drugs. Different from previously reported polyelectrolyte–protein nanoclusters, the polyethylene glycol polyelectrolyte–protein nanoclusters avoid organic solvent and protein modification, and the structure and bioactivity of proteins are well preserved. Moreover, the polyethylene glycol polyelectrolyte–protein nanoclusters have good hemocompatibility and biocompatibility. These novel polyethylene glycol polyelectrolyte–protein nanoclusters would provide a potent tool for fabrication of versatile protein drug carriers.
Introduction
Protein drugs have achieved rapid development in various fields of medicine, such as tumor treatment, inflammatory disease treatment, vaccines, and disease diagnosis.1,2 Compared with small-molecule drugs, protein drugs have the characteristics of high activity, strong specificity, low toxicity, clear biological function and beneficial clinical application. However, as biological macromolecules, proteins have complex advanced structures, resulting in low bioavailability and poor stability of structure and biological function.3,4 At present, the clinical application of protein drugs is mainly through intravenous administration. But short half-life and easy degradation are the main problems of free protein drugs.5,6 Compared with free drugs, polymer nanocarriers have many advantages, such as slow drug release, prolonged blood circulation time, enhanced permeation and retention effect, and responsive release.7–9 Therefore, the use of polymer nanocarriers to carry protein drugs has been widely studied and developed in recent years.There are two main methods for combining protein drugs with polymer nanocarriers: physical encapsulation and chemical bonding. Physical encapsulation method: proteins are encapsulated in the polymer nanoparticles in the process of preparation of the particles by copolymer self-assembly and emulsion method.10–12 Chemical bonding method: protein drug molecules are covalently bonded to polymers through mild and efficient chemical reactions between protein molecules and polymers.13,14 However, both of these methods carry the risk of causing structural and functional abnormalities of proteins when in the presence of organic solvents, chemical modification or mechanical force.15,16 Proteins are assembled by polypeptide chains composed of different amino acids. So, due to the differences in the structure and functional groups of amino acid residues, the proteins are amphiphilic and zwitterionic.17 Therefore, many researchers have tried to use the interaction between polyelectrolytes and the surface charge of proteins to form electrostatic complexes to prepare protein drug carriers. The method of electrostatic complexation is carried out entirely in the aqueous phase and the reaction conditions are mild: no heat, no chemical modification, no mechanical force and so on.18–20 However, common polyelectrolytes, such as polyacid electrolytes (polyacrylic acid, polystyrene sulfonic acid, etc.); polyalkali electrolytes (polyethylene imine, polyethylene pyridine, etc.); and inorganic polyelectrolytes (polyphosphate, etc.), all have problems of biocompatibility and metabolism in vivo.21 Poly(ethylene glycol) (PEG) is currently the most frequently used polymer in the biomedical field and the only polymeric therapeutic that has market approval for different drugs, such as small-molecule drugs,22,23 protein drugs,24 metal drugs,25–27 and nanocomposite drugs (organic–inorganic hybrids).28 Now, PEG with functional side chains can be synthesized by copolymerization of ethylene oxide and epoxy monomer with functional groups. PEG with functionalized side chains can allow the introduction of ionizable groups on these side chains, with this PEG electrolyte maintaining the excellent performance of traditional PEG.29,30 Therefore, preparation of protein-carrying nanoparticles using electrostatic complexation of protein with PEG polyelectrolyte is a novel and promising method.
As an epoxide monomer with an allyl group as pro-reactive functional group, allyl glycidyl ether (AGE) can be copolymerized with ethylene oxide (EO) to form side-chain-functionalized PEG derivatives (PEG-PAGE).12 In this study, PEG-PAGE was reacted with cysteamine, thioglycolic acid or mercaptoethanol to introduce various contents of functional groups on the side chains to obtain PEG polyelectrolyte. The PEG polyelectrolyte has different functional groups with different pKa values, and can complex proteins with different properties (acidic proteins, basic proteins, especially neutral proteins) (Scheme 1). To optimize the polymer–protein electrostatic complexation nanoclusters as a protein drug delivery vehicle, the electrostatic complexation behavior, stability, bioactivity, pharmacokinetics and biocompatibility were systematically investigated. Different from previously reported polyelectrolyte–protein nanoclusters, the PEG polyelectrolyte–protein nanoclusters avoid organic solvents and protein modification, and the structure and bioactivity of the proteins are well preserved. Additionally, the PEG polyelectrolyte–protein nanoclusters have good hemocompatibility and biocompatibility. The PEG electrolyte–protein complexation nanoclusters can be supplemented as new protein drug carriers.
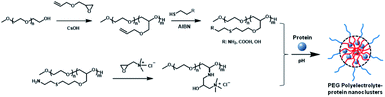 | ||
| Scheme 1 Synthesis of PEG-PAGE(R) polyelectrolyte and preparation of PEG polyelectrolyte–protein nanoclusters. | ||
Materials and methods
Materials
Caesium hydroxide monohydrate, β-mercaptoethanol, mercaptoacetic acid, and cysteamine were purchased from Aldrich; AGE (99%) from Acros; acetoxyacetyl chloride (97%) from Alfa Aesar; and tin(II) 2-ethylhexanoate (Sn(Oct)2, 90% in 2-ethylhexanoic acid) from Strem Chemicals. EO was from Sinopharm Chemical Reagent Beijing Co. Ltd (Beijing, China), and was dried over sodium hydroxide before use. Dichloromethane and triethylamine were dried by distillation from CaH2 under nitrogen. Toluene was purified by distillation from sodium with benzophenone. Dimethyl sulfoxide (DMSO) was stored over CaH2 and distilled under reduced pressure. Bovine hemoglobin (Hb), bovine serum albumin (BSA), bovine insulin and lysozyme were purchased from Shanghai Kayon Biological Technology Co. Ltd. Sodium ascorbate was from Aladdin. CO (99.95%) was from Dalian Date Gas Co. Ltd. Other solvents were of analytical grade and used as received.Characterization
1H NMR and 12C NMR spectra were recorded with a Bruker AV 400 MHz in CDCl3 (Aldrich) at 25 °C. UV spectra were recorded with a UV-visible spectrophotometer (Shimadzu UV-2450) at room temperature. Dynamic light scattering (DLS) experiments were conducted using a DAMN EOS instrument equipped with a He–Ne laser at a scattering angle of 90°. Transmission electron microscopy (TEM) measurements were performed using a JEOL JEM-1011 electron microscope operating at an acceleration voltage of 100 kV. Circular dichroism (CD) spectra of proteins were obtained with a JASCO J-600 spectropolarimeter at room temperature.Synthesis and characterization of copolymer poly(ethylene oxide-co-allyl glycidyl ether) with functional groups (PEG-PAGE(R))
Synthesis of PEG-PAGE was carried out according to the literature with some modifications:12 into a flame-dried flask was added 5 g of PEG5k in 40 mL of toluene and boiled to dehydrate for 2 h. Then, 0.168 g of caesium hydroxide monohydrate (1 mmol) was added under nitrogen atmosphere, and the mixture was stirred at 60 °C for 60 min, followed by evacuation at 90 °C for 3 h. 40 mL of THF and 10 mL of DMSO were then added to the evacuated flask, and the mixture was dispersed by ultrasound. AGE (2.05 g, 18 mmol) was added to the flask using a syringe, and the reaction system was kept stirring for 24 h at 40 °C. The reaction was stopped by adding a 3 mL of methanol. Finally, the solvent was drained, and the product was dissolved in water and placed in a dialysis bag (MWCO = 3500) for dialysis for 3 days. Product was freeze-dried and collected, with 97% yield. PEG-PAGE: 1H NMR (400 MHz, CDCl3): d 3.4–3.8 (m, –O–CH2–CH2–O– and –O–CH2–CH(CH2–O–CH2–CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH2)–O–), 4.0 (s, –O–CH2–CH
CH2)–O–), 4.0 (s, –O–CH2–CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH2), 4.56 (s, Ph–CH2–O–), 5.15–5.28 (dd, –O–CH2–CH
CH2), 4.56 (s, Ph–CH2–O–), 5.15–5.28 (dd, –O–CH2–CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH2), 5.84–5.97 (m, –O–CH2–CH
CH2), 5.84–5.97 (m, –O–CH2–CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH2), 7.32–7.35 (d, Ph–CH2–O–) ppm. 12C NMR (100 MHz, d6-DMSO): d 70.2, 71.3, 78.1 ppm.
CH2), 7.32–7.35 (d, Ph–CH2–O–) ppm. 12C NMR (100 MHz, d6-DMSO): d 70.2, 71.3, 78.1 ppm.
Radical-mediated thiol–ene reactions of PEG-PAGE with β-mercaptoethanol, mercaptoacetic acid, cysteamine: modifications of PEG-PAGE with pendent hydroxyl groups (PEG-PAGE(OH)), carboxyl groups (PEG-PAGE(COOH)) and amino groups (PEG-PAGE(NH2)) were achieved according to the literature. Briefly, the copolymer PEG-PAGE (1.2 g, 0.2 mmol, 18 AGE units per chain) and β-mercaptoethanol (0.94 g, 12 mmol) were dissolved in 30 mL of THF in a 250 mL round-bottom quartz flask, followed by N2 bubbling with a gentle flow for 30 min to eliminate dissolved oxygen. Then the mixture was stirred at room temperature under UV light (254 nm, 1.29 mW cm−2). After 2 h, the mixture was concentrated by evaporating part of the solvent. Residues were poured into large amounts of cold diethyl ether to afford precipitates. The precipitates were collected and redissolved in 10 mL of distilled water. Then the solution was placed in a dialysis bag (MWCO = 3500 Da) and dialyzed against distilled water for 3 days. The solution outside the bag was replaced with fresh water every 12 h. Finally, the mixture in the dialysis bag was freeze-dried to give a white product, yield 90%. PEG-PAGE(OH): 1H NMR (400 MHz, CDCl3): d 1.81 (quint, –O–CH2–CH2–CH2–S–), 2.59 (t, –S–CH2–CH2–OH), 2.69 (t, –CH2–S–CH2–CH2–OH), 3.35–3.9 (m, backbone and –O–CH2–CH2–CH2–S–CH2–CH2–OH) ppm. PEG-PAGE(COOH): 2.74 (t, –O–CH2–CH2–CH2–S–CH2–CH2–COOH) ppm. PEG-PAGE(NH2): 2.91 (t, –O–CH2–CH2–CH2–S–CH2–CH2–COOH) ppm.
Addition reaction of PEG-PAGE(NH2) with glycidyltrimethylammonium chloride (GTAC): briefly, the copolymer PEG-PAGE(NH2) (0.4 g, 0.025 mmol) was dissolved in 20 mL of PB, and GTAC (0.2 mL, 70% w/v, 1.05 mmol) was slowly dripped into the polymer solution. Then the mixture was stirred at room temperature. After 24 h, the reaction was stopped by adding a trace amount of hydrochloric acid. Then the solution was placed in a dialysis bag (MWCO = 3500 Da) and dialyzed against distilled water for 3 days. Finally, the mixture in the dialysis bag was freeze-dried to give a white product, yield 95%. PEG-PAGE(GTAC): 3.08 (t, CH2–N–(CH3)3) ppm.
Preparation of polyelectrolyte–protein nanoclusters
Protein solution (BSA and Hb, 3 g L−1) was mixed with polymer solution (PEG-PAGE(OH), PEG-PAGE(COOH) or PEG-PAGE(GTAC), 3 g L−1) in different proportions. Then the mixture was stirred at room temperature for 2 h. The hydrodynamic dimensions of the resulting PEG polyelectrolyte–protein nanoclusters were determined by DLS.Protein loading content (PLC) and protein loading efficiency (PLE) were calculated according to the following formulas:
| PLC (wt%) = (weight of loaded protein/weight of PEG polyelectrolyte–protein nanoclusters) × 100% |
| PLE (%) = (weight of loaded protein/weight of feeding protein) × 100% |
Cell lines and animals
Mouse fibroblast cells, L929, were obtained from the Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences, and cultured with DMEM (10% fetal bovine serum (FBS); 5% CO2 at 37 °C).Staphylococcus xylosus (ATCC 700404) was obtained from Guangdong Microbial Species Preservation Center, and grown overnight in tryptic soy broth (TSB, Oxoid) medium at 37 °C with constant shaking.
Male Wistar rats (100–150 g and 4–6 weeks old) and male KM mice (6 weeks, 20–25 g) were provided by the Animal Laboratory Center, Southern Medical University (Changchun, China). All mice received required care conditions throughout the experiments. All animal experiments were approved by the local institution review board and performed according to the Guidelines of the Committee on Animal Use and Care of Southern Medical University.
Hemocompatibility and biocompatibility of PEG polyelectrolyte–hemoglobin nanoclusters
Hemocompatibility of PEG polyelectrolyte–Hb nanoclusters was assayed against fresh rat whole blood. 3 mL of fresh whole blood was drawn from a Wistar rat and stored in heparinized Eppendorf tubes. 0.5 mL of blood was mixed with 0.5 mL of PEH (Hb concentration: 5 mg mL−1) dispersion or 0.5 mL of normal saline (0.9% NaCl). Afterwards, the whole blood and the mixture were incubated immediately at 37 °C in a water bath incubator with constant shaking. At 0 h, 3 h and 6 h, blood cell counting was performed using an ABX Micros 60 counter (ABX Diagnostics, Montpellier, France). After incubation for 6 h, the blood cell morphology of three samples was observed using an inverted microscope.
Results and discussion
Synthesis of PEG-PAGE(R) copolymer
The formation of electrostatic complexes through the interaction of surface charges of biomolecules and polyelectrolytes is a common method for preparing protein and polypeptide drug carriers. Due to the large particle size and low charge density of proteins, it is necessary to use polyelectrolyte chains with high charge density in order to improve the stability of the complexes.31 However, the biocompatibility of polyelectrolytes limits their application in vivo. PEG is currently the most frequently used polymer in the biomedical field and the only polymeric therapeutic that has market approval for different drugs. As an epoxide monomer with an allyl group as pro-reactive functional group, AGE can be copolymerized with EO to form side-chain-functionalized PEG derivatives (PEG-PAGE). As shown in Fig. 1A, the resonance signal peaks at δc (3.97 ppm), δd (5.23 ppm) and δe (5.88 ppm) are attributed to the double bond and its adjacent protons, respectively. The numerical average molecular weight of the PAGE chain segment can be calculated from the integral area ratio of methyl hydrogen δa (3.37 ppm) to δb (PEG5k-PAGE18). The 13C NMR spectra showed signals δa and δb (∼70.2 ppm), δc (∼71.3 ppm) and δd (∼78.1 ppm), attributed to the methylene carbons of the EO, and the secondary carbons and tertiary carbons of the AGE backbone, respectively, and they were all detected as a single signal due to the block structure (Fig. S1†).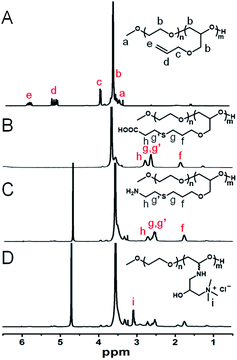 | ||
| Fig. 1 1H NMR spectra (400 MHz, CDCl3) of (A) PEG-PAGE, (B) PEG-PAGE(COOH), (C) PEG-PAGE(NH2), and (D) PEG-PAGE(GTAC). | ||
The double bond of PAGE can be used to introduce various functional groups through free radical reaction with sulfhydryl reagents under the conditions of thermal initiation, light and so on. As shown in Fig. 1B–D, after reacting with β-mercaptoethanol, mercaptoacetic acid, or cysteamine, the resonance signal peaks of double bond at δd (5.23 ppm) and δe (5.88 ppm) disappeared. Meanwhile, a corresponding new signal peak appears at 1.5 ppm to 3.0 ppm proving the successful introduction of functional groups in the side chain. Among them, the amino-modified polymers have poor water solubility. In order to improve the water solubility and charge density of the polymer, GTAC was used to modify the amino-functionalized polymers to convert them into quaternary ammonium salt ions, and the signal peak of aminomethyl appeared at 3.07 ppm (Fig. 1D). Meanwhile, UV-visible spectrometry showed PEG5k-PAGE18 has no characteristic absorption peak, while after reacting with cysteamine or GTAC, the characteristic absorption peak of amino group appeared at about 300 nm, further proving the successful synthesis of PEG-PAGE(NH2) and PEG-PAGE(GTAC) (Fig. S2†).
PEG is hydrophilic and crystalline, and the characteristic diffraction peaks (2θ) for PEG reported in the literature are 19.3° and 23.5°. As shown in Fig. S3,† the XRD curves of freeze-dried PEG-PAGE (2θ = 19.10°, 22.26°), PEG-PAGE(NH2) (2θ = 19.04°, 23.12°) and PEG-PAGE(GTAC) (2θ = 19.10°, 23.36°) powders show clear diffraction peaks, proving that the PEG-PAGE(R) were still crystalline.
Preparation of PEG polyelectrolyte–protein nanoclusters
First, the pKa values of the PEG-PAGE(R) copolymers were measured. The pKa of a polyelectrolyte represents its ability to dissociate hydrogen ions.32 A higher pKa value means a stronger alkalinity (positive charge) of the polyelectrolyte. In order to realize electrostatic complexation between copolymer and protein, the pH value of solution must be between the pKa value of copolymer and the isoelectric point of protein. The pH–V curves of PEG-PAGE(R) copolymers containing different dissociated groups were determined by potentiometric titration. The pH corresponding to the extreme value of the first derivative curve d(pH)/dV is the pKa value of the copolymers. As shown in Fig. 2, the pKa of PEG5k-PAGE18(COOH), PEG5k-PAGE18(OH) and PEG5k-PAGE18(NH2) were 4.95, 7.27 and 7.21, respectively. By chemically modifying the amino group, the pKa of PEG5k-PAGE18(GTAC) increases to 10.09. Here, copolymer polyelectrolytes with pKa in the range of 5–10 were obtained by introducing different functional groups into the copolymer side chains.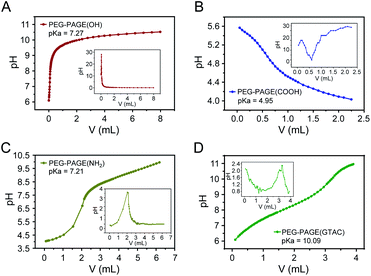 | ||
| Fig. 2 The potentiometric titration curves (pH–V curves) of PEG5k-PAGE18(R) copolymers containing different dissociated groups. | ||
Proteins with different isoelectric points (PI) were selected for the preparation of protein–polyelectrolyte electrostatic complexes. BSA and insulin are acidic proteins (PI < 7.4), and were mixed with the various polymers in different weight ratios in PBS (pH = 7.4). As shown in Fig. 3, mPEG5k is a non-ionic water-soluble polymer, and after mixing with BSA or insulin in different ratios, the particle size does not change and the solution shows a monomolecular distribution. At pH 7.4, PEG5k-PAGE18(COOH) and BSA or insulin have the same charge and will not form composite aggregates. Meanwhile, compared with PEG5k, the mixture had a larger particle size, which may be due to the irregular distribution of the surface charge of the protein resulting in the formation of a water-soluble complex with the polyelectrolyte. However, when BSA was mixed with PEG5k-PAGE18(GTAC), the particle size increases with increasing ratio of the polymer. The formation of nanoparticles is because of the proteins and polymers having different charges, and more free counterbalance ions are released after electrostatic complexation to form stable assemblies of nanoparticles. Moreover, with an increasing ratio of polymer, more proteins are involved in the complexation process, and the protein–polymer complex gradually aggregates, resulting in the increase of the particle size of nanoparticles. With the same principles, Hb is a neutral protein, and can form protein–polyelectrolyte electrostatic complexes with PEG5k-PAGE18(GTAC) in PBS (pH = 7.4); lysozyme is an alkaline protein, and can form protein–polyelectrolyte electrostatic complexes with PEG5k-PAGE18(COOH) in neutral solution (PBS, pH = 7.4).
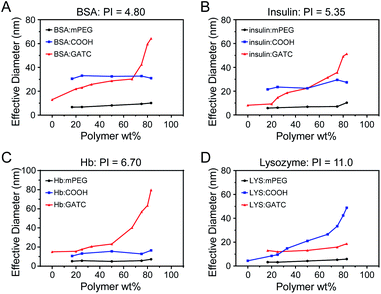 | ||
| Fig. 3 DLS results of protein: (A) bovine serum albumin, (B) insulin, (C) hemoglobin, and (D) lysozyme mixed with polymer in different proportions. | ||
As shown in Fig. 4, BSA, insulin, Hb and lysozyme all showed spherical morphology and quantitative analysis performed by using the TEM images revealed the average particle size of the BSA nanoclusters, insulin nanoclusters, Hb nanoclusters and lysozyme nanoclusters to be around 67.7 nm, 39.5 nm, 90.7 nm and 50.9 nm, respectively, as determined by histograms fitted by the Lorentzian function (Fig. S4†).33 These data confirmed the successful fabrication of polymer–protein nanoclusters through electrostatic complexing.
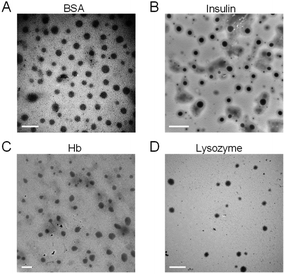 | ||
| Fig. 4 TEM images of (A) bovine serum albumin nanoclusters, (B) insulin nanoclusters, (C) hemoglobin nanoclusters and (D) lysozyme nanoclusters. Scale bar = 200 nm. | ||
Stability of protein in PEG polyelectrolyte–protein nanoclusters
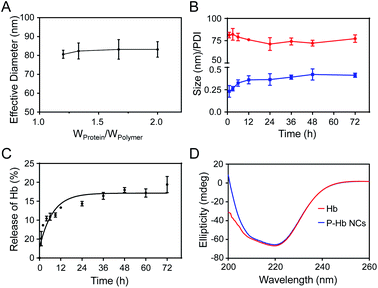
The PI of neutral functional protein is close to 7.0, so the electrostatic interaction between the protein and the polyelectrolyte is weak. So, Hb (PI = 6.7) was chosen as a model protein to investigate the stability of PEG polyelectrolyte–protein nanoclusters. In order to test the stability of complexation nanoclusters in an in vivo environment, we first measured the effect of adding protein to the final nanocluster solution (polymer = 83 wt%; also Wprotein/Wpolymer = 1.2) on the nanoparticle size. As shown in Fig. 5A, there were almost no significant changes in the size of nanoclusters after the gradual addition of protein. It is proved that the polyelectrolyte can fully complex with the protein at this ratio (polymer = 83 wt%; also Wprotein/Wpolymer = 1.2), so the complexation nanoclusters will not continue to be complexed with other protein in vivo. Meanwhile, the stability of complexation nanoclusters in 10% FBS was tested. The result showed there were no significant changes in size and PDI of PEG polyelectrolyte–Hb nanoclusters within 72 h (Fig. 5B), further proving the good stability of the complexation nanoclusters in vivo.Hb and insulin have a therapeutic effect in the blood, so we tested the protein release profile of PEG polyelectrolyte–Hb nanoclusters and PEG polyelectrolyte–insulin nanoclusters in PBS (pH 7.4, 37 °C). Less than 20% of Hb and 10% of insulin leaked out after 72 h (Fig. 5C and S5†), demonstrating the good stability of the nanoclusters in blood circulation. Meanwhile, the protein release profile of PEG polyelectrolyte–lysozyme nanoclusters was tested in PBS (pH 6.0, 37 °C), which simulates the acidic environment at the site of bacterial infection. In an acidic environment, the ability of the acid to dissociate hydrogen ions of polymer is reduced, resulting in a decreased electrostatic complexation capacity between the polymer and protein. So, almost 40% of lysozyme leaked out after 72 h (Fig. S5†).
The stability of the secondary structure of a protein affects its function. Here, CD spectra were obtained to evaluate the secondary structure of Hb. As shown in Fig. 5D, Hb in PEG polyelectrolyte–Hb nanoclusters had almost the same CD spectrum as native Hb, indicating that the electrostatic complexation process had no effect on the secondary structure of Hb.
Bioactivity of polymer–protein nanoclusters
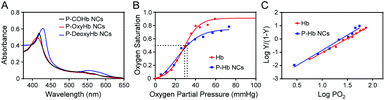
Hb is an iron-rich protein that reversibly binds to four oxygen molecules to deliver O2 to tissues.34 Hb in different gas-binding states shows different absorption spectra, and hence the gas-binding capacity of PEG polyelectrolyte–Hb nanoclusters could be monitored via UV-visible spectrophotometry. As shown in Fig. 6A, different gas-binding states of PEG polyelectrolyte–Hb nanoclusters could convert freely similar to native Hb with a change of the maximum absorption peaks. Furthermore, the calculated P50 and Hill coefficient values of PEG polyelectrolyte–Hb nanoclusters (25.9 mmHg and 2.287) were close to those of native Hb (26.6 mmHg and 2.129) (Fig. 6B and C) and of PEG-based copolymer Hb encapsulation vesicles in previous research (P50 = 29.5 mmHg, Hill coefficient = 2.507).12As another model protein, lysozyme has the function of killing bacteria.35 We next explored the antibacterial ability of PEG polyelectrolyte–lysozyme nanoclusters towards Staphylococcus xylosus by the bacteriological plate-counting method. As shown in Fig. S6,† the killing efficiency of PEG polyelectrolyte–lysozyme nanoclusters for Staphylococcus xylosus was close to that of free lysozyme and up to 61%. All the above results demonstrate that biological activities of Hb (transport gas) and lysozyme (antibacterial) were well preserved in the PEG polyelectrolyte–protein nanoclusters.
Biocompatibility of PEG polyelectrolyte–hemoglobin nanoclusters
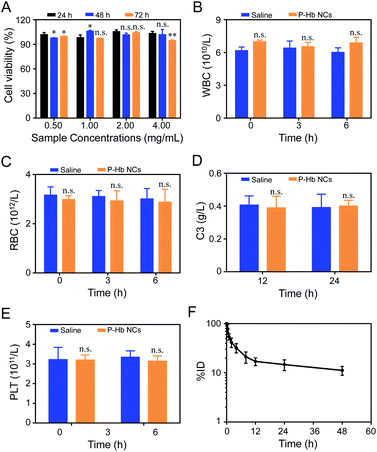
To assess the safety of PEG polyelectrolyte–protein nanoclusters in the biomedical field, the biocompatibility of PEG polyelectrolyte–Hb nanoclusters was firstly evaluated against L929 cells by MTT assay. As seen in Fig. 7A, no apparent toxicity was observed in cells treated by PEG polyelectrolyte–Hb nanoclusters. The cell viability in all groups exceeded 90%, confirming the good biocompatibility of PEG polyelectrolyte–Hb nanoclusters.When the PEG polyelectrolyte–Hb nanoclusters are applied in vivo, their blood compatibility is critical. The fresh blood of rats was mixed with PEG polyelectrolyte–Hb nanoclusters dispersed in normal saline and co-cultured in vitro, and the results of blood cell count at different times were obtained. As shown in Fig. 7B and C, normal saline has good blood compatibility and will not cause changes in the osmotic pressure of cells after mixing with whole blood—the number of blood cells only fluctuates slightly. Compared with the normal saline group, the addition of PEG polyelectrolyte–Hb nanoclusters did not cause changes in the number of blood cells (white blood cells and red blood cells), indicating that the nanoclusters would not cause damage to blood cells. At the same time, no adverse reactions such as erythrocyte aggregation and deformation were observed by optical microscopy (Fig. S7†).
The biocompatibility of PEG polyelectrolyte–Hb nanoclusters in vivo was carefully assessed. The level of C3 and platelets in blood were monitored after i.v. injection of PEG polyelectrolyte–Hb nanoclusters (20 mg mL−1, 3 mL) to evaluate the effects on complement activation and coagulation. As shown in Fig. 7D, compared to the saline group, the C3 concentration showed almost no fluctuation after i.v. injection of PEG polyelectrolyte–Hb nanoclusters, demonstrating no complement activation. Meanwhile, the platelet concentrations were almost the same as those of the saline group (Fig. 7E), indicating no obvious platelet aggregations. Furthermore, alterations of clinical chemical parameters ALT, AST, UA, and CREA were determined. These parameters are associated with the function of liver (AST, ALT) and kidney (UA, CREA) for human beings. The biochemical markers of mice injected with PEG polyelectrolyte–Hb nanoclusters were also within the normal ranges, indicating the low side effects (Fig. S8†). All these results indicate that the PEG polyelectrolyte–Hb nanoclusters have good compatibility with cells and blood, which ensures the safety of the nanoclusters in vivo.
The circulating half-life (T1/2) of rhodamine B-labeled PEG polyelectrolyte–Hb nanoclusters was about 5.72 ± 0.29 h, also very close to T1/2 of PEG-based copolymer Hb encapsulation vesicles (5.18 ± 0.54–6.80 ± 0.31 h) of previous research (Fig. 7F),12 confirming the slow clearance rate of PEG polyelectrolyte–Hb nanoclusters from blood.
Conclusions
Nanocarrier technology plays an important role in protein drug delivery. Green, safe and effective are the basic requirements of protein drug nanocarriers. In this work, side-chain-functionalized PEG-derived copolymers PEG-PAGE were synthesized by anion ring-opening polymerization. By introduction of various contents of functional groups on the side chains, we obtained PEG polyelectrolyte with different ionization properties, which can complex proteins with different properties (acidic proteins, basic proteins, especially neutral proteins). The preparation of PEG polyelectrolyte–protein nanoclusters was green and simple, avoiding organic solvent and protein modification, guaranteeing the stability of the structure and bioactivity of proteins. The PEG polyelectrolyte–protein nanoclusters were made of safe and non-toxic materials, having good hemocompatibility and biocompatibility. Meanwhile, the PEG polyelectrolyte–protein nanoclusters also have a good therapeutic effect (e.g. oxygen supply and antimicrobial). So, this method would provide a potent tool for achieving efficient protein drug delivery.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors acknowledge the financial support from China Postdoctoral Science Foundation (2020M672543).Notes and references
- B. J. Bruno, G. D. Miller and C. S. Lim, Ther. Delivery, 2013, 4, 1443–1467 CrossRef CAS PubMed.
- H. A. Daniel Lagassé, A. Alexaki, V. L. Simhadri, N. H. Katagiri, W. Jankowski, Z. E. Sauna and C. Kimchi-Sarfatyb, F1000Research, 2017, 6, 113 Search PubMed.
- D. K. Malik, S. Baboota, A. Ahuja, H. Sohail and A. Javed, Curr. Drug Delivery, 2007, 4, 141–151 CrossRef CAS.
- M. Matasci, D. L. Hacker, L. Baldi and F. M. Wurm, Drug Discovery Today: Technol., 2008, 5, 37–42 CrossRef.
- T. Vermonden, R. Censi and W. E. Hennink, Chem. Rev., 2012, 112, 2853–2888 CrossRef CAS PubMed.
- J. D. Pachioni-Vasconcelos, A. M. Lopes, A. C. Apolinario, J. K. Valenzuela-Oses, J. S. Ribeiro Costa, L. O. Nascimento, A. Pessoa, L. R. S. Barbosa and C. Rangel-Yagui, Biomater. Sci., 2016, 4, 205–218 RSC.
- S. Mura, J. Nicolas and P. Couvreur, Nat. Mater., 2013, 12, 991–1003 CrossRef CAS.
- J. Yin, Y. Chen, Z. H. Zhang and X. Han, Polymers, 2016, 8, 268 CrossRef PubMed.
- Z. H. Tang, C. L. He, H. Y. Tian, J. X. Ding, B. S. Hsiao, B. Chu and X. S. Chen, Prog. Polym. Sci., 2016, 60, 86–128 CrossRef CAS.
- D. A. Christian, S. S. Cai, D. M. Bowen, Y. Kim, D. Pajerowski and D. E. Discher, Eur. J. Pharm. Biopharm., 2009, 71, 463–474 CrossRef CAS PubMed.
- B. Li, S. S. He, Y. X. Qi, Z. G. Xie, X. S. Chen, X. B. Jing and Y. B. Huang, Int. J. Pharm., 2014, 468, 75–82 CrossRef CAS PubMed.
- Y. P. Wang, L. N. Wang, B. Li, Y. X. Cheng, D. F. Zhou, X. S. Chen, X. B. Jing and Y. B. Huang, ACS Macro Lett., 2017, 6, 1186–1190 CrossRef CAS.
- C. Boyer, X. Huang, M. R. Whittaker, V. Bulmus and T. P. Davis, Soft Matter, 2011, 7, 1599–1614 RSC.
- C. G. Palivan, O. Fischer-Onaca, M. Delcea, F. Itela and W. Meier, Chem. Soc. Rev., 2012, 41, 2800–2823 RSC.
- Q. Zhang, M. X. Li, C. Y. Zhu, G. Nurumbetov, Z. D. Li, P. Wilson, K. Kempe and D. M. Haddleton, J. Am. Chem. Soc., 2015, 137, 9344–9353 CrossRef CAS PubMed.
- Y. Z. Wu, T. Wang, Y. W. Ng. David and T. Weil, Macromol. Rapid Commun., 2012, 33, 1474–1481 CrossRef CAS.
- Y. P. Wang, L. S. Yan, S. S. He, D. F. Zhou, Y. X. Cheng, X. S. Chen, X. B. Jing and Y. B. Huang, Macromol. Biosci., 2018, 18, 1700282 CrossRef PubMed.
- C. L. Cooper, P. L. Dubin, A. B. Kayitmazer and S. Turksen, Curr. Opin. Colloid Interface Sci., 2005, 10, 52–78 CrossRef CAS.
- V. Bourganis, T. Karamanidou, O. Kammon and C. Kiparissides, Eur. J. Pharm. Biopharm., 2014, 11, 44–60 Search PubMed.
- M. Ishihara, S. Kishimoto, S. Nakamura, Y. Sato and H. Hattori, Polymers, 2019, 11, 672 CrossRef CAS.
- V. S. Meka, M. K. G. Sing, M. R. Pichika, S. R. Nali, V. R. M. Kolapalli and P. Kesharwani, Drug Discovery Today, 2017, 22, 1697–1706 CrossRef CAS PubMed.
- Y. P. Wang, Z. J. Luo, D. F. Zhou, X. F. Wang, J. J. Chen, S. P. Gong and Z. Q. Yu, Biomater. Sci., 2021, 9, 4110–4119 RSC.
- Q. H. Qian, L. J. Zhu, X. Y. Zhu, M. Sun and D. Y. Yan, Matter, 2019, 1, 1618–1630 CrossRef.
- S. N. S. Alconcel, A. S. Baas and H. D. Maynard, Polym. Chem., 2011, 2, 1442–1448 RSC.
- V. Nayak, K. R. B. Singh, A. K. Singh and R. P. Singh, New J. Chem., 2021, 45, 2849–2878 RSC.
- K. R. B. Singh, V. Nayak, T. Sarkar and R. P. Singh, RSC Adv., 2020, 10, 27194–27214 RSC.
- M. Fernandes, K. R. B. Singh, T. Sarkar, P. Singh and R. Pratap Singh, Adv. Mater. Lett., 2020, 11, 20081543 CrossRef CAS.
- R. P. Singh, P. Singh and K. R. B. Singh, Compos. Mater., 2021, 1–28 Search PubMed.
- B. Obermeier, F. Wurm, C. Mangold and H. Frey, Angew. Chem., Int. Ed., 2011, 50, 7988–7997 CrossRef CAS PubMed.
- C. Mangold, F. Wurm and H. Frey, Polym. Chem., 2012, 3, 1714–1721 RSC.
- A. L. Becker, K. Henzler, N. Welsch, M. Ballauff and O. Borisovb, Curr. Opin. Colloid Interface Sci., 2012, 17, 90–96 CrossRef CAS.
- J. Kötz, M. Hahn, B. Philipp, E. A. Bekturov and S. E. Kudaibergenov, Die Makromolekulare Chem., 1993, 194, 397–410 CrossRef.
- P. Singh, K. R. B. Singh, J. Singh, S. N. Das and R. P. Singh, RSC Adv., 2021, 11, 18050–18060 RSC.
- Y. Jia, L. Duan and J. B. Li, Adv. Mater., 2016, 28, 1312–1318 CrossRef CAS PubMed.
- Y. M. Ye, S. Klimchuk, M. W. Shang and J. J. Niu, RSC Adv., 2019, 9, 20169–20173 RSC.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1ra05055a |
| This journal is © The Royal Society of Chemistry 2021 |