Open Access Article
Open Access ArticleUptake of carbon nanodots into human AML cells in comparison to primary hematopoietic cells†
Cathrin Nollmanna,
Christian Wimmenauera,
Stefan Fasbender a,
Saskia Mayerb,
Ron-Patrick Caddedub,
Paul Jägerb,
Thomas Heinzel
a,
Saskia Mayerb,
Ron-Patrick Caddedub,
Paul Jägerb,
Thomas Heinzel *a and
Rainer Haas*b
*a and
Rainer Haas*b
aCondensed Matter Physics Laboratory, Heinrich-Heine-University, 40204 Düsseldorf, Germany. E-mail: Thomas.Heinzel@hhu.de
bDepartment of Haematology, Oncology and Clinical Immunology, Heinrich-Heine-University, 40204 Düsseldorf, Germany. E-mail: Haas.med@uni-duesseldorf.de
First published on 2nd August 2021
Abstract
Carbon nanodots (CNDs) comprise a class of next generation nanomaterials with a wide variety of potential applications. Here, we report on their uptake into primary hematopoietic cells from three normal donors and malignant cells from five patients with de novo acute myeloid leukemia (AML). A significant CND uptake was observed in all cell types of the normal and leukemic cells. Still, the uptake was significantly smaller for the CD34+ and CD33+ myeloid subsets of the malignant cell population as compared to the normal blood-derived CD34+ and CD33+ cells. For the T and B lymphoid cell populations as defined by CD3 and CD19 within the leukemic and normal samples a similar uptake was observed. The CNDs accumulate preferentially in small clusters in the periphery of the nucleus as already shown in previous studies for CD34+ progenitor/stem cells and human breast cancer cells. This particular subcellular localization could be useful for targeting the lysosomal compartment, which plays a pivotal role in the context of autophagy associated survival of AML cells. Our results demonstrate the usability of CNDs beyond their application for in vitro and in vivo fluorescence labeling or drug delivery into normal and malignant cells.
1. Introduction
Carbon nanodots, the family of carbon based, nanometer-sized particles which includes graphene quantum dots as well as small graphitic crystallites, have a large surface to volume ratio and excellent biocompatibility,1,2 and studying new ways for their production is still an active field of research.3–6 Since they also show fluorescence with advantageous properties in an aqueous environment, CNDs have been widely used in biomedical studies,3,7–9 although the origin of the fluorescence is still under debate.10–13 As far as living cells are concerned, CNDs enter the cytoplasm of many human cell lines as well as of primary human blood cells, without significant effects on the cell viability.14–19 As for other nanoparticles, CNDs have been used in studies for cancer diagnosis or drug delivery.20–26 In this context, the question arises whether the uptake of CNDs by malignant primary cells differs from that observed for healthy cells.We therefore investigated the uptake of CNDs into leukemic cells that were freshly obtained from patients with de novo acute myeloid leukemia. In the majority of patients with this kind of leukemia, the pathological blasts resemble their normal counterparts to some extent, in particular with respect to the expression of particular lineage- and differentiation associated surface molecules. Between those, CD34 and CD33 are prominent representatives reflecting an early stemness phenotype and myeloid differentiation, respectively. Our focus was on the aspect whether there is a differential uptake between primary human blood cells and leukemic cells, which could be of translational relevance.
2. Experimental section
2.1 Patients according to AML classification
The AML can be divided into different subtypes. The most commonly used classification schemes are the French–American–British (FAB)27 and the World Health Organization (WHO)28 system. The FAB classification is based on cytomorphological and cytochemical criteria, while the more recent WHO classification combines the FAB classification with immunological, cyto-as well as molecular genetic alterations. Table 1 lists the WHO and FAB classifications of the AML samples used in our study, as well as the percentage of blasts in the bone marrow and peripheral blood. The AML samples fall into different FAB categories permitting to some extent a subtype related assessment of the uptake.| Sample | WHO classification | FAB classification | Percentage of blasts |
|---|---|---|---|
| AML 1 | AML with MDS-associated modifications | AML M0 | PB: 39% BM: 32% |
| AML 2 | AML with MDS-associated modifications | AML M2 | PB: 65% BM: 69% |
| AML 3 | Acute leukemia, assignment unclear | AML M4/5 | PB: 10% BM: 40% |
| AML 4 | AML without further cytometric or molecular genetic specification | AML M2 | PB: 1% BM: 57% |
| AML 5 | AML without further cytometric or molecular genetic specification | AML M1 | PB: 77% BM: 85% |
2.2 Materials
Citric acid (ACS reagent, ≥99.5%), Diethylentriamine (DETA, 99%), Dulbecco's Phosphate Buffered Saline (DPBS), Lysis Buffer, Float-A-Lyzer dialysis devices (100–500 Da). Antibodies against CD45-PE-Cy7, CD34-PerCP-Cy5.5, CD33-PE, CD19-APC-R700 and CD3-APC-H7 were purchased from BD biosciences. Stem SPAN™ SFEM medium was bought at STEMCELL™ Technologies and microwave reaction vessels were obtained from CEM GmbH. NucBlue™ Live ReadyProbes™ Reagent (Invitrogen™), Poly-L-Lysine coated 8 well μ-slide was obtained from Ibidi.2.3 Preparation and characterization of CNDs
The CNDs were prepared and characterized exactly as described in detail elsewhere.18 In brief, fluorescent CNDs were synthesized according to the method of Qu et al.29 with slight modifications. 210 mg citric acid and 340 mg Diethylentriamine (DETA) were mixed and heated to 180 °C for 150 s in a closed microwave reaction chamber (CEM Discover). The resulting viscous liquid was dissolved in 10 ml DI water. Citric acid, DETA and very small particles were removed by dialysis for 72 h, using a 100–500 Da membrane, with two water exchanges every 24 h. Afterwards, the CND solution was freeze-dried and dissolved for further use. The CNDs were characterized by scanning probe microscopy, transmission electron microscopy as well as by Raman- and XPS spectroscopy. These measurements indicate that the CNDs are composed of the mass fractions 40% C, 33% O, 19% N and 8% H. About 30% of the carbon bonds are C–C bonds, and the Raman spectra reveal their mixture of sp2 with sp3 hybridization. The average size of the CNDs was (3.3 ± 0.6) nm. Some particles showed a hexagonal crystal symmetry in the TEM with a lattice constant of approximately 220 pm, which is 10% smaller than that one of free-standing graphene. Hence, we conclude that the CNDs are particles with mixed crystal structures of graphene-, graphite- and possible diamond-type sections. Their photoluminescence properties are most relevant for the present study. They were measured using a Horiba FluoroMax®-4 spectrofluorometer, while the absorbance spectra were taken with an Agilent Cary 4000 spectrophotometer. The absorbance shows a peak around 360 nm wavelength of approximately 80 nm linewidth (full width at half maximum-FWHM). To avoid UV exposure of the cells, we excited the CNDs with light of λ = 400 nm, yielding a fluorescence spectrum centered at λ = 460 nm (FWHM ≈ 100 nm), which fits well into the V450 channel of the flow cytometer (see below), which we used for their detection. The quantum yield of the CNDs was 23%.20 Since our particles do not show a fluorescence wavelength that differs from the values expected for size quantization and furthermore contain structural elements, like sp3 hybridizations, that should be absent in graphene, we refer to them as carbon nanodots. These nanoparticles are selected for the present study for several reasons. First of all, it has been shown earlier that they have a small toxicity and almost no influence on the gene expression of the exposed cells.19 Second, they have a small mass and are thus expected to exert only a marginal influence on the dynamics of attached macromolecules of interest. Finally, they are relatively simple to prepare, show a competitive quantum yield and have a long shelf life of several months.2.4 Collection of leukapheresis derived blood samples from normal donors
Primary hematopoietic cells were obtained from leukapheresis products (LP) of three healthy individuals who served as HLA-identical donors for an allogeneic blood stem cell transplantation using the granulocyte colony stimulating factor (G-CSF) at a dose of 480 μg per day over a period of five days. This increases the number of circulating human progenitor and stem cells (HSCs) – as characterised by the expression of CD34 on the cell surface – in the peripheral blood. This kind of LP samples are furthermore enriched with mononuclear blood cells (MNCs) including T and B cells, monocytes and CD34+ HSCs.30–32 The samples also contain a significant percentage of granulocytes which are activated due to the exposure to G-CSF for five days.332.5 Cell preparation
Blood samples from LPs of three healthy donors and AML samples from five patients de novo AML were used for the in vitro studies. In order to remove the erythrocytes, the samples from the donors and from the patients were lysed with 50 ml ammonium chloride, once and twice, respectively. For all samples, the remaining leukocytes were resuspended in 50 ml PBS and centrifuged for 5 min at 300 g. Cell pellets were resuspended in 2 ml PBS. 3 ml per well of StemSpan™ Serum-Free Expansion Medium (SFEM) were dispended in six well plates and appropriate amounts of cell suspension were added, resulting in a final concentration of 2 × 106 cells per ml.2.6 Cultivation of the cells for CND uptake studies
CNDs were dissolved in PBS at concentrations of 20 mg ml−1. The obtained solutions were sterile filtered. 75 μl of CND solution, corresponding to a concentration of 500 μg ml−1, was added to the cell culture. The same amount of PBS without CNDs was added to the wells serving as negative control. The cells were cultivated in a Heracell™ 150i incubator in a humidified atmosphere at 5% CO2 and 37 °C. After 24 h, all samples were washed twice with PBS (centrifugation for 5 minutes at 300 g) and stained with antibodies as described in detail below.2.7 Flow cytometry workflow
To study the differential uptake of CNDs in the various subpopulations of primary human blood cells and leukemic cells, monoclonal antibodies directed against lineage and differentiation specific antigens, i.e., CD45-PE-Cy7, CD34-PerCP-Cy5.5, CD33-PE, CD19-APC-R700 and CD3-APC-H7 were used. The cells were incubated for 15 minutes in the dark with 2 μl diluted CD45 (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
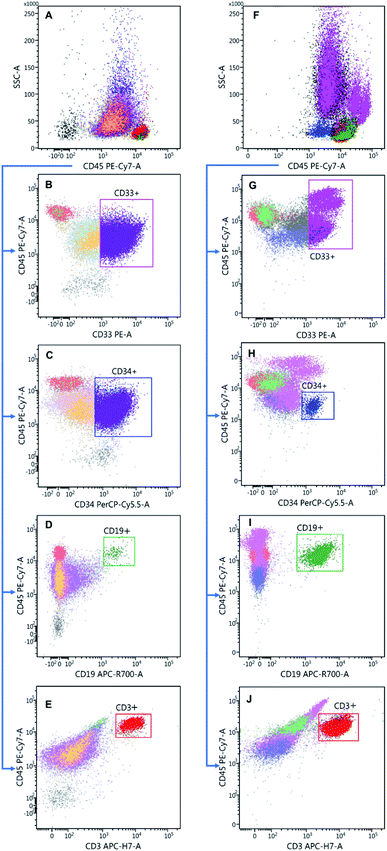
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 with PBS), 2.5 μl CD34, 2.5 μl CD33, 1 μl CD19 and 1 μl CD3 per sample. Afterwards, they were washed with 2 ml PBS (centrifugation for 5 minutes at 300 g) and fixed with 200 μl 0.5% formaldehyde. To exclude results that do not originate from viable cells (e.g., cell fragments or clumps), a gate was set in a forward vs. side scatter plot (FSC vs. SSC, see Fig. S1† in the ESI for examples). The FSC strength allows the discrimination of the cells by their size, while the SSC signal distinguishes the cell types by their granularity. Our gating strategy is exemplified for one AML sample and one donor sample each in Fig. 1. For an overall characterization of the sample composition, all viable cells from each sample are represented in a CD45 vs. SSC scatter plot (Fig. 1A and F). This also allows us to set blast gates for the AML samples later on.34–36 Afterwards, the CD33+ cells were gated out in a CD33 vs. CD45 plot, see Fig. 1B and G. Since the AML samples were extracted from the bone marrow (BM), the CD33+ gate of the AML samples contains both myeloid progenitor cells and malignant blasts. The donor samples, on the other hand, are collected from the peripheral blood (PB), and hence the CD33+ cells are mainly monocytes, mixed with some basophils (a subtype of granulocytes). In the next step, the stem and progenitor cells (CD45+/CD34+) were selected with a CD34 vs. CD45 plot as shown in Fig. 1C and H, respectively. The CD34+ cells of the AML samples include the malignant blasts. Even though some of the CD33+ cells of the donor samples have a high fluorescence intensity in the CD34 PerCp-Cy5.5 channel, they were not included in the CD34+ gate since CD34+/CD33+ cells are absent in the peripheral blood. To distinguish between the lymphocyte subpopulations, CD3+/CD19− cells (T cells) as well as CD19+/CD3− cells (B cells) were gated out in a CD45 vs. CD3 respectively CD45 vs. CD19 plot. In order to distinguish between the different types of blasts, further gates were set which are detailed in Section 2.7.
10 with PBS), 2.5 μl CD34, 2.5 μl CD33, 1 μl CD19 and 1 μl CD3 per sample. Afterwards, they were washed with 2 ml PBS (centrifugation for 5 minutes at 300 g) and fixed with 200 μl 0.5% formaldehyde. To exclude results that do not originate from viable cells (e.g., cell fragments or clumps), a gate was set in a forward vs. side scatter plot (FSC vs. SSC, see Fig. S1† in the ESI for examples). The FSC strength allows the discrimination of the cells by their size, while the SSC signal distinguishes the cell types by their granularity. Our gating strategy is exemplified for one AML sample and one donor sample each in Fig. 1. For an overall characterization of the sample composition, all viable cells from each sample are represented in a CD45 vs. SSC scatter plot (Fig. 1A and F). This also allows us to set blast gates for the AML samples later on.34–36 Afterwards, the CD33+ cells were gated out in a CD33 vs. CD45 plot, see Fig. 1B and G. Since the AML samples were extracted from the bone marrow (BM), the CD33+ gate of the AML samples contains both myeloid progenitor cells and malignant blasts. The donor samples, on the other hand, are collected from the peripheral blood (PB), and hence the CD33+ cells are mainly monocytes, mixed with some basophils (a subtype of granulocytes). In the next step, the stem and progenitor cells (CD45+/CD34+) were selected with a CD34 vs. CD45 plot as shown in Fig. 1C and H, respectively. The CD34+ cells of the AML samples include the malignant blasts. Even though some of the CD33+ cells of the donor samples have a high fluorescence intensity in the CD34 PerCp-Cy5.5 channel, they were not included in the CD34+ gate since CD34+/CD33+ cells are absent in the peripheral blood. To distinguish between the lymphocyte subpopulations, CD3+/CD19− cells (T cells) as well as CD19+/CD3− cells (B cells) were gated out in a CD45 vs. CD3 respectively CD45 vs. CD19 plot. In order to distinguish between the different types of blasts, further gates were set which are detailed in Section 2.7.
FACS analysis was performed using a BD FACSLyric™ flow cytometer. It is equipped with a 488 nm and a 640 nm laser to measure the fluorescence intensities in the PE, PerCP-Cy5.5, PE-Cy7, APC-R700 and APC-H7 channels and a laser with an excitation wavelength of 405 nm, allowing the measurement of the CND induced fluorescence in the V450 channel. For each sample, at least 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 events were recorded. The analysis was carried out using the BD FACSuite™ software.
000 events were recorded. The analysis was carried out using the BD FACSuite™ software.
The ratio of the mean V450 fluorescence intensity measured for the samples with CNDs to that one in the control samples was calculated, resulting in the uptake factor as the parameter, which quantifies the cellular uptake of CNDs for each specific subpopulation. We thereby postulate that the CND fluorescence intensity represents a suitable parameter reflecting the local particle number. This implies that the intensity is not concentration-or pH-dependent. Concentration-dependent studies have shown that for the CND concentrations used here, the fluorescence intensity is linear as a function of the concentrations, while a significant pH dependence is observed only for unphysiologically low of high pH values (not shown).
2.8 Confocal fluorescence microscopy
The AML cell line HL-60 (passage number 18) was selected for the microscopy experiments. The cells were incubated in appropriate nutrition medium with a concentration of 500 μg ml−1 CNDs at 37 °C and 5% CO2 for 48 h in a Poly-L-Lysine coated 8 well μ-slide, the nuclei were stained using NucBlue™ Live ReadyProbes™ Reagent (Invitrogen™) and the nutrition medium was exchanged for fresh medium without CNDs. The cells were imaged using a Zeiss LSM 710 confocal microscope evaluating the Hoechst 33342 channel (excitation 405 nm, emission 410–495 nm), the CND channel (excitation 488 nm, emission 495–530 nm) in framewise acquisition mode. A 63× oil objective with NA 1.40 was used.2.9 t-SNE representation of the ensembles
Visualization of the multi-labelled cell ensembles in two dimensions by t-distributed stochastic neighbour embedding (t-SNE) has been carried out.37,38 The FlowJo™ software has been used for this purpose. The perplexity was set to 30 and the number of iterations to 1000, respectively. The learning rate was automatically adjusted for every sample by FlowJo™ software. In our t-SNE plots, the cell subtypes appear in clusters, while a colour scale represents the CND fluorescence intensity. In order to attribute partially overlapping clusters to the corresponding cell types in the AML ensembles, overlays with the gated CD33+, CD34+, CD19+ and CD3+ populations are created, and the thereby identified populations are framed in the t-SNE plots accordingly.2.10 Ethical statement
All experiments were performed in compliance with the relevant laws and institutional guidelines and have been approved by the ethical committee of the Heinrich Heine University (Study-no.: 2018-50_1). All donors had given their informed consent according to the guidelines of the ethical committee specified above.3. Results and discussion
3.1 Subset analysis of AML samples
To investigate the CND uptake by the blasts depending on their maturity level, four categories were defined based on the expression level of CD33 and CD34. Undifferentiated blasts almost only express CD34 antigens on their surface, while CD33 gradually emerges at a later stage of maturation when CD34 is vanishing. To investigate the influence of the maturity level on the CND uptake, the blasts were gated within a CD45 vs. SSC plot for every AML sample (Fig. 2A and B). Afterwards, the blasts were classified according to the expression of CD33 and CD34 in four subsets depending on whether they were positive or negative for the respective antigen.The resulting distribution varied significantly between the different AML samples (Fig. 2C). While the CD33+/CD34−, CD34+/CD33− or CD33+/CD34+ blasts are nearly equally distributed in AML 2, AML 4 and AML 5, CD34+/CD33− blasts predominated in AML 1 and CD33+/CD34− blasts in AML 3, reflecting the degree of relative maturity of the blasts within their pathological boundaries. The results correspond to some degree with the classifications of the AMLs (Table 1). AML 1 was classified as M0, i.e., a predominantly undifferentiated acute myeloblastic leukemia (CD34+/CD33−), whereas AML 2 and 4 belong to the M2 class (AML with maturation). AML 5 belongs to M1, an acute myeloblastic leukemia with some maturation, as indicated by the increase of the CD33+/CD34+ and CD33+/CD34− blasts. Finally, in AML 3, the CD33+/CD34− blasts were dominant reflecting the M4/M5 classification (acute myelomonocytic/monocytic leukemia).
3.2 Cellular uptake of CNDs
We proceed by examining whether there is a differential uptake between primary human blood cells and the leukemic cells collected from the bone marrow, as well as between different subpopulations of the samples. In order to quantify and compare the uptake, we defined the uptake factor as the ratio of the mean fluorescence signal after CND exposure to that one of the negative control.First, healthy and leukemic cells show an uptake of CNDs, reflected by a significant increase in signal intensity in the cells cultivated in the presence of CNDs as compared to the controls. The mean fluorescence signal in the V450 channel is increased by at least a factor of four. Representative examples are given in Fig. 3 for AML 2 (C) and for donor 3 (D). In more detail, the uptake factor for the CD34+ cells (HSCs) from the donor samples is 1.7 – fold greater as compared to that one of the AML samples. The uptake factors of the CD33+ populations differ even more between AML and donor samples, as the mean uptake factor for the donor samples is increased by 3.2. These findings indicate that the leukemic cells CD33+ and CD34+ leukemic cells have an apparently reduced ability to take up small compounds such as CNDs from the extracellular space.
Next, we have studied whether one of the four blast subpopulations (as obtained from the gating protocol shown in Fig. 2) shows a selective uptake behaviour. The resulting uptake factors do not show any differences between the various blast categories (see Fig. S2† of the ESI). This finding indicates that the degree of differentiation of the blasts is not related to the uptake capability of the CNDs, suggesting that a subset specific targeting of blasts without further modifications of the CNDs is not feasible.
Having a closer look on the intensity histogram of the CD33+ population (Fig. 3(B) and (C)) it becomes apparent that the intensity of the donor CD33+ cells show two peaks, one around 2 × 103 counts and a second one around 104 counts. This split was not found in the negative control, which represents the autofluorescence. Hence, the splitting indicates that two different CD33+ cell types are present which differ with regard to their CND uptake behaviour, which can be related to CD33+ monocytes only present in the LP products of the normal donors. This cell type is a prototype for a phagocytic cell implying that the CNDs are engulfed by vesicles related to the endolysosomal pathway, as suggested in earlier work.19 On the other hand, the first peak is probably related to a small proportion of CD33+ progenitor cells contained within the population of mobilized CD34+ cells (see Fig. S3† in the ESI).
For the CD19+ and CD3+ populations, there was no significant difference between the uptake factors of AML and donor samples. Still, for both, AML as well as donor samples, the uptake factor for the CD19+ subpopulation is significantly greater in comparison to the CD3+ cells. For the AML and the normal donor samples, the mean uptake factors of the CD19+ cells are about 1.38 and 1.76 times greater, respectively. A smaller uptake avidity of CD3+ T and T helper cells compared to CD19+ B cells from healthy donors was already observed in our previous study.19 The difference between these two types of lymphoid cells may relate to the phagocytic ability of activated B cells.39
The t-SNE plots (Fig. 3(D) (for AML 2) and (E) (for donor 3)) permit a very informative illustration of our findings, as the fluorescence intensity of the CND signal in the V450 channel is color-coded. With regard to the AML samples, the relatively homogeneous intensity of colour across all subpopulations represents the similar uptake behaviour of the various cell types with a strong overlap of the CD33+ and the CD34+ cells. In contrast, the populations of the healthy samples are distinguishable in the t-SNE map, each of them characterized by a particular colour-coded uptake activity, which corresponds to the distinct peaks shown in Fig. 3C.
3.3 Subcellular distribution of CNDs in AML cells
In order to compare the intracellular distribution of the CNDs in AML cells, confocal microscopy images of HL-60 cells (corresponding to AML FAB M2 cells) were taken after 48 h of incubation with CNDs (Fig. 4). The CNDs accumulate preferentially in small clusters in the periphery of the nucleus. Based on the results of previous studies in HSCs and human breast cancer cells using a counterstaining method these clusters could be localized to lysosomes.19,40 It is therefore conceivable that the CNDs following ingestion into the leukemic blasts are stored in the lysosomes, suggesting that the endolysosomal pathway is also effective in AML cells. In the light of this finding the results of Folkerts et al. are interesting and of potential therapeutic relevance.41 They could show decreased survival upon HCQ 20 μM hydroxychloroquine (HCQ) treatment for leukemic cell lines as well as primary sorted AML CD34+ cells (n = 36) compared to normal bone marrow CD34+ cells (N = 6; NBM CD34+: 41.7% ± 7.1 vs. AML CD34+: 21.3% ± 3.2, p = <0.05). | ||
| Fig. 4 Microscopy image of HL-60 cells taken 48 h after incubation with 500 μg ml−1 CNDs taken with a Zeiss LSM 710 confocal microscope (63× oil, NA 1.40). The nucleus was stained with Hoechst 33342, which was exited with a 405 nm UV diode laser and emission light was detected between 410 nm and 495 nm (cyan). The CNDs were exited with a 488 nm line from a multiline argon laser and the fluorescence was detected between 495 nm and 530 nm (yellow). The images have been acquired framewise. Image (A) displays only the fluorescence channels, while an overlay of the transmitted light from the CND channel and the fluorescence channels is shown in (B). Images of the control samples, taken with the same imaging parameters, are shown in the ESI.† | ||
Microscopy images of the control samples are contained within in the ESI (see Fig. S4† and S5 in the ESI).
4. Conclusions
We compared the cellular uptake of small graphene quantum dots into normal blood cells with that into primary leukemic cells from patients with AML. Based on the intensity of the CNDs related autofluorescence recorded following a 24 h exposure time in an in vitro culture, a significantly smaller uptake was noted into leukemic cells compared to normal cells. This was true for both, the CD34+ as well as CD33+ subset. With regard to the uptake into lymphoid cells, a similar degree of uptake was observed for normal and leukemic cells, while a significant difference was only found between CD19+ B cells and CD3+ T cells irrespective of the sample source. This decreased differential uptake by the malignant cells studied here in comparison to their healthy counterparts forms a challenge for a selective addressing of those cells to which, e.g., a drug should be delivered. Suitable drug delivery systems based on our CNDs therefore may require some functionalization which increases the uptake by the target cells, like antigens or sugars, for example by preferential binding to the target cells. Alternatively, one might imagine selective inhibitor schemes where the CNDs protect the healthy cells from the impact of a drug. These issues are, however, beyond our scope here and will be the topic of future studies. It should be noted in this context that the reduced uptake of quantum dots in the malignant cell type studied here cannot be generalized to malignant cells of other organs, such as solid tumors of the breast or lung. The latter ones are epithelial in nature by their germline affiliation and thus may well differ with regard to their uptake properties from leukemia cells of mesenchymal origin.Following their uptake, the CNDs reside in close proximity to endosomal–lysosomal machinery, which is involved in the uptake of extracellular particles via endocytosis. This subcellular location could be useful for therapeutic targeting involving the lysosomal compartment, which plays a pivotal role in the context of autophagy.41 Watson and colleagues demonstrated the dual function of autophagy for the balance between cell death and cell survival.42 They found that the complete blockade of autophagy induced the death of leukemic cells, while a reduction of this pathway increased their proliferation, which was associated with a significantly reduced latency of the disease. The dual role of autophagy for cancer progression and resistance is complex and therefore challenging when a targeted therapy is envisaged. Therefore, one direction of further work may be geared towards methods mediating a more specific leukemic uptake to assess functional effects on autophagy related processes in a dose-dependent manner.
List of abbreviations
| AML | Acute myeloid leukemia |
| t-SNE | t-Distributed stochastic neighbour embedding |
| CNDs | Carbon nanodots |
| LP | Leukapheresis products |
Author contributions
CN prepared the carbon nanodots, collected the AML and apheresate cytometry data, participated in the data analysis, wrote the initial draft and prepared the figures. CW took the confocal microscopy pictures, participated in the preparation and characterization of the CNDs and the data analysis. He furthermore initiated the t-SNE representation. SF contributed to the CND preparation setup and the flow cytometry. SM contributed to the recording of the cytometric data. RPD guided and supervised the cytometry and the gating of the cell ensembles. PJ collected the samples and evaluated the medical aspects of the AML data. TH co-designed the experiments, participated in the interpretation of the data and co-wrote the manuscript. RH initiated the study, interpreted the data and co-wrote the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
Access to the confocal microscope at the Center for Advanced Imaging (CAi) of HHU Düsseldorf is gratefully acknowledged. C. N. and C. W. gratefully acknowledge the support by the Jürgen Manchot Stiftung. The authors thank Cornelia Monzel for providing access to her laboratories.Notes and references
- Y. Yan, J. Gong, J. Chen, Z. Zeng, W. Huang, K. Pu, J. Liu and P. Chen, Adv. Mater., 2019, 31, e1808283 CrossRef PubMed.
- H. Singh, S. Sreedharan, K. Tiwari, N. H. Green, C. Smythe, S. K. Pramanik, J. A. Thomas and A. Das, Chem. Commun., 2019, 55, 521 RSC.
- K. Yadav, M. Das, N. Hassan, A. Mishra, J. Lahiri, A. K. Dubey, S. K. Yadav and A. S. Parmar, RSC Adv., 2021, 11, 976 RSC.
- B. H. Lee, R. L. McKinney, M. T. Hasan and A. V. Naumov, Materials, 2021, 14, 616 CrossRef CAS PubMed.
- M. J. Deka, P. Dutta, S. Sarnia, O. K. Medhi, N. C. Talukdar and D. Chowdhury, Heliyon, 2019, 5, e01985 CrossRef PubMed.
- P. Roy, P.-C. Chen, A. P. Periasamy, Y.-N. Chen and H.-T. Chang, Mater. Today, 2015, 18, 447 CrossRef CAS.
- N. Panwar, A. M. Soehartono, K. K. Chan, S. Zeng, G. Xu, J. Qu, P. Coquet, K. Yong and X. Chen, Chem. Rev., 2019, 119, 9559 CrossRef CAS PubMed.
- J. Du, N. Xu, J. Fan, W. Sun and X. Peng, Small, 2019, 15, 1805087 CrossRef PubMed.
- W. Su, H. Wu, H. Xu, Y. Zhang, Y. Li, X. Li and L. Fan, Mater. Chem. Front., 2020, 4, 821 RSC.
- R. Ludmerczki, S. Mura, C. M. Carbonaro, I. M. Mandity, M. Carraro, N. Senes, S. Garroni, G. Granozzi, L. Calvillo, S. Marras, L. Malfatti and P. Innocenzi, Chem.–Eur. J., 2019, 25, 11963 CrossRef CAS PubMed.
- J. Schneider, C. J. Reckmeier, Y. Xiong, M. von Seckendorff, A. S. Susha, P. Kasák and A. L. Rogach, J. Phys. Chem. C, 2017, 121, 2014 CrossRef CAS.
- W. Kasprzyk, T. Świergosz, S. Bednarz, K. Walas, N. V. Bashmakova and D. Bogdał, Nanoscale, 2018, 10, 13889 RSC.
- W. Wang, B. Wang, H. Embrechts, C. Damm, A. Cadranel, V. Strauss, M. Distaso, V. Hinterberger, D. M. Guldi and W. Peukert, RSC Adv., 2017, 7, 24771 RSC.
- L. Cao, X. Wang, M. J. Meziani, F. Lu, H. Wang, P. G. Luo, Y. Lin, B. A. Harruff, L. M. Veca, D. Murray, S. Xie and Y. Sun, J. Am. Chem. Soc., 2007, 129, 11318 CrossRef CAS PubMed.
- C. Liu, P. Zhang, F. Tian, W. Li, F. Lib and W. Liu, J. Mater. Chem., 2011, 21, 13163 RSC.
- S. Zhu, J. Zhang, C. Qiao, S. Tang, Y. Li, W. Yuan, B. Li, L. Tian, F. Liu, R. Hu, H. Gao, H. Wei, H. Zhang, H. Sun and B. Yang, Chem. Commun., 2011, 47, 6858 RSC.
- W. Shang, X. Zhang, M. Zhang, Z. Fan, Y. Sun, M. Han and L. Fan, Nanoscale, 2014, 6, 5799 RSC.
- S. Fasbender, S. Allani, C. Wimmenauer, R. P. Cadeddu, K. Raba, J. C. Fischer, B. Bulat, M. Luysberg, C. A. M. Seidel, T. Heinzel and R. Haas, RSC Adv., 2017, 7, 12208 RSC.
- S. Fasbender, L. Zimmermann, R. P. Cadeddu, M. Luysberg, B. Moll, C. Janiak, T. Heinzel and R. Haas, Sci. Rep., 2019, 9, 12018 CrossRef PubMed.
- C. Zhao, X. Song, Y. Liu, Y. Fu, L. Ye, N. Wang, F. Wang, L. Li, M. Mohammadniaei, M. Zhang, Q. Zhang and J. Liu, J. Nanobiotechnol., 2020, 18, 142 CrossRef CAS PubMed.
- Q. Liu and A. Atrens, Corros. Rev., 2013, 31, 85 CAS.
- M. Zhang, L. Bai, W. Shang, W. Xie, H. Ma, Y. Fu, D. Fang, H. Sun, L. Fan, M. Han, C. Liu and S. Yang, J. Mater. Chem., 2012, 22, 7461 RSC.
- I. Al-Ogaidi, H. Gou, Z. P. Aguilar, S. Guo, A. K. Melconian, A. K. A. Al-Kazaz, F. Meng and N. Wu, Chem. Commun., 2014, 50, 1344 RSC.
- Z. L. Wu, M. X. Gao, T. T. Wang, X. Y. Wan, L. L. Zhenga and C. Z. Huang, Nanoscale, 2014, 6, 3868 RSC.
- X. Sui, C. Luo, C. Wang, F. Zhang, J. Zhang and S. Guo, Nanomedicine, 2016, 12, 1997 CrossRef CAS PubMed.
- X. Gong, Q. Zhang, Y. Gao, S. Shuang, M. M. F. Choi and C. Dong, ACS Appl. Mater. Interfaces, 2016, 8, 11288 CrossRef CAS PubMed.
- J. M. Bennett, D. Catovsky, M. T. Daniel, G. Flandrin, D. A. Galton, H. R. Gralnick and C. Sultan, Ann. Intern. Med., 1985, 103, 620 CrossRef CAS PubMed.
- S. H. Swerdlow, E. Campo, N. L. Harris, E. S. Jaffe, S. A. Pileri, H. Stein, J. Thiele and J. W. Vardiman, WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues, WHO Press, Geneva, Switzerland, 2008 Search PubMed.
- D. Qu, M. Zheng, J. Li, Z. Xie and Z. Sun, Light: Sci. Appl., 2015, 4, e364 CrossRef CAS.
- R. Möhle, M. Pförsich, S. Fruehauf, B. Witt, A. Krämer and R. Haas, Bone Marrow Transplant., 1994, 14, 827 Search PubMed.
- K. Theilgaard-Mönch, K. Raaschou-Jensen, H. Palm, K. Schødt, C. Heilmann, L. Vindeløv, N. Jacobsen and E. Dickmeiss, Bone Marrow Transplant., 2001, 28, 1073 CrossRef PubMed.
- M. D'Aveni, J. Rossignol, T. Coman, S. Sivakumaran, S. Henderson, T. Manzo, P. S. E. Sousa, J. Bruneau, G. Fouquet, F. Zavala, O. Alegria-Prvot, M. Garfa-Traor, F. Suarez, H. Trebeden-Ngre, M. Mohty, C. L. Bennett, R. Chakraverty, O. Hermine and M. T. Rubio, Sci. Transl. Med., 2015, 7, 281ra42 CrossRef PubMed.
- S. Murea, S. Fruehauf, W. J. Zeller and R. Haas, J. Hematother., 1996, 5, 351 CrossRef CAS PubMed.
- M. J. Borowitz, K. L. Guenther, K. E. Shults and G. T. Stelzer, Am. J. Clin. Pathol., 1993, 100, 534 CrossRef CAS PubMed.
- G. T. Stelzer, K. E. Shults and M. R. Loken, Ann. N. Y. Acad. Sci., 1993, 677, 265 CrossRef CAS PubMed.
- F. Lacombe, F. Durrieu, A. Briais, P. Dumain, F. Belloc, E. Bascans, J. Reiffers, M. R. Boisseau and P. Bernard, Leukemia, 1997, 11, 1878 CrossRef CAS PubMed.
- L. van der Maaten and G. Hilton, J. Mach. Learn. Res., 2008, 9, 2579 Search PubMed.
- E. D. Amir, K. L. Davis, M. D. Tadmor, E. F. Simonds, J. H. Levine, S. C. Bendall, D. K. Shenfeld, S. Krishnaswamy, G. P. Nolan and D. Pe'er, Nat. Biotechnol., 2013, 31, 545 CrossRef CAS.
- Q. Zhu, M. Zhang, M. Shi, Y. Liu, Q. Zhao, W. Wang, G. Zhang, L. Yang, J. Zhi, L. Zhang, G. Hu, P. Chen, Y. Yang, W. Dai, T. Liu, Y. He, G. Feng and G. Zhao, Immunobiology, 2016, 221, 558 CrossRef CAS.
- D. Kersting, S. Fasbender, R. Pilch, J. Kurth, A. Franken, M. Ludescher, J. Naskou, A. Hallenberger, C. von Gall, C. J. Mohr, R. Lukowski, K. Raba, S. Jaschinski, I. Esposito, J. C. Fischer, T. Fehm, D. Niederacher, H. Neubauer and T. Heinzel, Nanotechnology, 2019, 30, 39 CrossRef PubMed.
- H. Folkerts, B.-J. Wieringa, P. J. Coffer, J. J. Schuringa and E. Vellenga, Blood, 2015, 126, 3831 CrossRef.
- A. S. Watson, T. Riffelmacher, A. Stranks, O. Williams, J. de Boer, K. Cain, M. MacFarlane, J. McGouran, B. Kessler, S. Khandwala, O. Chowdhury, D. Puleston, K. Phadwal, M. Mortensen, D. Ferguson, E. Soilleux, P. Woll, S. E. W. Jacobsen and A. K. Simon, Cell Death Discovery, 2015, 1, 15008 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1ra05033h |
| This journal is © The Royal Society of Chemistry 2021 |