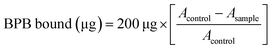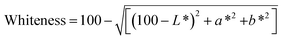Open Access Article
Open Access ArticleBiochemical property and gel-forming ability of mackerel (Auxis thazard) surimi prepared by ultrasonic assisted washing
Panumas Somjid a,
Worawan Panpipat
a,
Worawan Panpipat a,
Tanyamon Petcharatab and
Manat Chaijan
a,
Tanyamon Petcharatab and
Manat Chaijan *a
*a
aFood Technology and Innovation Research Center of Excellence, Department of Food Science and Innovation, School of Agricultural Technology and Food Industry, Walailak University, Thasala, Nakhon Si Thammarat 80161, Thailand. E-mail: cmanat@wu.ac.th; Fax: +66 75 672302; Tel: +66 75 672384
bProfessional Culinary Arts Programme, School of Management, Walailak University, Nakhon Si Thammarat 80161, Thailand
First published on 10th November 2021
Abstract
A low gel-forming ability is needed to be encountered using the dark-fleshed fish as a raw material. Optimal washing process can be a principled way of improving the gelling properties because washing is the most important step for surimi production. This study aimed to investigate the effect of ultrasonic-assisted washing (UAW) on the biochemical properties and gel-forming ability of frigate mackerel (Auxis thazard) surimi. Unwashed mince and conventional washing (CW) with 3-cycle of water (10 min per cycle) were compared to UAW for 5 and 10 min per cycle. UAW tended to improve the lipid removal, reduce the TCA-soluble peptide, and increase the surface hydrophobicity of surimi, but it had no influence on the Ca2+-ATPase activity, reactive sulfhydryl content, haem protein content, and lipid oxidation. UAW for 5 min per cycle rendered the surimi with the highest gel strength, whiteness, and water holding capacity as well as a regular aggregated network. With this method, the washing time can be reduced to 50% compared to the CW. Therefore, UAW for 5 min per cycle was an alternative approach for the production of mackerel surimi.
1. Introduction
Surimi, a concentrated form of fish myofibrillar proteins, is obtained by repeatedly washing the minced fish with water. It is commonly used for the production of various kinds of gelled products such as kamaboko, crab stick, fish tofu, and chikuwa.1 Surimi-based product is becoming increasingly popular as a healthy food choice because they contain high protein and low fat. Although the white fish are intensively used, the dark-fleshed fish species can be applied for transforming to surimi.2,3 The use of dark-fleshed fish can prevent limited white muscle fish resources due to overexploitation. Frigate mackerel (Auxis thazard), one of the dark-fleshed fish species wieldy distributed in Southern Thailand, can be used for surimi production.4 However, this kind of fish rendered the surimi with a poor gel-forming ability associated with large amounts of muscle lipids and haem proteins.4 Generally, washing is an essential step during surimi production. Most of the unwanted components (i.e., lipids, blood, enzymes, sarcoplasmic proteins, and undesirable debris) can be eliminated from the minced fish by means of washing. CW is not appropriate for all dark-fleshed fish species, including frigate mackerel.4 Still, it is a challenge to develop an appropriate washing process for dark-fleshed fish surimi production.Ultrasound technology has been progressively used as a highly effective alternate in the food processing industry.5 The ultrasound can be conducted in an efficient way with respect to the processing cost, reduced processing time, and lower energy consumption without a negative impact on the product quality.6 Ultrasound has been successfully employed in the meat processing industry to improve marination and tenderization as well as to inactivate various microorganisms.7 It was also used to improve the textural, binding, and microstructural properties of chicken myofibrillar protein composite gel.8 In surimi, the application of ultrasound has been established in the chopping step for gel preparation to enhance the gelling property of surimi.9,10 The strategy in gel improving is based on the cavitation phenomenon, which could be advantageous to structural modifications and promote the protein–protein interactions to form elastic gels.10 McClements11 suggested that cavitation can alter the physicochemical properties and functionalities of the treated protein molecules, e.g., promoting the unfolding/disaggregation of proteins and improving the texture/viscoelasticity of protein gels. However, the aid of ultrasound in a washing process has not been reported for mackerel surimi. The cavitation bubbles, typically form during sonication, may have helped to enhance the washing efficiency as well as to improve the gelation capability of the resulting surimi. Therefore, the objective of this study was to compare the effects of ultrasonic-assisted washing (UAW) and CW on the biochemical and gelling properties of frigate mackerel (A. thazard) surimi.
2. Materials and methods
2.1 Chemicals
All chemicals such as sodium dodecyl sulfate (SDS), β-mercaptoethanol (βME), adenosine triphosphate (ATP), trichloroacetic acid (TCA), thiobarbituric acid (TBA), 5,5-dithio-bis(2-nitrobenzoic acid) (DTNB), and 2,4-dinitrophenylhydrazine (DNPH) were purchased from Sigma Aldrich (St. Louis, MO, USA).2.2 Fish sample
Mackerel (A. thazard) with a size of 90–100 g were purchased from Thasala market, Nakhon Si Thammarat, Thailand. The fish was transported in ice (a fish/ice ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2, w/w) to the laboratory within 30 min. The fish were immediately washed, fileted, and the whole muscles were collected. Then, the mince was made using a meat grinder (4 mm hole diameter; Panasonic MK-G20MR, Japan).
2, w/w) to the laboratory within 30 min. The fish were immediately washed, fileted, and the whole muscles were collected. Then, the mince was made using a meat grinder (4 mm hole diameter; Panasonic MK-G20MR, Japan).
2.3 Surimi preparation by CW and UAW
To prepare surimi by CW, mince was washed thrice (10 min per cycle) with cold water (4 °C) using a washing medium/mince ratio of 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (v/w).2 After filtering through a layer of nylon screen, dewatering was carried out by a hydraulic press. To produce the surimi by UAW, the procedure was similar to the CW process, but the ultrasound (40 kHz) was applied for 5 or 10 min per cycle during washing (Ultrasons-H model 3000841 JP Selecta, Barcelona, Spain). The maximum sonication time was set at 10 min per cycle to avoid excessive denaturation/modification of proteins caused by the cavitation, heat, and free radicals generated by ultrasound. Coldwater was used as a washing medium to prevent the excessive heat generated during ultrasound treatment. Unwashed mince was used as a control. The final moisture contents of unwashed mince and surimi were adjusted to be equal to 80 g/100 g.
1 (v/w).2 After filtering through a layer of nylon screen, dewatering was carried out by a hydraulic press. To produce the surimi by UAW, the procedure was similar to the CW process, but the ultrasound (40 kHz) was applied for 5 or 10 min per cycle during washing (Ultrasons-H model 3000841 JP Selecta, Barcelona, Spain). The maximum sonication time was set at 10 min per cycle to avoid excessive denaturation/modification of proteins caused by the cavitation, heat, and free radicals generated by ultrasound. Coldwater was used as a washing medium to prevent the excessive heat generated during ultrasound treatment. Unwashed mince was used as a control. The final moisture contents of unwashed mince and surimi were adjusted to be equal to 80 g/100 g.
2.4 Determination of Ca2+-ATPase activity, reactive sulfhydryl (SH) content, and protein surface hydrophobicity
The Ca2+-ATPase activity, reactive SH content, and protein surface hydrophobicity of natural actomyosin (NAM) from surimi and unwashed mince was determined using the method of Benjakul et al.,12 Ellman,13 and Somjid et al.,3 respectively.For the Ca2+-ATPase activity, NAM was diluted to 2.5–8 mg mL−1 with 0.6 M KCl (pH 7.0). 1 mL NAM solution was combined with 0.6 mL 0.5 M Tris–maleate (pH 7.0) and 1 mL 0.1 M CaCl2. To make up a total volume of 9.5 mL, deionized water was added. Then, a 0.5 mL solution of 20 mM ATP was added. After an 8 min incubation at 25 °C, the reaction was stopped by adding 5 mL cold 15% (w/v) TCA. After that, centrifugation (3500 × g/5 min/25 °C) was performed using an RC-5B plus centrifuge (Sorvall, Norwalk, CT, USA), and the inorganic phosphate liberated in the supernatant was quantified. The activity of the Ca2+-ATPase was measured in moles of inorganic phosphate released/mg protein/min. A blank solution was prepared by adding chilled TCA prior to the addition of ATP.
For the reactive SH content, the NAM sample (0.5 mL, 4 mg mL−1) was combined with 4.5 mL of 0.2 M Tris–HCl buffer (pH 6.8) and 0.5 mL of 0.1% DTNB solution in a total volume of 7.5 mL. The absorbance was measured at 412 nm using a Shimadzu UV-2100 spectrophotometer (Shimadzu Scientific Instruments Inc., Columbia, MD, USA) after a 25 min incubation at 40 °C. By substituting 0.6 M KCl (pH 7.0) for the sample, a blank was created. The absorbance was used to calculate the SH content, which was reported as mol/108 g protein using the molar extinction of 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 600 M−1 cm−1.
600 M−1 cm−1.
The hydrophobicity of NAM dispersed in 20 mM phosphate buffer (pH 6) was measured using bromophenol blue sodium salt (BPB) for electrophoresis. To 1 mL of NAM suspension, 200 μL of 1 mg mL−1 BPB (in distilled water) was added and mixed well. A control group was created without NAM. For 10 min, samples and controls were agitated at room temperature (26–28 °C). The absorbance of the supernatant was measured at 595 nm (A) using a Shimadzu UV-2100 spectrophotometer against a blank of phosphate buffer after centrifugation (2000 × g/15 min/room temperature). The following formula was used to calculate the amount of BPB bound:
2.5 Determination of TCA-soluble peptide
The finely chopped sample (2 g) was mixed with 18 mL of 5 g/100 mL TCA and homogenized for 2 min at 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm using an IKA® homogenizer (Model T25 digital Ultra-Turrax®, Staufen, Germany). Then, the incubation was made at 4 °C for 1 h and the centrifugation was performed (8000 × g/5 min) using an RC-5B plus centrifuge. TCA-soluble peptides were measured in the supernatant using the Lowry method14 and reported as μmol/tyrosine/g sample.15
000 rpm using an IKA® homogenizer (Model T25 digital Ultra-Turrax®, Staufen, Germany). Then, the incubation was made at 4 °C for 1 h and the centrifugation was performed (8000 × g/5 min) using an RC-5B plus centrifuge. TCA-soluble peptides were measured in the supernatant using the Lowry method14 and reported as μmol/tyrosine/g sample.15
2.6 Determination of total haem protein
A sample was homogenized with 3 vol of 0.1 mol L−1 phosphate buffer (pH 7.0) containing 5 g/100 mL SDS at 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 20 s and subsequently incubated at 85 °C for 1 h. After cooling for 10 min, the centrifugation was performed (5000 × g/15 min/room temperature) using an RC-5B plus centrifuge and the absorbance was read at 535 nm using a Shimadzu UV-2100 spectrophotometer. Total haem protein content was estimated using a standard curve of bovine hemoglobin (Hb) and reported as μmol Hb per kg sample.16
000 rpm for 20 s and subsequently incubated at 85 °C for 1 h. After cooling for 10 min, the centrifugation was performed (5000 × g/15 min/room temperature) using an RC-5B plus centrifuge and the absorbance was read at 535 nm using a Shimadzu UV-2100 spectrophotometer. Total haem protein content was estimated using a standard curve of bovine hemoglobin (Hb) and reported as μmol Hb per kg sample.16
2.7 Determination of total lipid
The lipid was extracted by the method of Bligh and Dyer.17 The sample (25 g) was homogenized with 200 mL of chloroform![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) methanol
methanol![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) distilled water mixture (50
distilled water mixture (50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100
100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50) at a speed of 9500 rpm for 2 min at 4 °C using an IKA Labortechnik homogenizer. The homogenate was treated with 50 mL of chloroform and homogenized at 9500 rpm for 1 min. Thereafter, 25 mL of distilled water were added and homogenized again for 30 s. The homogenate was centrifuged at 3000 × g at 4 °C for 15 min using an RC-5B plus centrifuge, and transferred into a separating flask. The chloroform phase was drained into the 125 mL Erlenmeyer flask containing about 2–5 g of anhydrous sodium sulfate, shaken well, and decanted into a round-bottom flask through Whatman no. 4 filter paper. The solvent was evaporated at 25 °C using an EYELA rotary evaporator N-100 (Tokyo, Japan), and the residual solvent was removed by flushing with nitrogen. The total lipid content was reported as a g/100 g sample.
50) at a speed of 9500 rpm for 2 min at 4 °C using an IKA Labortechnik homogenizer. The homogenate was treated with 50 mL of chloroform and homogenized at 9500 rpm for 1 min. Thereafter, 25 mL of distilled water were added and homogenized again for 30 s. The homogenate was centrifuged at 3000 × g at 4 °C for 15 min using an RC-5B plus centrifuge, and transferred into a separating flask. The chloroform phase was drained into the 125 mL Erlenmeyer flask containing about 2–5 g of anhydrous sodium sulfate, shaken well, and decanted into a round-bottom flask through Whatman no. 4 filter paper. The solvent was evaporated at 25 °C using an EYELA rotary evaporator N-100 (Tokyo, Japan), and the residual solvent was removed by flushing with nitrogen. The total lipid content was reported as a g/100 g sample.
2.8 Determination of thiobarbituric acid-reactive substances (TBARS)
TBARS was analyzed by the method of Buege and Aust.18 A ground sample (0.5 g) was homogenized with 2.5 mL of a solution containing 0.375 g/100 mL (w/v) TBA, 15 g/100 mL (w/v) TCA and 0.25 M HCl. The mixture was heated in a boiling water bath (95–100 °C) for 10 min to develop a pink color, then cooled with running tap water and centrifuged at 3600 × g at 25 °C for 20 min. The absorbance of the supernatant was measured at 532 nm using a Shimadzu UV-2100 spectrophotometer. A standard curve was prepared using 1,1,3,3-tetramethoxypropane at concentrations ranging from 0 to 10 ppm. The TBARS content was reported as mg malondialdehyde equivalent per kg sample.2.9 Gel preparation and analysis
Unwashed mince or surimi with 80 g/100 g moisture content was chopped with dry NaCl (2.5 g/100 g) for 5 min to obtain the homogeneous sol. Then, the sol was filled into polyvinylidene casing (dia. 2.5 cm), sealed tightly, and subjected to a 2-step gelation (40 °C/30 min and 90 °C/20 min).2 After cooling in iced water and keeping them at 4 °C for 24 h, the gels were subjected to analyses including gel strength, whiteness, expressible drip, microstructure, and sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE).The breaking force (g) and deformation (mm) were determined by a spherical head plunger (dia. 5 mm) to press into the specimen (5 cm high/6 cm per min/60% compression) using a texture analyzer (Stable Micro Systems, Godalming, Surrey, UK).2 The gel strength (g cm) was obtained by multiplying the breaking force with the deformation.
For the expressible drip, a gel sample (0.5 cm thick) was weighed and placed between layers of Whatman filter paper no. 1 (two pieces at the top and three pieces at the bottom). After compressing with the standard weight (5 kg) for 2 min, the sample was reweighed. Expressible drip was reported as the percentage of the sample weight.2
The gel color (L*, a*, and b*) was determined using a Hunterlab Miniscan/EX instrument and the whiteness was calculated as follows:
The microstructure of gel was visualized using a scanning electron microscopy (SEM) (GeminiSEM, Carl Zeiss Microscopy, Germany) as described by Somjid et al.3 The gels, cut into 2–3 mm cubes, were fixed with 2.5 mL/100 mL glutaraldehyde in 0.2 M phosphate buffer (pH 7.2) for 2 h at room temperature, then rinsed with 0.1 M phosphate buffer 2 times. The gels were fixed with 1 g/100 mL osmium tetroxide, followed by further washing with 0.1 M phosphate buffer and distilled water. The samples were then dehydrated in ethanol with serial concentrations of 50, 70, 80, 90, and 100 mL/100 mL. Samples were critical-point dried using CO2 as the transition fluid. Dried samples were mounted on a bronze stub and sputter-coated with gold. The specimens were observed using an SEM at an acceleration voltage of 3 kV.
Protein patterns of surimi and surimi gels were visualized using SDS-PAGE,19 under non-reducing and reducing conditions. A protein standard (Bio-Rad Laboratories, Inc., Richmond, CA, USA) was used as a reference. The samples were mixed with a sample buffer (4 mL of 10 g/100 mL SDS, 2 mL of glycerol, 2.5 mL of 0.5 M Tris–HCl (pH 6.8), and 0.03 g BPB) at 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio (v/v). The sample buffer without and with 1 mL of βME were referred to as non-reducing and reducing conditions, respectively. The samples (15 μg protein) were loaded onto the polyacrylamide gel made of 10% running gel and 4% stacking gel and subjected to electrophoresis at a constant current of 15 mA per gel using a Mini Protein II unit (Bio-Rad Laboratories Inc., Richmond, CA, USA). After separation, the proteins were stained with 0.02% (w/v) Coomassie Brilliant Blue R-250 in 50 mL/100 mL methanol and 7.5 mL/100 mL acetic acid and destained with 50 mL/100 mL methanol and 7.5 mL/100 mL acetic acid, followed by 5 mL/100 mL methanol and 7.5 mL/100 mL acetic acid.
1 ratio (v/v). The sample buffer without and with 1 mL of βME were referred to as non-reducing and reducing conditions, respectively. The samples (15 μg protein) were loaded onto the polyacrylamide gel made of 10% running gel and 4% stacking gel and subjected to electrophoresis at a constant current of 15 mA per gel using a Mini Protein II unit (Bio-Rad Laboratories Inc., Richmond, CA, USA). After separation, the proteins were stained with 0.02% (w/v) Coomassie Brilliant Blue R-250 in 50 mL/100 mL methanol and 7.5 mL/100 mL acetic acid and destained with 50 mL/100 mL methanol and 7.5 mL/100 mL acetic acid, followed by 5 mL/100 mL methanol and 7.5 mL/100 mL acetic acid.
2.10 Statistical analysis
A completely randomized design was used in this study. The data were expressed as means ± standard deviations (SD) of three replications (n = 3) for all analyses. Data were subjected to analysis of variance (ANOVA). Comparison of means was performed by Duncan's multiple range test. Statistical analysis was conducted using SPSS 8.0 for Windows (SPSS Inc., Chicago, IL).3. Results and discussion
3.1 Ca2+-ATPase activity, reactive SH content, and protein surface hydrophobicity
Ca2+-ATPase activity, reactive SH content, and protein surface hydrophobicity of mackerel unwashed mince and surimi prepared using CW and UAW processes are shown in Table 1. From the results, the Ca2+-ATPase activity of NAM from unwashed mince was higher than all surimi (p < 0.05). A decrease in the activity of Ca2+-ATPase might be due to the denaturation of myosin during washing, regardless of the washing method. Zhang et al.20 reported that Ca2+-ATPase, mainly existing in the myosin head, is a sensitive indicator of myosin denaturation and is essential to maintain muscle structure, integrity, and function. Some research findings stated that Ca2+-ATPase activity of actomyosin exposed to high-intensity ultrasound decreased with increasing ultrasonic power and time.21–23 In this research, the Ca2+-ATPase activity of NAM extracted from UAW surimi remained constant with increasing ultrasonic time (p > 0.05). Results indicated that myosin from frigate mackerel was resistant to the ultrasound power applied in this study. However, a reduction in the Ca2+-ATPase activity suggested conformational changes or unfolding of the myosin molecule, which may have helped to facilitate the gelation. Since the gelation of myofibrillar proteins starts with partial denaturation and continues through cross-linking to form gel networks.24| Sample | Unwashed mince | Conventional surimi | Ultrasound (5 min per cycle) | Ultrasound (10 min per cycle) |
|---|---|---|---|---|
| a Values are given as mean ± standard deviation from triplicate determinations. Different letters in the same row indicate significant differences (p < 0.05). BPB = bromophenol blue, TCA = trichloroacetic acid. | ||||
| Ca2+-ATPase activity (μmol per mg protein per min) | 3.05 ± 0.22a | 1.79 ± 0.09b | 1.75 ± 0.20b | 1.72 ± 0.06b |
| Reactive sulfhydryl content (mol per 108 g protein) | 10.68 ± 0.94a | 6.80 ± 0.60b | 6.46 ± 0.37b | 6.50 ± 0.50b |
| Hydrophobicity (BPB bound (μg)) | 91.30 ± 0.24 ab | 84.64 ± 2.41c | 92.75 ± 0.84a | 88.23 ± 0.72bc |
| TCA-soluble peptide (μmol tyrosine per g sample) | 3.21 ± 0.08a | 1.72 ± 0.10b | 1.62 ± 0.04b | 1.36 ± 0.02c |
| Total lipid (g/100 g sample) | 6.50 ± 0.10a | 5.21 ± 0.20bc | 5.60 ± 0.00b | 4.92 ± 0.20c |
| Total haem protein (μmol kg−1) | 45.85 ± 1.46a | 25.40 ± 0.06b | 25.59 ± 0.10b | 25.51 ± 0.37b |
| Thiobarbituric acid reactive substances (mg malondialdehyde equivalent per kg) | 2.58 ± 0.59a | 1.66 ± 0.03b | 1.71 ± 0.39b | 1.85 ± 0.27b |
The content of reactive SH in NAM decreased significantly after washing (p < 0.05) (Table 1). Regardless of the washing method, the decrease of SH content might be due to the oxidation of SH to form disulfide interactions during washing. Although the cavitation can generate some temporal radicals (OH˙ and H˙),25 the reactive SH contents of NAM from UAW surimi and CW surimi were not different (p > 0.05). The result was coincidental with that of Chandrapala et al.26 who found that ultrasound treatment had no impact on the content of the free SH group of whey protein concentrate.
Surface hydrophobicity can be used to assess protein conformational changes.27 From the result, the highest protein surface hydrophobicity was found in unwashed mince and surimi prepared using UAW for 5 min per cycle, followed by UAW for 10 min per cycle, and CW, (p < 0.05) (Table 1). The washing process may cause the protein to unfold and subsequently aggregation via disulfide and hydrophobic interactions. All the NAM from surimi was denatured and exposed to the reactive groups during washing. Some of those groups can be interacting with adjacent molecules to form aggregates via disulfide bonds and hydrophobic interactions. As a result, the degree of unfolding and aggregation influenced the residual content of the SH group and the surface hydrophobicity of NAM. For UAW, the cavitation could reduce the void volume of proteins leading to the compression and conformational changes of proteins. Moreover, the air bubbles produced could enhance the conformational changes of proteins due to the surface-active ability of proteins at the air–water interface. Short time UAW (5 min per cycle) may cause partial denaturation of the proteins resulting from minor structural changes caused by cavitation. However, the surface hydrophobicity of surimi markedly decreased at a longer UAW treatment, indicating the aggregation of the proteins. In accordance with the finding of Chandrapala et al.,26 the surface hydrophobicity of whey protein concentrate increased for up to 5 min. However, the surface hydrophobicity tended to decrease when the sonication time was extended. Hu et al.28 reported that the surface hydrophobicity of the isolated soy protein increased when it was treated with ultrasound.
3.2 TCA-soluble peptide
The TCA-soluble peptide is an index of autolysis of unwashed mince/surimi by endogenous proteases. From the results (Table 1), the highest TCA-soluble peptide was found in the unwashed mince, indicating the occurrence of natural peptides and the proteolytic degradation products. Unwashed mince of mackerel consists of several sarcoplasmic proteins, including endogenous proteinases. Some proteinases can be eliminated during the surimi production process, resulting in a decrease in the amount and activity of these enzymes, which reduces protein degradation. Furthermore, the washing water may be used to eliminate any soluble peptides that have accumulated in the mince. Among the surimi, the lowest TCA-soluble peptide was found in surimi prepared using UAW for 10 min per cycle (p < 0.05). UAW especially for 10 min per cycle has the potential to reduce TCA-soluble peptide content from mackerel mince. It is possible that the ultrasound has inhibitory activity against endogenous proteases from mackerel mince due to elevated temperature, free radicals, and shear forces produced by cavitation.29 Zhang et al.30 reported that endogenous proteinases in tilapia surimi were inhibited by ultrasound of 100 W for 30 min during gel formation. Moreover, Gao et al.10 also stated that ultrasonic treatment can decrease TCA-soluble peptides of low-salt surimi gel from silver carp.3.3 Lipid content, total haem protein content, and lipid oxidation
Lipid is one of the impurities in surimi because it can impair the gel-forming ability and initiate gel rancidity.2 The lipid was significantly removed by both CW and UAW (p < 0.05) (Table 1). Although most of the lipid would be removed by washing with water, some lipid groups were also attached to the cell membrane, which was difficult to remove. The UAW for 10 min per cycle tended to produce the surimi with the lowest lipid content (Table 1). Prolonged sonication may induce phase separation of lipid to the top phase, which can be removed by dewatering. This is because ultrasonication favors the penetration of washing media into the internal structure and therefore can enhance mass transfer, resulting in the increase of lipid extractability.Residual haem proteins, particularly hemoglobin and myoglobin had a detrimental effect on the quality aspects of surimi (e.g., whiteness and lipid oxidation).16 From the results (Table 1), the total haem protein of unwashed mackerel mince was 45.85 μmol kg−1. The total haem protein of surimi was lower than unwashed mince (p < 0.05). Unlike the lipid removal, however, UAW for 5 and 10 min per cycle had no effect on haem protein removal. About 40% of total haem proteins were eliminated after washing, regardless of the washing method. This was feasibly due to the fact that haem proteins are sarcoplasmic proteins and the extractability would be dependent on their water solubility. The power/contact time of the ultrasound used in this experiment might be insufficient to release the bound haem proteins during washing. However, prolonged sonication may have a negative impact on the gelling ability of the resulting surimi.
TBARS values of unwashed mince and surimi prepared using CW and UAW are presented in Table 1. The highest TBARS content was found in the unwashed mince (p < 0.05), indicating the highest lipid oxidation. Normally, washing process could remove the lipid from fish muscle, resulting in reducing lipid oxidation. However, UAW had no effect on lipid oxidation, as indicated by comparable TBARS compared with CW surimi (p > 0.05). It can be implied that, based on the lipid content, the degree of lipid oxidation in surimi prepared by UAW for 10 min per cycle was higher than the others. Kang et al.31 reported that the ultrasound-induced lipid oxidation in beef during curing due to the generation of OH˙ by sonolysis. Also, the temperature may be elevated by the cavitation, which can readily enhance lipid oxidation.32 In this study, it was noticed that ultrasound was not significant to promote lipid oxidation because the washing was performed under cold temperature.
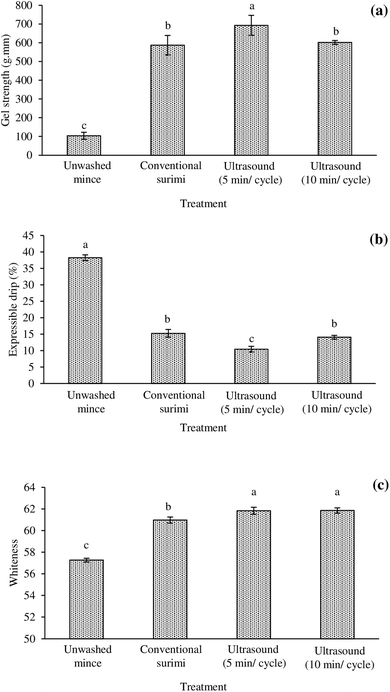
3.4 Gel strength
Gel strength is the prime quality factor of surimi reflecting the ability to form gels upon heating. The gel strengths of unwashed mince and surimi produced using CW and UAW are shown in Fig. 1a. Unwashed mince had the poorest gel-forming ability, as evidenced by the lowest gel strength. Although unwashed mince still had the highest Ca2+-ATPase activity, SH content, and hydrophobicity (Table 1), its gel strength was found to be the lowest. This was due to the presence of impurities such as lipid, sarcoplasmic proteins, haem proteins, proteases, lipid oxidation products, which can prevent the formation of three-dimensional networks of myosin. Also, unwashed mince had the highest TCA-soluble peptides, which were difficult to form cross-links. Morrissey et al.15 stated that protein degradation in fish has a negative connotation on the gel-forming ability of surimi. Proteolytic degradation of myosin resulted in the loss of structural domains, which are essential for molecular cross-linking. Myosin can be cleaved into small fragments. The myosin head sub-fragment still had the Ca2+-ATPase activity and some fragments should carry the reactive SH group and hydrophobic residues. However, the cross-linking ability of those small fragments was limited and thus, the gel was much poorer than that of intact myosin. The results showed that the gel strength of surimi prepared using CW and UAW was higher than unwashed mince (p < 0.05). In particular, surimi prepared using UAW for 5 min per cycle displayed the highest gel strength (p < 0.05). The UAW for 5 min per cycle may stabilize the endogenous transglutaminase (TGase) and suppress endogenous protease. Thus, it can facilitate the formation of intermolecular forces during setting the surimi sol at 40 °C. In addition, proper exposure of certain functional groups such as hydrophobic patches and SH domains can promote the protein–protein and protein–water interactions. Among surimi, the highest surface hydrophobicity found in surimi prepared using UAW for 5 min per cycle (Table 1), which may be transformed into the intra and inter-hydrophobic cross-links to form gel networks during gelation. Zhang et al.30 reported that ultrasound could activate the endogenous TGase and deactivate the endogenous protease during the gel formation as indicated by the improved gel strength. Fan et al.33 suggested that the use of ultrasound before setting and heating could increase the gel strength of the surimi from silver carp. However, an extended sonication time of up to 10 min per cycle for washing can cause an adverse effect on the gel strength of the resulting surimi. This may be due to the denaturation and aggregation of myofibrillar proteins as well as the loss of TGase activity. The formation of aggregates during UAW for 10 min per cycle may have resulted in a decrease in surface hydrophobicity (Table 1), and these aggregates were unable to form the ordered gel structure.3.5 Expressible drip
During thermal gelation, proteins are denatured and subsequently re-associated to form a gel network, where water is imbibed within.1 From the results, the lowest expressible drip was found in the surimi gel prepared using UAW for 5 min per cycle, suggesting the highest water holding capacity (WHC) of this gel (p < 0.05; Fig. 1b). Different expressible drip suggested the variation in WHC of gel. Generally, the lower expressible drip was in a relation to the increased gel strength (Fig. 1a). The highest expressible drip was observed in the gel of unwashed mince (p < 0.05; Fig. 1b). It was likely due to the presence of interferences (e.g., lipids and haem proteins) in the networks, which can prevent the protein–water interaction. Surimi prepared using UAW for 5 min per cycle had the highest gel strength and lowest expressible drip. The denser and homogenous gel cavities could be formed and water molecules can be firmly trapped in the gel network. Protein gels with homogeneous structures showed better WHC than those with non-homogeneous structures.34 With increasing the UAW up to 10 min per cycle, an increased expressible drip was noticeable. Long time ultrasound may cause the formation of aggregates, which can be localized in the gel networks and prevent the optimal water binding of the gel. Thus, UAW at the appropriate time can improve the WHC of surimi gel. Riener et al.35 stated that ultrasound (24 kHz/10 min) could enhance the WHC of yoghurts. Zhang et al.36 reported that moderate high-intensity ultrasound (≤600 W) treatments rendered the myofibrillar protein gel with a denser and uniform microstructure associated with an improved WHC, while stronger high-intensity ultrasound (>600 W) gave the gel with a larger and irregular microstructure, along with a decreased WHC.3.6 Whiteness
Gels from all surimi had higher whiteness than unwashed mince (p < 0.05; Fig. 1c). The whitest gel was found in surimi prepared by the UAW (p < 0.05). The lowest whiteness of gel from the unwashed mince may be due to the redox instability of haem proteins to form met-derivatives. Upon thermal gelation, not only the metmyoglobin/methemoglobin formation caused the darkening of gel but the Maillard reaction was also affected. In the presence of aldehydic lipid oxidation products, the Maillard reaction can occur.37 Although the total haem protein and TBARS contents were similar among surimi, the whiteness of surimi gel prepared using UAW was better than that in CW surimi (p < 0.05). It might be described by the denser and more uniform structure of the gel from surimi previously washed by the aid of ultrasound. Thus, the more the water molecules are retained, the better the glossiness and transparency of the gel. Wen et al.38 found that ultrasound could improve the whiteness of myofibrillar protein gel from grass carp.3.7 Gel microstructure
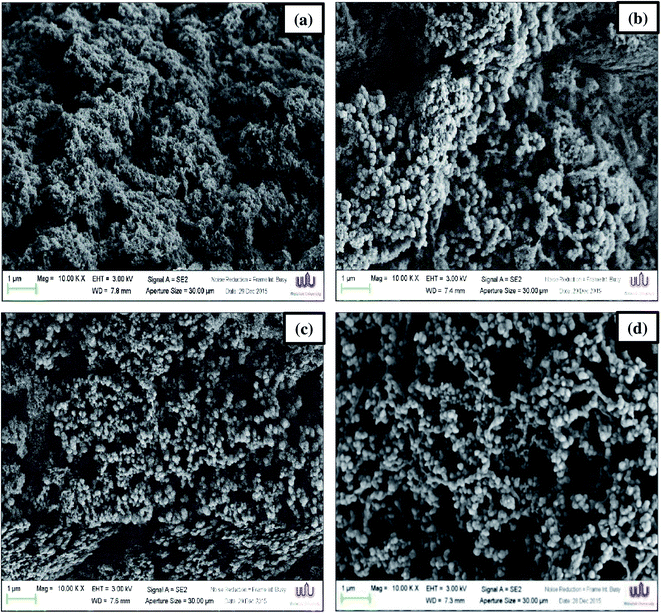
From these results, densely packed structures and irregular clots were observed in unwashed mince gel (Fig. 2a). CW surimi gel had a structure with aggregates of packed spherical proteins arranged in clusters with irregular pores (Fig. 2b). On the other hand, surimi gels prepared using UAW had a more uniform gel network with homogeneous pores (Fig. 2c and d). UAW may have aided in the dissociation of myosin assemblies, resulting in better dispersion and the creation of a compact structure.10 Especially, a well-organized structure and a finer cross-link can be observed in the gel prepared from surimi washed with UAW for 5 min per cycle (Fig. 2c). However, a longer UAW (10 min per cycle) yielded a gel with larger pores (Fig. 2d). The use of high-intensity ultrasound prior to chopping silver carp surimi can improve myosin solubility and facilitate protein dispersion, resulting in improved gel microstructures.10,393.8 Protein patterns
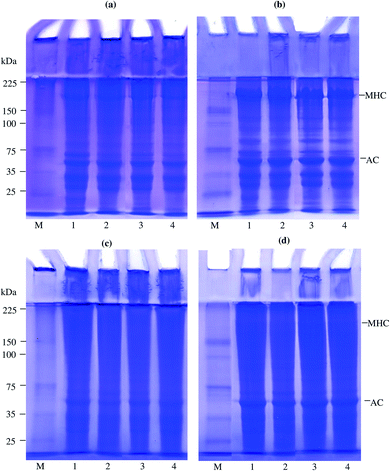
Protein patterns of samples before and after gelation under non-reducing and reducing conditions are depicted in Fig. 3a–d. The MHC and actin (AC) are the main proteins in surimi. UAW for 5 and 10 min per cycle markedly decreased the intensity of the MHC (lanes 3 and 4) compared to CW (lane 2) (Fig. 3a and b). Kang et al.31 stated that the reduction of MHC of beef muscle occurred during the ultrasonic process. Moreover, Li et al.40 reported that the use of ultrasound treatment (20 kHz/450 W/3–6 min) mainly decreased the MHC intensity of the chicken breast. The decrease in MHC of surimi prepared by UAW was due to the unfolding of proteins and the formation of protein aggregates stabilized by disulfide bonds as evidenced by the different patterns between non-reducing and reducing conditions (Fig. 3a and b). However, some of the protein aggregates were still retained in the stacking gel in the presence of βME (Fig. 3b), suggesting the presence of other bonds, e.g., hydrophobic interactions. Those aggregates were found in all surimi and unwashed mince, indicating the susceptibility to denaturation and aggregation of proteins during handling and surimi production. After gelation (Fig. 3c and d), the MHC bands of all gelled samples were thinner than non-gelled samples. The decrease in MHC after heating was due to polymerization and/or degradation depending on heating conditions. The decrease in MHC bands was considered to be due to the cross-linking of proteins during thermal gelation.4,41 The result was coincidental with an increase in the gel strength. Polymerized proteins hung on the stacking gel for all gels (Fig. 3c), but the intensities were decreased in the presence of βME (Fig. 3d). Results suggested that all gels were stabilized by both the disulfide bond and other bonds such as hydrophobic interaction. In silver carp surimi gels, Gao et al.10 found that high-intensity ultrasound enhanced protein aggregation was linked by disulfide bonds, hydrophobic interactions, and non-disulfide covalent interactions.4. Conclusion
Here, the effect of UAW on biochemical and gelling properties of frigate mackerel mince was carried out. UAW had no effect on the Ca2+-ATPase activity, reactive SH content, haem protein content, and lipid oxidation, but it seemed to improve lipid removal, reduce the TCA-soluble peptide, and increase the surface hydrophobicity. Overall, UAW for 5 min per cycle can improve the quality of mackerel surimi gel and the washing time can be shortened by 50%. Since the CW took 30 min (10 min per cycle) for washing, only 15 min can be done for UAW (5 min per cycle). This condition rendered surimi with superior gel strength, WHC, and whiteness. Moreover, uniform gel networks with irregular pores were formed. Therefore, UAW was an alternative process to improve the quality of frigate mackerel surimi. This could provide a foundation for the development of surimi that requires less washing time. In order to establish an eco-efficient surimi production, an ultrasound-assisted washing method with a reduced number of washing cycles was recommended to be investigated in the future.Author contributions
Panumas Somjid: conceptualization (equal); investigation (lead); writing – original draft (equal); writing – review and editing (equal). Worawan Panpipat: funding acquisition (equal); conceptualization (equal); writing – original draft (equal); writing – review and editing (equal). Tanyamon Petcharat: conceptualization (equal); writing – original draft (equal); writing – review and editing (equal). Manat Chaijan: funding acquisition (equal); conceptualization (lead); methodology (lead); writing – original draft (equal); writing – review and editing (equal).Conflicts of interest
The authors declare that they have no conflict of interest.Acknowledgements
This project was funded by National Research Council of Thailand (NRCT) (NRCT5-RSA63019-01). This research was financially supported by the new strategic research project (P2P), Walailak University, Thailand.References
- F. Liang, Y. Zhu, T. Ye, S. Jiang, L. Lin and J. Lu, LWT--Food Sci. Technol., 2020, 133, 110098 CrossRef CAS.
- M. Chaijan, S. Benjakul, W. Visessanguan and C. Faustman, Food Res. Int., 2004, 37, 1021–1030 CrossRef CAS.
- P. Somjid, W. Panpipat and M. Chaijan, J. Food Sci. Technol., 2017, 54, 3979–3988 CrossRef CAS PubMed.
- M. Chaijan, W. Panpipat and S. Benjakul, Food Chem., 2010, 121, 85–92 CrossRef CAS.
- M. Gallo, L. Ferrara and D. Naviglio, Foods, 2018, 7, 164 CrossRef CAS PubMed.
- C. Gambuteanu, B. Daniela and P. Alexe, J. Agroaliment. Processes Technol., 2013, 19, 88–93 Search PubMed.
- A. D. Alarcon-Rojo, L. M. Carrillo-Lopez, R. Reyes-Villagrana, M. Huerta-Jiménez and I. A. Garcia-Galicia, Ultrason. Sonochem., 2019, 55, 369–382 CrossRef CAS PubMed.
- Y. Y. Zhao, P. Wang, Y. F. Zou, K. Li, Z. L. Kang, X. L. Xu and G. H. Zhou, Food Res. Int., 2014, 58, 98–104 CrossRef CAS.
- Y. Zhang, Q. X. Zeng and Z. W. Zhu, J. Food Process Eng., 2009, 34, 1695–1713 CrossRef.
- X. Gao, Y. Xie, T. Yin, Y. Hu, J. You, S. Xiong and R. Liu, Ultrason. Sonochem., 2021, 70, 105326 CrossRef CAS PubMed.
- D. J. McClements, Trends Food Sci. Technol., 1995, 6, 293–299 CrossRef CAS.
- S. Benjakul, T. A. Seymour, M. T. Morrissey and H. An, J. Food Sci., 1997, 62, 729–733 CrossRef CAS.
- G. L. Ellman, Arch. Biochem. Biophys., 1959, 82, 70–77 CrossRef CAS PubMed.
- O. H. Lowry, N. J. Rosebrough, A. L. Farr and R. J. Randall, J. Biol. Chem., 1951, 193, 265–275 CrossRef CAS.
- M. T. Morrissey, J. W. Wu, D. Lin and H. An, J. Food Sci., 1993, 58, 1050–1054 CrossRef CAS.
- M. Chaijan and I. Undeland, Food Chem., 2015, 173, 1133–1141 CrossRef CAS PubMed.
- E. G. Bligh and W. J. Dyer, Can. J. Biochem. Physiol., 1959, 37, 911–917 CrossRef CAS PubMed.
- J. A. Buege and S. D. Aust, Methods Enzymol., 1978, 52, 302–310 CAS.
- U. K. Laemmli, Nature, 1970, 227, 680–685 CrossRef CAS PubMed.
- L. Zhang, Q. Li, H. Hong and Y. Luo, Food Chem., 2020, 316, 126343 CrossRef CAS PubMed.
- L. Tang and J. Yongsawatdigul, Ultrason. Sonochem., 2020, 63, 104922 CrossRef CAS PubMed.
- R. Liu, Q. Liu, S. Xiong, Y. Fu and L. Chen, Ultrason. Sonochem., 2017, 37, 150–157 CrossRef CAS PubMed.
- R. Saleem and R. Ahmad, Food Chem., 2016, 205, 43–51 CrossRef CAS PubMed.
- X. D. Sun and R. A. Holley, Compr. Rev. Food Sci. Food Saf., 2011, 10, 33–51 CrossRef CAS.
- İ. Gülseren, D. Güzey, B. D. Bruce and J. Weiss, Ultrason. Sonochem., 2007, 14, 173–183 CrossRef PubMed.
- J. Chandrapala, B. Zisu, M. Palmer, S. Kentish and M. Ashokkumar, Ultrason. Sonochem., 2011, 18, 951–957 CrossRef CAS PubMed.
- A. R. Jambrak, T. J. Mason, V. Lelas, L. Paniwnyk and Z. Herceg, J. Food Eng., 2014, 121, 15–23 CrossRef CAS.
- H. Hu, J. Wu, E. C. Li-Chan, L. E. Zhu, F. Zhang, X. Xu, G. Fan, L. Wang, X. Huang and S. Pan, Food Hydrocolloids, 2013, 30, 647–655 CrossRef CAS.
- A. Vercet, J. Burgos, S. Crelier and P. Lopez-Buesa, Innovative Food Sci. Emerging Technol., 2001, 2, 139–150 CrossRef CAS.
- Y. Zhang, Q. X. Zeng, Z. W. Zhu and R. Zhou, J. Food Process Eng., 2011, 34, 533–548 CrossRef.
- D. C. Kang, Y. H. Zou, Y. P. Cheng, L. J. Xing, G. H. Zhou and W. G. Zhang, Ultrason. Sonochem., 2016, 33, 47–53 CrossRef CAS PubMed.
- J. M. Lorenzo, A. Cittadini, P. E. Munekata and R. Domínguez, Meat Sci., 2015, 108, 50–54 CrossRef CAS PubMed.
- D. Fan, L. Huang, B. Li, J. Huang, J. Zhao, B. Yan and H. Zhang, LWT--Food Sci. Technol., 2017, 75, 497–504 CrossRef CAS.
- F. J. Kao, N. W. Su and M. H. Lee, J. Agric. Food Chem., 2003, 51, 6211–6216 CrossRef CAS PubMed.
- J. Riener, F. Noci, D. A. Cronin, D. J. Morgan and J. G. Lyng, Food Chem., 2009, 114, 905–911 CrossRef CAS.
- Z. Zhang, J. M. Regenstein, P. Zhou and Y. Yang, Ultrason. Sonochem., 2017, 34, 960–967 CrossRef CAS PubMed.
- M. Chaijan, S. Benjakul, W. Visessanguan, S. Lee and C. Faustman, J. Agric. Food Chem., 2007, 55, 4562–4568 CrossRef CAS PubMed.
- Q. H. Wen, Z. C. Tu, L. Zhang, H. Wang and H. X. Chang, J. Food Biochem., 2017, 41, 12288 CrossRef.
- X. Gao, J. Yongsawatdigul, R. Wu, J. You, S. Xiong, H. Du and R. Liu, LWT, 2021, 150, 111945 CrossRef CAS.
- K. Li, Z. L. Kang, Y. Y. Zhao, X. L. Xu and G. H. Zhou, Food Bioprocess Technol., 2014, 7, 3466–3477 CrossRef CAS.
- S. Benjakul, C. Chantarasuwan and W. Visessanguan, Food Chem., 2003, 82, 567–574 CrossRef CAS.
| This journal is © The Royal Society of Chemistry 2021 |