Open Access Article
Open Access ArticleFerritin-catalyzed synthesis of ferrihydrite nanoparticles with high mimetic peroxidase activity for biomolecule detection†
Hai Chen *a,
Liang Maa and
Yuhao Zhang*ab
*a,
Liang Maa and
Yuhao Zhang*ab
aCollege of Food Science, Southwest University, Chongqing, 400715, China. E-mail: chenhai2509@swu.edu.cn
bKey Laboratory of Luminescence Analysis and Molecular Sensing (Southwest University), Ministry of Education, Chongqing, 400715, China
First published on 30th July 2021
Abstract
Natural enzymes are generally sophisticated structural proteins that catalyze biological reactions with high specificity and efficiency and thus offer great potential in various disciplines, but intrinsic proteinic features such as easy denaturation and digestion restrict their practical application. So far, many functional nanomaterials with robust structures have been advanced as enzyme-mimetic catalysts to replace natural enzymes, however, their synthesis processes are generally complicated and require harsh experimental conditions such as high temperature and pressure. Herein, we report the facile synthesis of nanoparticles with high peroxidase-like activity by enzymatic catalysis. Specifically, by utilizing the intrinsic ferroxidase activity and mineralization capability of ferritin, ferrihydrite nanoparticles can be easily prepared within the apoferritin cavity at room temperature in aqueous solution (pH ∼ 7.0). Notably, reconstituted ferrihydrite nanoparticles exhibited comparative catalytic activity with a natural enzyme, horse radish peroxidase. Notably, when coupled with another oxidase, a dual-enzyme sensor system can be constructed for the detection of biomolecules such as glucose, and xanthine. Due to the biocompatibility and easy modification of the ferritin shell using well-established chemical and genetic techniques, the nanoparticles encapsulated by a protein shell possess great potential application in theranostics and immunoassays.
1. Introduction
Natural enzymes, most of which are proteins with a sophisticated three-dimensional structure, possess the ability to enhance the rate of interaction between biomolecules with astonishing catalytic efficiency and substrate specificity, thus not only efficiently regulating many biological processes in living organisms, but also exhibiting great potential application in biosensing, diagnosis, energy transformation, product formation and immunoassays. Generally, the sophisticated structure of natural enzymes is maintained through weak, non-covalent forces, thus it can be easily disrupted by environmental stimulation, resulting in the loss of their catalytic activity.1,2 ‘Artificial enzymes’ are the concept of imitating the general and essential principles of natural enzymes by using alternative materials, aiming to overcome the intrinsic drawbacks of the natural enzymes, and have received considerable attention from scientists in the field of nanotechnology. The conventional design of an artificial enzyme is to reproduce or integrate the structure of enzyme active sites, yielding ideal enzyme-implanted scaffold architectures.3,4 In recent decades, beyond understanding the complex interplay between the surface structures, morphology, and physicochemical properties of inorganic nanomaterials at the atomic level, further exploration of their functionality allows one to capture the intrinsic enzymatic activity of enzymes such as peroxidase, oxidase, and catalase in various nanomaterials. Importantly, these nanomaterials exhibit high tolerance to environmental conditions coupled with high catalytic efficiency, thus promoting the development of artificial enzymes through the synthesis of specific nanostructured materials.5–7 So far, a series of nanostructured materials including metal nanoparticles,8–13 nanoclusters,14,15 graphene oxide,16 carbon nanoparticles,17 metal–organic frameworks,18 have been synthesized and demonstrated to possess mimetic catalytic activity. As compared with the natural protein enzymes, these nanomaterials-based enzyme mimics exhibit great advantages in stability, cost, and even catalytic activity. In spite of great achievements, the fabrication of nanomaterials-based enzyme mimics is an active area of development and still remains challenges in addressing the problems such as biocompatibility, cytotoxicity, homogeneity, and complex synthetic methods.Ferritin is a conglomerate of 24 similar subunits that self-assemble into a hollow protein,19,20 which provides itself a great opportunity for acting as biotemplate in constrained nanomaterial synthesis, as well as nanocarrier in drug encapsulation and delivery.21–24 So far, the structural features of ferritin as biotemplate or nanocarrier have received considerable attention, but unfortunately the intrinsic enzymatic characteristic of ferritin for its application in the field of nanoscience and nanotechnology seems to be ignored. It has been well-established that ferritin is a natural ferroxidase enzyme that can catalyze the fast oxidation of Fe(II) by O2 or H2O2 into Fe(III) at the ferroxidase center, followed by the transfer of Fe(III) products into the central cavity, thereby promoting the formation of iron cores.25–27 However, to the best of our knowledge, the functionality and application of these formed iron nanoparticles as enzyme mimetics are yet to be fully investigated prior to this study.
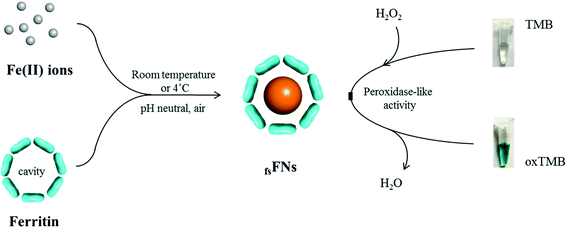
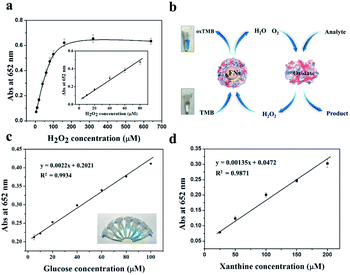
Previous study revealed that the formed ferric iron cores within the ferritin cavity can induce the decomposition of H2O2 molecules to produce hydroxyl radical, and that the amount of produced hydroxyl radical is closely associated with the iron loading amount within the protein cavity.28 Since the formation of hydroxyl radicals is a prerequisite for the peroxidase-like catalytic activity of reported nanoparticles,15 we envisioned that these iron nanoparticles might have peroxidase-like activity. To confirm this idea, we catalytically synthesized iron nanoparticles (which were identified as ferrihydrites) at room temperature by incubation of apo human H chain ferritin (HuHF) with Fe(II) solutions in aqueous solution (pH 7.0). Interestingly, these ferrihydrite nanoparticles possess high mimetic peroxidase activity, which can efficiently catalyze the conversion of the chromogen substrate into a colored product in the presence of H2O2 (Fig. 1). Notably, when coupled with another oxidase, a straight-forward dual-enzyme sensor system was fabricated for the detection of both glucose and xanthine.
2. Experimental
2.1 Preparation of rHuHF and ferrihydrite nanoparticles
cDNA encoding the full-length amino acid sequence of HuHF was cloned into the pET-3a vector (Novagen) and verified by DNA sequencing. The expression plasmids were transformed into the E. coli strain BL21 (DE3). Cells were grown at 37 °C on LB medium supplemented with 50 mg L−1 ampicillin, and protein expression was induced with 1 mM isopropyl-β-D-1-thiogalactopyranoside (IPTG) when the cell density reached an absorbance of OD600nm = 0.6. The cells were harvested by centrifugation, followed by disruption by sonication. The supernatant of the resulting crude extract was collected by centrifugation and fractionated by 60% saturation of ammonium sulfate. Finally, rHuHF was purified by ion exchange chromatography and Sephacryl S-300 gel filtration chromatography. Purified protein was characterized by SDS-PAGE and Native-PAGE.28 Ferrihydrite nanoparticles with different sizes were prepared as follows: different amounts of freshly prepared FeSO4 solution dissolved in pH 2.0 ddH2O were aerobically added to a series of ferritin solutions (100 mM Mops, 0.15 M NaCl, pH 7.0) in each increment of 100 Fe(II)/shell at intervals of 10 minutes to make the iron/protein ratio increase from 200/1 to 1000/1, respectively, and then resulting solutions were kept overnight at 4 °C.2.2 Transmission electron microscopy
Transmission electron microscopy (TEM) was conducted using a Hitachi S-5500 transmission electron microscope operating at 30 kV. Samples were diluted to get the required concentration prior to being placed on carbon-coated copper grids. After the excess solution was removed with filter paper, the samples were further stained using 2% uranyl acetate for 10 min, except for the observation of iron core. X-ray powder diffraction analysis was performed with the X-ray powder diffractometer on finely powdered samples using Cu Kα radiation.2.3 Peroxidase-like activity assays
The catalytic oxidation was carried out by using the substrates of 3,3′,5,5′-tetramethylbenzidine (TMB) in the presence or absence of H2O2 with the protein concentration of 0.25 μM in 0.2 M HOAc–NaOAc buffer either Mops buffer. The color formation was monitored at 652 nm using a Varian Cary 50 spectrophotometer (Varian, USA). The effect of iron content on the peroxidase-like activity was measured at 652 with a kinetic model using a Varian Cary 50 spectrophotometer (Varian, USA). The effect of pH and temperature on the catalytic activity was also studied under similar conditions.2.4 Reaction mechanism and kinetic analysis
The reaction kinetics for the catalytic oxidation of TMB was conducted by monitoring the absorbance at 652 nm on a Varian Cary 50 spectrophotometer (Varian, USA). Experiments were performed using 0.25 μM of holoferritin in 1 mL of 0.2 M NaAc buffer solution (pH 4.0) at a fixed concentration of H2O2 while varying the concentration of TMB. Similarly, the kinetic analyses with H2O2 as substrate were performed using 0.25 μM of holoferritin at a fixed concentration of TMB while varying concentrations of H2O2. Calculation of kinetic parameters was conducted based on the Michaelis equation: V = Vmax × 1/(Km + [S]), where V is the initial velocity, Vmax is the maximal reaction velocity, [S] is the concentration of substrate and Km is the Michaelis constant. Km and Vmax were calculated by the Lineweaver–Burk plot method. Absorption values at 652 nm were converted to the concentration of TMB derived oxidation products by Beer–Lambert Law with the wavelength-dependent absorptivity coefficient of 39![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 M−1 cm−1.
000 M−1 cm−1.
2.5 Glucose and xanthine detection
Glucose detection was carried out as follows: 10 μL of 20 mg mL−1 glucose oxidase and 190 μL of glucose with different concentrations in 10 mM PBS (pH 7.0) were mixed, followed by incubation at 37 °C for 40 min. Then, 800 μL of 0.2 M NaAc buffer solution (pH 4.0) containing 1.0 mM TMB and 0.25 μM of ferrihydrite nanoparticles was added into the above reaction solution. The mixed solution was incubated at 37 °C for 40 min. Finally, the solution was used to perform the absorption spectroscopy measurement at 652 nm. To test the specificity of the present method, maltose, lactose, fructose, and sucrose instead of glucose were used as controls under identical experimental conditions. Xanthine detection was performed by using nearly the same procedures as above except that xanthine oxidase was used instead of glucose oxidase.3. Results and discussion
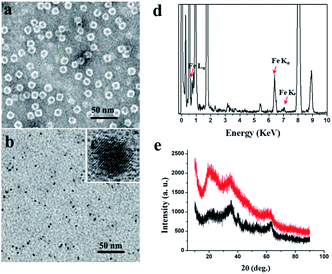
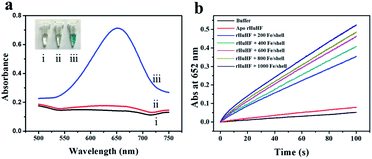
We chose recombinant human H type ferritin (rHuHF) as a platform for the synthesis of iron nanoparticles because (1) it can be easily purified and obtained through prokaryotic expression system; (2) the mechanism of iron oxidative deposition has been well illustrated; and (3) it has become a popular vehicle for drug or bioactive nutrients and also an excellent platform for the design of diagnostic kits.29–31 Since the hydrophilic channels of ferritin are the major pathway for iron atom access, thus the synthesis of iron nanoparticles can be achieved by directly aerobic incubation of the protein with Fe(II) solutions without disrupting the quaternary structure of ferritin shell.19–21 After purification and characterization of rHuHF (Fig. S1†), Fe(II) ions were aerobically added into protein solution (1.0 μM, 100 mM Mops, 0.15 M NaCl, pH 7.0) at room temperature in eight successive increments of 100 Fe(II)/protein at intervals of 10 min to prepare iron nanoparticles with a final ratio of iron to protein as 800/1. Transmission electron microscope (TEM) analyses showed that the in vitro reconstituted iron cores were encapsulated by intact protein shell with diameters of 2–4 nm, and energy-dispersive X-ray (EDX) analyses confirmed the presence of iron in these formed cores (Fig. 2), indicating that iron nanoparticles were successfully synthesized within the cavity of ferritin nanocages. Further high resolution TEM (HRTEM) analyses revealed that well-defined iron nano-particle crystallites can be generated within the protein shell, the lattice spacing of the nanoparticles is 2.6 ± 0.2 Å. Moreover, X-ray diffraction analyses found that the chemical composition of reconstituted iron nanoparticles within rHuHF is nearly identical to that of natural holo horse spleen ferritin which has been identified as ferrihydrites (Fig. 2e).27,32 SDS and native PAGE results further confirmed that the formation of the ferrihydrite nanoparticles within the ferritin cavity did not damage the structure of ferritin subunit and its shell (Fig. S1†), which can provide a great platform for bioconjugation and modification. Since the ferrihydrite nanoparticles were coated by ferritin shell, they were named as fsFNs.To determine the possible peroxidase-like catalytic activity of fsFNs, we investigate the oxidation of the peroxidase substrate 3,3′,5,5′-tetramethylbenzidine (TMB) by H2O2 in the presence of fsFNs. Enzymatic oxidation of TMB is marked by a color change from transparent and colorless to blue. As expected, two control experiments in the absence of either fsFNs or H2O2 exhibited negligible color changes. In contrast, fsFNs can catalyze the oxidation of TMB by H2O2 to produce a blue color (Fig. 3a). Such solution gave a maximum absorbance peak at 652 nm, which originates from the oxidation product of TMB.16 Thus it means that fsFNs possess peroxidase-like catalytic activity. Furthermore, kinetic measurements show that apo rHuHF proteins mixed with TMB and H2O2 exhibit negligible catalytic activity, indicating that the catalytic activity of fsFNs should stem from the ferrihydrite nanoparticles. In addition, once 200 Fe(II)/shell, which was thought to be the marginal content for initial iron core formation within the cavity of ferritin cage,25,26 was aerobically added to apo ferritin solution to prepare reconstituted ferrihydrite nanoparticle, the resulting sample exhibits a dramatic increase in the absorption at 652 nm, indicating that iron nanoparticles with a size of 200 iron/protein shell have the peroxidase-like activity. Moreover, with an increase in iron loading content within the ferritin cavity, the peroxidase-like catalytic activity also increased with all samples, and notably such increase exhibited in a dose-dependent manner (Fig. 3b and S2†). These findings demonstrated that the peroxidase-like activity of the reconstituted ferrihydrite nanoparticles is proportional to their size. However, we found that some ferritin aggregation occurred beyond the iron/protein ratio of 800/1, therefore we chose the ferrihydrite nanoparticles with 800 iron/protein shell to investigate their functionality and applications. Natural iron cores within ferritin were reported to contain ∼3000 atoms of iron per molecule or higher in organism because their iron accumulation process was very slow and well-controlled.27 To elucidate whether a much higher catalytic activity would be observed with natural iron cores, the peroxidase activity of natural holo horse spleen ferritin (HoSF) was investigated. As shown in Fig. S3,† buffer solution almost has no peroxidase-like activity and natural holo HoSF exhibits weak peroxidase-like activity. In contrast, fsFNs show the strongest activity among these three samples, the activity of which is about 5 fold higher than that of holo HoSF under the same experimental conditions. The reason might be that the reconstituted iron cores within rHuHF appear to be more polymorphic and loose as compared to those in natural holo HoSF, (Fig. S3†), which is beneficial for the contact of the nanoparticles with substrates.
It has been established that the relative catalytic activity of an enzyme is highly dependent on the pH and temperature. Therefore, we examined the peroxidase-like activity of fsFNs over the pH range from 2.0 to 9.0 at the temperature from 20 °C to 80 °C. As shown in Fig. S4,† the catalytic oxidation activity of TMB with H2O2 was much higher in acidic solution than that in neutral or basic solution, and fsFNs exhibit the high peroxidase-like activity at pH 3.0–4.0, These findings are in good agreement with the structure of fsFNs. Bear in mind that fsFNs consist of two parts in structure: protein shell and ferrihydrite nanoparticles. At pH 3.0–4.0, the structure of ferritin shell is very loose and began to undergo partial disassociation,33 which enables these nanoparticles to contact with more substrates, thereby enhancing their catalytic activity. In addition, the catalytic activity of fsFNs increased as increasing the temperature over the range of 20 °C to 80 °C. Such temperature-dependent manner can be ascribed as the high stability of ferritin cages which can resist the treatment of high temperature (∼80 °C) without significant disruption of its quaternary structure, as well as the flexibility of the channel pores which can expand upon thermal treatment.34 As a new mimic peroxidase, the stability of fsFNs is expected to be higher than that of the natural enzyme. To test this idea, we carried out the stability experiment by measuring the catalytic activity of fsFNs as a function of storage time at 4 °C. As shown in Fig. S4,† upon storage for 20 d, fsFNs still exhibited nearly the same peroxidase catalytic activity as the freshly prepared one, indicative of their high storage stability.
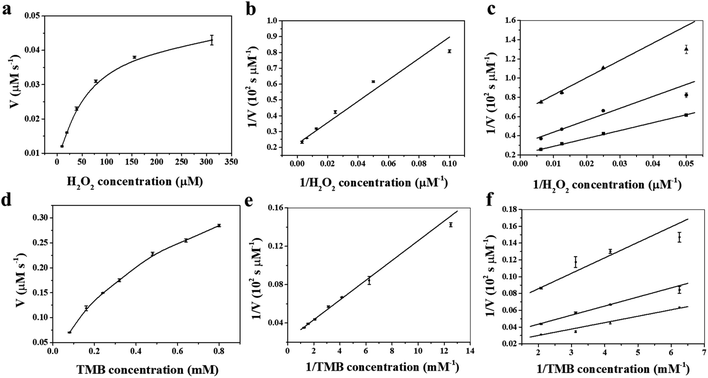
To shed light on the catalytic mechanism of fsFNs as peroxidase-like enzymes, the steady-state kinetic assays were measured with H2O2 as substrate and TMB as chromogen. As shown in Fig. 4a and b, typical Michaelis–Menten curves were obtained for fsFNs with TMB or H2O2 as substrate, and the initial reaction rate exhibited obvious dependence on the substrate concentration. Then, the Lineweaver–Burk plot method was applied to evaluate whether the catalytic reaction of fsFNs follows the Michaelis–Menten behavior. It can be seen from Fig. 4c and d that the reciprocal of initial rate was directly proportional to that of the substrate concentration of TMB or H2O2 just like natural HRP, demonstrating that such peroxidase-like catalytic reaction obeys the typical Michaelis–Menten behavior. The maximum initial velocity (Vmax) and Michaelis–Menten constant (Km) were calculated using Lineweaver–Burk plot and the data were displayed in Table S1.† The apparent Km value of fsFNs was about two times lower than that of HRP towards H2O2, indicating that the higher affinity of fsFNs for H2O2 than HRP.18 This finding is not surprising based on the fact that, except for oxygen, H2O2 is also a natural substrate for HuHF and acts as an oxidizing agent responsible for the fast oxidation of ferrous ions during iron mineralization.28 It is reasonable to believe that H2O2 just like O2 can diffuse fast into ferritin shells through unidentified pathways to fulfill its oxidizing function. The apparent Km value of fsFNs with TMB as the substrate was little higher than that of HRP, suggesting that fsFNs have slightly weaker affinity for TMB as compared to HRP. A possible reason for this phenomenon could come from the small pore size of the ferritin channels (0.3–0.4 nm),24–27 which delays the entrance of TMB into the inner cavity to interact with iron nanoparticles due to the steric hindrance. Moreover, compared to other metal nanoparticle synthesized within ferritin nanocage, such as gold clusters, platinum nanoparticles, M-HFn-CoxFe3−xO4 nanoparticles, and Fe3O4 MNPs, the fsFNs shows a comparable catalytic activity.35–37 Notably, the synthesis process of fsFNs is much more simple and convenient. For further investigation of the catalytic mechanism of fsFNs, measurements on the catalytic activity of fsFNs were conducted by varying concentrations of H2O2 at a fixed concentration of TMB and vice versa. Fig. 4e and f illustrate the double reciprocal plots of initial velocity against the concentration of one substrate with varying concentration of another substrate, these double reciprocal plots show the characteristic parallel lines of a ping–pong mechanism, as has been observed for HRP and some other nanomaterials.11,15 These results demonstrated that fsFNs bind and react with the first substrate during the catalysis procedure, and then releasing the first product before reacting with the second substrate.
On the basis of the intrinsic peroxidase catalytic activity of fsFNs, the absorbance of the colored oxidized products is highly dependent on the concentration of the H2O2 substrate, thus, the relationship between the absorbance and H2O2 concentration was evaluated. As shown in Fig. 5a, the absorbance at 652 nm to H2O2 concentration was linear in the range 5 × 10−6 M to 8 × 10−5 M, and the limit of detection and limit of quantitation are experimentally determined to be 4.85 × 10−6 M and 14.71 × 10−6 M, respectively, demonstrating that the change of the absorbance at 652 nm can be applied for the detection of H2O2. Since hydrogen peroxide is the main product of glucose oxidation by glucose oxidase, therefore, it means that fsFNs can be explored for replacement of the traditionally used HRP to determine the glucose when coupling with the glucose oxidase (Fig. 5b). The glucose determination was performed in two steps: (1) glucose oxidase catalyzes the oxidation of glucose to gluconic acid, accompanied with the conversion of the oxygen substrate into H2O2 in a neutral pH buffer solution. (2) The produced H2O2 will be reduced by fsFNs in the presence of TMB co-substrate at pH 4.0, which produces a color change at 652 nm. Fig. 5c shows a standard absorbance response curve to glucose concentration, and the linear range is from 5 × 10−6 M to 1 × 10−4 M with the limit of detection of 14.0 × 10−6 M. To determine the specificity of such biosensing method, control experiments were conducted by using lactose, fructose, sucrose, and maltose. As shown in Fig. S5,† the signal values still remained very low as compared with glucose, and thus the color difference can be distinguished even by naked eye. Thus, the demonstrated sensing system possesses high selectivity for glucose, which could be derived from the fact that glucose oxidase has high affinity for oxidized glucose. As a further example, we used xanthine oxidase instead of glucose oxidase to construct another dual-enzyme sensor system for the detection of xanthine. Fig. 5d shows that the absorbance at 652 nm was also in line with the concentration of xanthine, and the linear range is from 2.5 × 10−5 M to 2 × 10−4 M, giving the limit of detection and limit of quantitation of 3.47 × 10−5 M and 10.52 × 10−5 M, respectively, suggesting that fsFNs could be developed into biosensors for xanthine.
4. Conclusions
In conclusion, we demonstrated that fsFNs possess high peroxidase-like catalytic activity for the first time, which have the comparative catalytic efficiency and binding affinity for substrates TMB and H2O2 with HRP. Coupling with another enzyme such as glucose oxidase or xanthine oxidase, a dual-enzyme sensor system can be constructed for the detection of glucose and xanthine. Notably, the synthesis of fsFNs was carried out by taking advantage of the intrinsic ferroxidase activity and mineralization capability of ferritin itself. Different from reported methods for preparation of other nanoparticles, the fabrication of fsFNs catalyzed by ferritin is simple, environment friendly, condition mild, and highly effective, which offers new means to direct the synthesis of nanoparticles with high enzymatic activity. Moreover, compared with other inorganic materials, the reconstituted ferrihydrite nanoparticles were coated with the ferritin protein shell which provides a platform for the bioconjugation of other molecules, endowing fsFNs with great potential applications in theranostics and immunoassays.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the Fundamental Research Funds for the Central Universities (SWU120055, XDJK2019B028).Notes and references
- J. Barber, Chem. Soc. Rev., 2009, 38, 185–196 RSC.
- R. Dicosimo, J. Mcauliffe, A. J. Poulose and G. Bohlmann, Chem. Soc. Rev., 2013, 42, 6437–6474 RSC.
- D. W. Watkins, J. M. X. Jenkins, K. J. Grayson, N. Wood, J. W. Steventon, K. K. Le Vay, M. I. Goodwin, A. S. Mullen, H. J. Bailey, M. P. Crump, F. MacMillan, A. J. Mulholland, G. Cameron, R. B. Sessions, S. Mann and J. L. R. Anderson, Nat. Commun., 2017, 8, 358–367 CrossRef PubMed.
- F. Yu, V. M. Cangelosi, M. L. Zastrow, M. Tegoni, J. S. Plegaria, A. G. Tebo, C. S. Mocny, L. Ruckthong, H. Qayyum and V. L. Pecoraro, Chem. Rev., 2014, 114, 3495–3578 CrossRef CAS PubMed.
- H. Wei and E. Wang, Chem. Soc. Rev., 2013, 44, 6060–6093 RSC.
- L. Gao, K. Fan and X. Yan, Theranostics, 2017, 7, 3207–3227 CrossRef CAS PubMed.
- Z. Dong, Q. Luo and J. Liu, Chem. Soc. Rev., 2012, 41, 7890–7908 RSC.
- H. Wei and E. Wang, Anal. Chem., 2008, 80, 2250–2254 CrossRef CAS PubMed.
- X. Jiao, H. Song, H. Zhao, W. Bai, L. Zhang and Y. Lv, Anal. Methods, 2012, 4, 3261–3267 RSC.
- W. Chen, J. Chen, A. Liu, L. Wang, G. Li and X. Lin, ChemCatChem, 2011, 3, 1151–1154 CrossRef CAS.
- L. Gao, J. Zhuang, L. Nie, J. Zhang, Y. Zhang, N. Gu, T. Wang, J. Feng, D. Yang, S. Perrett and X. Yan, Nat. Nanotechnol., 2007, 2, 577–583 CrossRef CAS PubMed.
- L. Rastogi, D. Karunasagar, R. B. Sashidhar and A. Giri, Sens. Actuators, B, 2017, 240, 1182–1188 CrossRef CAS.
- Y. Jv, B. Li and R. Cao, Chem. Commun., 2010, 46, 8017–8019 RSC.
- X. X. Wang, Q. Wu, Z. Shan and Q. M. Huang, Biosens. Bioelectron., 2011, 26, 3614–3619 CrossRef CAS PubMed.
- X. Jiang, C. Sun, Y. Guo, G. Nie and L. Xu, Biosens. Bioelectron., 2015, 64, 165–170 CrossRef CAS PubMed.
- Y. Song, K. Qu, C. Zhao, J. Ren and X. Qu, Adv. Mater., 2010, 22, 2206–2210 CrossRef CAS PubMed.
- W. Shi, Q. Wang, Y. Long, Z. Long, Z. Cheng, S. Chen, H. Zheng and Y. Huang, Chem. Commun., 2011, 47, 6695–6697 RSC.
- Y. L. Liu, X. J. Zhao, X. X. Yang and Y. F. Li, Analyst, 2013, 138, 4526–4531 RSC.
- R. R. Crichton and J. P. Declercq, Biochim. Biophys. Acta, Gen. Subj., 2010, 1800, 706–718 CrossRef CAS PubMed.
- D. M. Lawson, P. J. Artymiuk, S. J. Yewdall, J. M. A. Smith, J. C. Livingstone, A. Treffry, A. Luzzago, S. Levi, P. Arosio, G. Cesareni, C. D. Thomas, W. V. Shaw and P. M. Harrison, Nature, 1991, 349, 541–544 CrossRef CAS PubMed.
- M. Uchida, S. Kang, C. Reichhardt, K. Harlen and T. Douglas, Biochim. Biophys. Acta, Gen. Subj., 2010, 1800, 834–845 CrossRef CAS PubMed.
- D. He and J. Marles-Wright, New Biotechnol., 2015, 32, 651 CrossRef CAS PubMed.
- G. Jutz, R. P. Van, M. B. Santos and A. Böker, Chem. Rev., 2015, 115, 1653–1701 CrossRef CAS PubMed.
- J. Zang, H. Chen, G. Zhao, F. Wang and F. Ren, Crit. Rev. Food Sci. Nutr., 2016, 57, 3673–3683 CrossRef PubMed.
- P. Arosio, R. Ingrassia and P. Cavadini, Biochim. Biophys. Acta, Gen. Subj., 2009, 1790, 589–599 CrossRef CAS PubMed.
- E. K. Honarmand, P. L. Hagedoorn and W. R. Hagen, Chem. Rev., 2015, 115, 295–326 CrossRef PubMed.
- N. D. Chasteen and P. M. Harrison, J. Struct. Biol., 1999, 126, 182–194 CrossRef CAS PubMed.
- G. Zhao, P. Arosio and N. D. Chasteen, Biochemistry, 2006, 45, 3429–3436 CrossRef CAS PubMed.
- J. Zhao, M. Liu, Y. Zhang, H. Li, Y. Lin and S. Yao, Anal. Chim. Acta, 2013, 759, 53–60 CrossRef CAS PubMed.
- D. Men, T. T. Zhang, L. W. Hou, J. Zhou, Z. P. Zhang, Y. Y. Shi, J. L. Zhang, Z. Q. Cui, J. Y. Deng, D. B. Wang and X. E. Zhang, ACS Nano, 2015, 9, 10852–10860 CrossRef CAS PubMed.
- K. Fan, C. Cao, Y. Pan, D. Lu, D. Yang, J. Feng, L. Song, M. Liang and X. Yan, Nat. Nanotechnol., 2012, 7, 459–464 CrossRef CAS PubMed.
- J. S. Stacey, P. Katharine, K. Hyunjeong, J. C. Branton, B. Juliana and F. W. Brian, Inorg. Chem., 2012, 51, 6421–6424 CrossRef PubMed.
- M. Kim, Y. Rho, K. S. Jin, B. Ahn, S. Jung, H. Kim and M. Ree, Biomacromolecules, 2011, 12, 1629–1640 CrossRef CAS PubMed.
- R. Yang, J. Tian, Y. Liu, Z. Yang, D. Wu and Z. Zhou, J. Agric. Food Chem., 2017, 65, 9950–9955 CrossRef CAS PubMed.
- F. Jia, J. J. Yin, N. Bo, X. Wu, H. Ye, M. Ferrari, G. J. Anderson, J. Wei, Y. Zhao and G. Nie, Biomaterials, 2011, 32, 1611 CrossRef PubMed.
- X. Jiang, C. Sun, Y. Guo, G. Nie and L. Xu, Biosens. Bioelectron., 2015, 64, 165–170 CrossRef CAS PubMed.
- L. Gao, J. Zhuang, L. Nie, J. Zhang, Y. Zhang, N. Gu, T. Wang, J. Feng, D. Yang, S. Perrett and X. Yan, Nat. Nanotechnol., 2007, 2, 577 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1ra03816h |
| This journal is © The Royal Society of Chemistry 2021 |