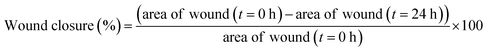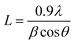Open Access Article
Open Access ArticleBiofabricated platinum nanoparticles: therapeutic evaluation as a potential nanodrug against breast cancer cells and drug-resistant bacteria
Saliha Manzoora,
Dar Junaid Bashira,
Khalid Imtiyazb,
M. Moshahid A. Rizvib,
Irshad Ahamadc,
Tasneem Fatmac,
Nidhi Bharal Agarwald,
Indu Arora*e and
Mohammed Samim *a
*a
aDepartment of Chemistry, School of Chemical and Life Sciences, Jamia Hamdard, New Delhi-110062, India. E-mail: shamim_chem@yahoo.co.in
bGenome Biology Lab, Department of Biosciences, Jamia Milia Islamia, New Delhi-110025, India
cCyanobacterial Biotechnology Lab, Department of Biosciences, Jamia Milia Islamia, New Delhi-110025, India
dCenter for Translational and Clinical Research, Jamia Hamdard, New Delhi-110062, India
eDepartment of Biomedical Sciences, Shaheed Rajguru College, Delhi University, New Delhi, India
First published on 16th July 2021
Abstract
Use of plant extracts for the synthesis of various metal nanoparticles has gained much importance recently because it is a simple, less hazardous, conservative and cost-effective method. In this research work, platinum nanoparticles were synthesized by treating platinum ions with the leaf extract of Psidium guajava and their structural properties were studied using various characterization techniques. The formation of platinum nanoparticles was confirmed by the disappearance of the absorbance peak at 261 nm in UV-visible spectra. The results of gas chromatography-mass spectrometry (GC-MS) and Fourier transform infrared spectroscopy (FT-IR) analysis showed functional moieties responsible for bio-reduction of metal ions and stabilization of platinum nanoparticles. The use of dynamic light scattering (DLS) imaging techniques confirmed the formation of stable monodispersed platinum nanoparticles showing a zeta potential of −23.4 mV. The morphological examination using high resolution transmission electron microscopy (HR-TEM) and Scanning electron microscopy (SEM) confirmed the formation of spherical platinum nanoparticles with an average diameter of 113.2 nm. X-ray powder diffraction (XRD) techniques showed the crystalline nature of biosynthesized platinum nanoparticles with a face-centered cubic structure. The results of energy-dispersive X-ray spectroscopy (EDAX) showed 100% platinum content by weight confirming the purity of the sample. The cytotoxic effect of biosynthesized platinum nanoparticles assessed in a breast cancer (MCF-7) cell-line by a 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay, revealed an IC50 of 167.2 μg ml−1. The results of a wound healing assay showed that treatment with platinum nanoparticles induced an anti-migratory effect on MCF-7 cells. In the cell cycle phase distribution, treatment with platinum nanoparticles inhibited cell proliferation as determined by flow cytometry with PI staining. Significant cell cycle arrest was detected at the G0/G1 phase with a notable decrease in the distribution of cells in the S and G2/M phases. The anti-bacterial activity of bio-synthesized platinum nanoparticles was evaluated against four pathogenic bacteria i.e. B. cereus (Gram positive), P. aeruginosa (Gram negative), K. pneumonia (Gram negative) and E. coli (Gram negative). The biosynthesized platinum nanoparticles were found to show dose-dependent inhibition against pathogenic bacteria with a significant effect on Gram-negative bacteria compared to Gram-positive bacteria. This synergistic blend of green and simplistic synthesis coupled with anti-proliferative and anti-bacterial properties makes these biogenic nanoparticles suitable in nanomedicine.
Introduction
Cancer is one of the major life threatening and deadly diseases worldwide, which causes most deaths.1 There were approximately 18 million cancer cases around the world in 2018. Breast cancer remains the most common cancer in women worldwide, with a contribution of 25.4% to the total number of new cases diagnosed in 2018.2 With this growing global burden, prevention of cancer is the utmost public health challenge of the 21st century. Options of treatment available for cancer are chemotherapy, radiotherapy and drugs with severe side-effects. These treatments are unbearable as they cause pain and strain and deteriorate the health of the patient. So, researchers are looking to develop some new and alternative methods for treating cancers.3 For many years, molecules based on platinum have received substantial attention because of their electrocatalytic properties.4,5 Derivatives of platinum, such as cisplatin, carboplatin and oxaliplatin have found use for treating cancer. However, some reports have suggested several shortcomings of cisplatin, such as neurotoxicity, hemolysis,6 nephrotoxicity,7 ototoxicity and toxicity to gametogenesis.8In last few decades, nanotechnology has emerged as a budding field with wide applications in nanomedicine and biomedical engineering. Platinum nanoparticles have brought a new revolution in the field of nanotechnology with numerous biomedical applications including diagnostics, drug delivery, photothermal therapy and medical implants.9–11 The synthesis of platinum nanoparticles for therapeutic applications has received considerable interest from both research and industrial fields because of their high stability, low toxicity and less side effects.12–15 In recent times, platinum nanoparticles have amassed growing interest in different biomedical applications such as targeted drug delivery, photoabalation therapy, biosensing, bioimaging, antimicrobial agent and anticancer agents.16
Biosynthesized nanoparticles play a crucial role in molecular interactions and can cross biological barriers without effecting normal cells. Platinum nanoparticles synthesized using Saccharomyces boulardii exhibited anticancer properties against squamouscarcinoma cell line (A431) and luminal breast cancer cell line (MCF7).17 Furthermore, apoptotic development in an early stage was detected in HeLa cells. The results of in vitro studies have revealed that platinum nanoparticles showed a dose-dependent inhibition on the growth of A549cells.18 Platinum nanoparticles can induce a potential anticancer activity compared to therapeutic drugs such as cis-platin by enhancing the activity of superoxide dismutase and increasing the levels of glutathione and malondialdehyde.19 In a study done earlier, it has been demonstrated that platinum nanoparticles leads to DNA strand breaks in human colon carcinoma cells (HT29).20 In the in vitro studies done on human breast adenocarcinoma cell-line (MCF-7), platinum nanoparticles induced apoptosis through G0/G1 cell cycle arrest. Thus, the authors concluded that DNA damage was due to the indexed oxidative pressure.21 In another study done on human bronchial epithelium (BEAS-2B) and human lung alveolar type II epithelioid cells (A549), it was found that 24 h after treatment with platinum nanoparticles A549 cells resulted in a loss of viability.22 The peptide stabilized platinum nanoparticles show significantly greater toxicity against hepatic cancer cells (HepG2) due to high cellular uptake and oxidative environment.23 The platinum nanoparticles also have potent anti-cancer activities against PA-1 cell line via induction of apoptosis and cell cycle arrest.24
In the current scenario, there is an urgent need to develop alternative therapeutics nontoxic to humans but toxic to pathogenic microorganisms due to the tendency of microorganism to attain resistance to antibiotics. Several studies done recently have focused on developing metallic nanoparticle-based therapeutics against pathogenic bacteria as these possess antibacterial properties against pathogens.25,26 The antibacterial activity of the bio-synthesized Pt NPs have also been extensively evaluated and the results observed are highly promising. Bio-synthesized Pt NPs using J. glandulifera and J. gossypifolia showed potent anti-bacterial activity against various pathogenic bacteria.27 Dates (Ajwa and Barni) based Pt NPs also showed immense anti-bacterial activity against B. subtilis and E. coli.28 Pt NPs using the leaf extract of X. strumarium havealso been reported to have excellent anti-bacterial activity against a panel of strains.29
Various methods developed for the synthesis of nanoparticles include physical and chemical methods. Physical methods such as laser ablation, vapor deposition, arc discharge, sputter deposition, ball milling and flame pyrolysis dissipates radiation and consumes enormous energy.30 Chemical methods such as wet chemical reduction,31 microemulsion,32 electrochemical process,33,34 photochemical reduction,35,36 hydrolysis,37 thermal decomposition,38 sono-decomposition,39,40 chemical vapor deposition41,42 use toxic chemicals not only hazardous to the living beings but also non-ecofriendly. Due to the limitations of these methods, the use of green synthesis approach based synthesis of nanoparticles is in great demand. The literature describes the use of various plants such as Ocimum santum,43 Brassica juncea,44 Diopyros Kaki45 and Asparagus racemosus Linn.46 in the biosynthesis of platinum nanoparticles by phytoreduction. The use of green synthesis as an alternate offers numerous advantages such as cost effectiveness, rapid synthesis, eco-friendly, monodispersity, large-scale production, reducing waste production and decreasing production cost.47–49 Moreover, nanoparticles with definite size and shape can be obtained by adjusting the temperature, pH and concentration of the reducing agent.50 There has been limited or almost nil research on the use of P. guajava extract in the green synthesis of platinum nanoparticles compared with the synthesis of nanoparticles of Ag, Au and other metals. Also, till date, there are no reports of the therapeutic application of platinum nanoparticles synthesized using the extract of P. guajava. This research has been encouraged by the reported prominent anti-cancer activity shown by the biosynthesized platinum nanoparticles.51 Given this background, here we report for the first time the anti-cancer and anti-bacterial activity of platinum nanoparticles synthesized using leaf extract of P. guajava. To obtain monodispersed platinum nanoparticles, various reaction conditions were optimized. The prepared platinum nanoparticles were further studied for their in vitro cytotoxicity and cell viability using the MCF-7 cell line and the biocompatibility of these platinum nanoparticles was evaluated. The antibacterial properties of the synthesized platinum nanoparticles were also investigated and they were generated as antibacterial agents against a broad range of Gram-positive and Gram-negative bacteria for controlling and preventing the spread of bacterial infections.
Material and method
Materials
Hexachloroplatinic acid hexahydrate was purchased from Molychem Pvt. Limited, Mumbai 400![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 002, India. Leaves of P. guajava were collected from (Herbal garden, Jamia Hamdard, New Delhi, India). MTT reagent was obtained from Sigma (USA). Dulbecco's modified Eagle medium (DMEM), fetal bovine serum (FBS), penicillin/streptomycin (PenStrep) solution and Trypsin–EDTA were provided by Gibco Life Technologies. Dimethyl sulfoxide (DMSO) was purchased from Qualigens Fine Chemicals (Mumbai, India). Propidium iodide (PI) from Sisco Research Laboratories. Pvt. Ltd, Taloja-410208, Maharashtra, India. Mueller Hinton agar (MHA) culture media, antibiotic streptomycin and Petri plates purchased from Hi-media (23, Vandhani Industrial Estate, L.B.S. Marg, Mumbai-400086, India).
002, India. Leaves of P. guajava were collected from (Herbal garden, Jamia Hamdard, New Delhi, India). MTT reagent was obtained from Sigma (USA). Dulbecco's modified Eagle medium (DMEM), fetal bovine serum (FBS), penicillin/streptomycin (PenStrep) solution and Trypsin–EDTA were provided by Gibco Life Technologies. Dimethyl sulfoxide (DMSO) was purchased from Qualigens Fine Chemicals (Mumbai, India). Propidium iodide (PI) from Sisco Research Laboratories. Pvt. Ltd, Taloja-410208, Maharashtra, India. Mueller Hinton agar (MHA) culture media, antibiotic streptomycin and Petri plates purchased from Hi-media (23, Vandhani Industrial Estate, L.B.S. Marg, Mumbai-400086, India).
Methods
Preparation of extract
Leaves of P. guajava as shown in (Fig. 1) were collected from the herbal garden at Jamia Hamdard and washed with distilled water to remove dust and other macroparticles. These washed leaves were then shade dried and fine powdered in a grinder. Then, 6 g of fine powder was taken in a beaker and about 100 ml of a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 mixture of methanol and deionized water was added to it. The above mixture was then kept on a hot-plate and after the temperature reached 90 °C, was heated for 10 min. The mixture was then removed from the hot-plate and allowed to cool at room temperature. The mixture was filtered using a Whatman filter paper and the filtrate was further used for the synthesis of platinum nanoparticles. The flask containing the plant extract was properly sealed and stored at 4 °C for further use.
1 mixture of methanol and deionized water was added to it. The above mixture was then kept on a hot-plate and after the temperature reached 90 °C, was heated for 10 min. The mixture was then removed from the hot-plate and allowed to cool at room temperature. The mixture was filtered using a Whatman filter paper and the filtrate was further used for the synthesis of platinum nanoparticles. The flask containing the plant extract was properly sealed and stored at 4 °C for further use.
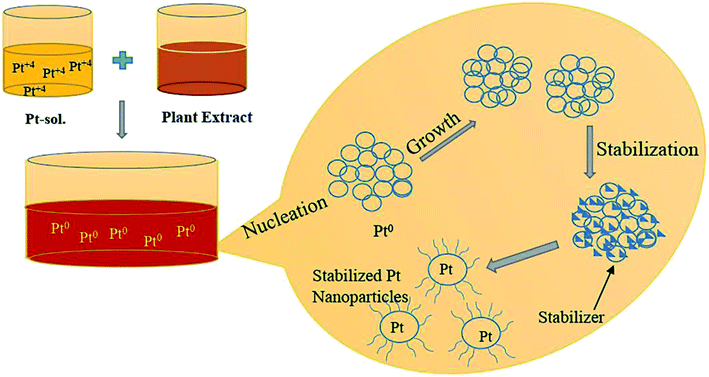
Biosynthesis of platinum nanoparticles using leaf extract of P. guajava
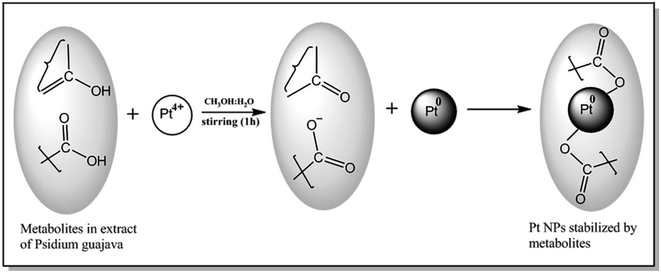
First, 1.0 mM aqueous solution of Hexachloroplatinic acid hexahydrate was prepared. 500 μl of the above prepared salt solution was added to 50 ml water in a flask and kept for continuous stirring of 1 h. Then, 4 ml of leaf extract was added drop-wise to above solution kept on stirring, followed by stirring for one more hour. Color change of the solution from light yellow to reddish brown indicated the formation of platinum nanoparticles. The platinum nanoparticle solution was then purified using the dialysis bag method for 72 h followed by lyophilization or freeze drying to increase the stability of platinum nanoparticles. Lastly, the final obtained powdered platinum nanoparticles were used for characterization and other activity purposes (Fig. 2).GC-MS analysis of extract of P. guajava
For GC-MS analysis, a Shimadzu QP-2010 ultra instrument was used to obtain the chromatograms. The GC-MS detection was done using an electron ionization system operated in the electron impact mode with ionization energy of 70 eV. The carrier gas used was helium (99.99%) with an injection volume of 1 μl and a constant flow rate of 1.21 ml min−1 at a pressure of 73.3 kPa with a split ratio of 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. The injector temperature was maintained at 260 °C, the ion source temperature at 220 °C and the column oven temperature at 60 °C. Mass spectra was obtained in the scan range (m/z 40–650). The calculation of relative percentage amount of each component was determined by comparing its average peak area to the total area. The extract components were identified by searching the NIST and WILEY libraries.
1. The injector temperature was maintained at 260 °C, the ion source temperature at 220 °C and the column oven temperature at 60 °C. Mass spectra was obtained in the scan range (m/z 40–650). The calculation of relative percentage amount of each component was determined by comparing its average peak area to the total area. The extract components were identified by searching the NIST and WILEY libraries.
Characterization of synthesized platinum nanoparticles
Synthesis of platinum nanoparticles was also analyzed using various characterization techniques. The reduction of platinum ions into platinum nanoparticles by using the leaf extract of P. guajava was first examined using UV-visible spectroscopy. A UV-visible spectrophotometer (U-2910, Hitachi instrument) at Jamia Hamdard, New Delhi, India was used. The functional groups present in the biomolecules comprising the leaf extract of P. guajava was analyzed by infra-red spectroscopy. FT-IR was done on a Bruker alpha instrument at Jamia Hamdard. To determine the stability of the platinum nanoparticles in the suspension, zeta potential analyzer by Nano ZS (Malvern, instrument corporation; Malvern, UK) was used at Jamia Milia Islamia, New Delhi, India. Distribution of platinum nanoparticles and their average size were determined at 25 °C by using a Zeta-sizer Nano ZS (Malvern, instrument corporation; Malvern, UK) analyzer at Jamia Hamdard, New Delhi, India. The shape and size of the synthesized platinum nanoparticles was determined by using small droplets of the dispersed nanoparticles, placed it on carbon coated grids and allowed it to dry at room temperature. Then, it was analyzed by using HR-TEM (JOEL JEM-1400) instrument at Indian Institute of Technology Delhi, India. The crystallinity of the lyophilized platinum nanoparticles was analyzed on (Rigaku D/max-2550 PC) X-ray diffractometer at Indian Institute of Technology Delhi, India. The surface morphology of synthesized platinum nanoparticles was determined by using SEM (Zeiss EVO 18) instrument at AIIMS, New Delhi, India. The quantity of the synthesized platinum nanoparticles was determined by using the SEM (Zeiss EVO 50) instrument at the Indian Institute of Technology Delhi, India.Investigation of anti-proliferative activity
Maintenance of breast cancer cell line
The in vitro cytotoxicity of platinum nanoparticles was determined using MCF-7 human breast adenocarcinoma cell line that was procured from the National Centre for Cell Science (NCSS), Pune, India. These cells were cultured by using T-25 culture flasks in DMEM, supplemented with 10% (v/v) heat-inactivated FBS and 1% (v/v) antibiotic solution (penicillin/streptomycin). The flasks were kept at 37 °C in atmosphere of 5% CO2 in an humidified chamber. The cytotoxicity of platinum nanoparticles was further studied by using MTT assay, which is considered as the most active assay for conduction of cell viability under in vitro conditions for any synthetic or biological materials.MTT assay
The in vitro toxicity of the biosynthesized platinum nanoparticles was evaluated by using MTT assay against MCF-7 cells. 96 well flat bottom plates were used in which cells were plated. Once the cells got attached to the plate, growth media was replaced with fresh media and incubated for 24 h with a series of dilution (7.81, 15.62, 31.25, 62.5, 125, 250 and 500 μg ml−1) of platinum nanoparticles. The cells which were left untreated act as control. Then, 20 μl of MTT in Phosphate buffer saline (PBS) with concentration 5 mg ml−1 was added to the plates of above respective concentrations. In order to facilitate the formation of formazan crystals, it was incubated again for 4 h. The media was carefully drawn out from each well and the formed MTT formazan crystals were dissolved in 200 μl DMSO and it was shaken well for 15 min to dissolve it completely. Then, optical density (O.D) of the solution was measured at 570 nm by using microplate reader iMark microplate (Bio-Rad, USA).52 The viable cells average was calculated and the experiments were carried out in triplicates. The cell viability percentage was calculated by using the following formula:Further, the graph was plotted b/w the dilutions and the cell viability by the use of GraphPad prism software.
Wound healing assay
The wound healing assay of platinum nanoparticles was performed according to the reported method on MCF-7 cells with slight modifications.53 MCF-7 cell lines (1 × 105 cells per well) were cultured in 12 well plate for about 24 h. Sterilized P10 pipette tip was used to scratch the convergent monolayers. To remove the detached cells, the cell monolayer was washed with PBS and maintained again at 37 °C with fresh media. The MCF-7 cells were treated with platinum nanoparticles free solution and platinum nanoparticles solution at respective IC50 concentrations. After treatment, the migrated cells were identified across the wound and images were captured with a phase contrast microscope (Nikon Instruments Inc. Japan) at 0 and 24 h. Using the following formula, the percentage of wound closure relative to that of the initial scratch was calculated:Cell cycle analysis
The cells were harvested and a single cell suspension in PBS + 2% FBS was prepared. The MCF-7 cells were then washed and spun at 300 × g for 5 min followed by resuspension at 3–6 × 106 cells per ml. An aliquot of 500 μl cells was taken in a 15 ml polypropylene, V-bottomed tube and fixed by chilled 70% ethanol overnight at 4 °C. The MCF-7 cells were incubated with 50 μl of RNase A (10 μg ml−1) at 37 °C for 1 h and then stained with propidium iodide staining solution for 30 min. The florescence intensity was analysed by performing flow cytometry at 488 nm using BD FACSDiva 8.0.2 software (BD Biosciences-US).54 All the experiments for cell cycle analysis were repeated 3 times.Investigation of anti-bacterial activity
The anti-bacterial activity of biosynthesized platinum nanoparticles was analyzed by using the agar plate disc diffusion method. Bacterial cultures B. cereus (MCC 2243) Gram positive, E. coli (MCC 2412) Gram negative, K. pneumonia (MCC 2716) Gram negative bacteria were procured from Microbial Culture Collection at National Centre for Cell Science, Pune, India and P. aeruginosa (MTCC 2453) Gram negative bacteria were acquired from Microbial Type Culture Collection (MTCC), Institute of Microbial Technology (IMTECH), Chandigarh India. Bacterial cultures were prepared by growing a single colony in nutrient broth medium and adjusting the turbidity to 0.5 McFarland standards. A 100 μl volume of bacterial culture was spread onto 25 ml Mueller–Hinton agar medium plates. Different concentrations (10, 20, 30 and 40 μg ml−1) of platinum nanoparticles were added on the sterilized disc of 5 mm size. Standard antibiotic (streptomycin) 10 μg ml−1 were used as a control. The inoculated plates were incubated at 37 °C in sterilized conditions and dark regions overnight. The results were expressed as the mean diameter of the inhibition zone in mm ± standard deviation. All experiments were concurrently performed in triplicate.55,56Statistical analysis
The data from individual groups were presented as the mean ± standard deviation (SD). For anti-bacterial activity data analysis, a two-way ANOVA was performed. The multiple t-test was performed for data analysis of wound healing assay and cell cycle analysis. For MTT assay data analysis was done using one-way ANOVA multiple comparison test. All the statistical analysis was performed on GraphPad Prism v8.4 (GraphPad Software, San Diego, CA, United States). The minimum criterion for statistical significance was set at p < 0.05 for all the comparisons.Results and discussion
The GC-MS analysis of P. guajavaextract
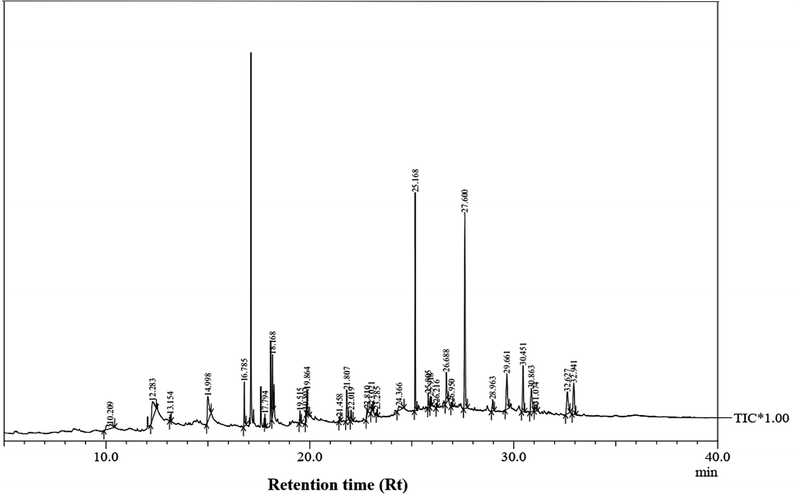
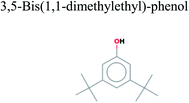
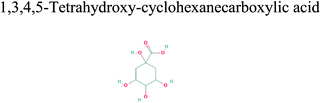
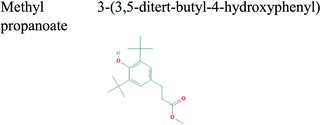
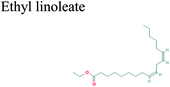
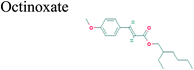
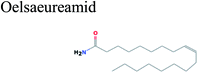
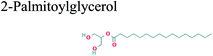
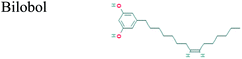
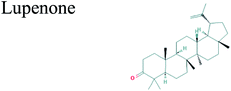
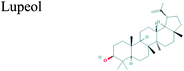
The GC-MS analysis of P. guajava extract was performed to determine the percentage of different metabolites and investigate their role in the reduction and stabilization of the synthesized platinum nanoparticles. The GC-MS chromatogram (Fig. 3) showed organic molecules like vitamin E (19.24%), 1,3,4,5-tetrahydroxy-cyclohexanecarboxylic acid (5.74%), lupenone (5.74%), γ-sitosterol (5.59%), 3-butyl-1,2,4-cyclopentanetrione (5.45%), lupeol (4.34%), n-hexadecanoic acid (4.13%), bilobol (3.60%), oelsaeureamid (3.06%), 2-ethylbutyric acid, eicosyl ester (1.34%), ethyl linoleate (0.52%), octadecanamide (0.97%), 2-palmitoylglycerol (0.75%), ethyl linolenate (0.24%), γ-tocopherol (0.46%), 3,5-bis(1,1-dimethylethyl)-phenol (0.39%). The metabolite composition of the extract showed vitamin E as the major component along with other phenols, carboxylic acids, sterols, esters, terpenoids and amides (Table 1). Thus, the results of GC-MS analysis indicate the reduction and stabilization of platinum nanoparticles by phenolic compounds and carboxylic acids, as shown in the mechanism (Fig. 4).57UV-visible spectra analysis
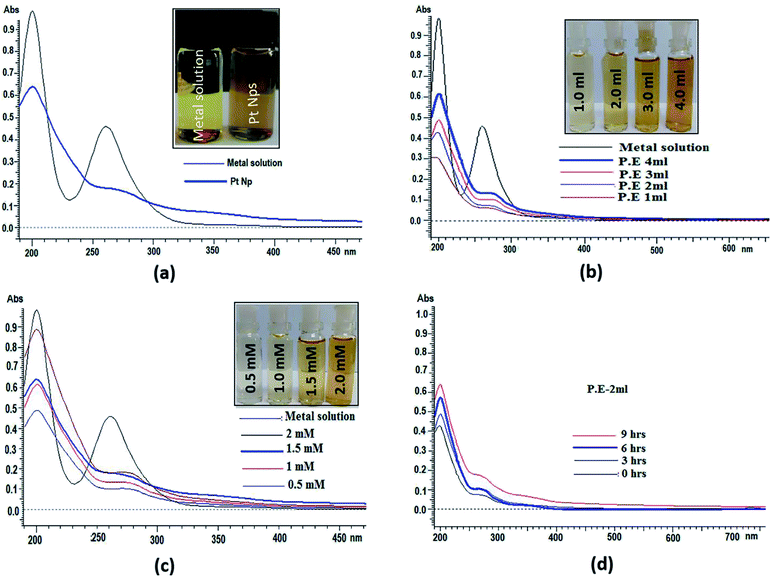
UV-visible spectroscopy is among the most widely used and easily available techniques to characterize the synthesized platinum nanoparticles. In UV-visible spectroscopy, absorption maxima were scanned between 190 and 600 nm. UV-visible spectra of both platinum salt solution and platinum nanoparticles were recorded in which two absorbance peaks at 201 nm and 261 nm appear in platinum salt solution. These are characteristic of the absorbance of platinum complexes due to ligand to metal charge transfer transition.58 In platinum nanoparticles, continuous spectrum and disappearance of a peak at 261 nm, as shown in (Fig. 5a) shows the complete reduction of platinum ions and formation of platinum nanoparticles. Initially, the reduction of Pt(IV) to Pt(0) was confirmed by the colour change due to surface plasmon resonance (SPR) found in metallic nanoparticles, which arise because of the combined vibrational effect of free electrons on the surface. UV-visible spectra were also determined using different concentrations of P. guajava extract, aqueous solution of hexachloroplatinic acid hexahydrate and the reaction time at a room temperature of 24–25 °C (Fig. 5b). shows the increase in the bio-reduction of platinum ions on increasing the concentration of P. guajava extract without any phenomenon of red or blue shift, which suggests the formation of stable platinum nanoparticles occurs from platinum ions and the size distribution of nanoparticles is uniform. Different concentrations of the salt solution (0.5, 1, 1.5 and 2 mM) were also prepared. In these reactions, the concentration of the P. guajavaextract used for the reduction of platinum ions was kept constant and the concentration of the salt solution was varied. The (Fig. 5c) shows bio-reduction increases without any change in wavelength, which confirms that the biosynthesized platinum nanoparticles were of uniform size distribution and stable. The effect of time interval has also been studied on the biosynthesis of platinum nanoparticles with the reaction mixture containing 2 ml P. guajavaextract. The (Fig. 5d) shows an increase in the bio-reduction with the increase in the reaction time without any shift in wavelength. However, with the increase in the time interval, no significant colour change was observed.FT-IR spectrum analysis
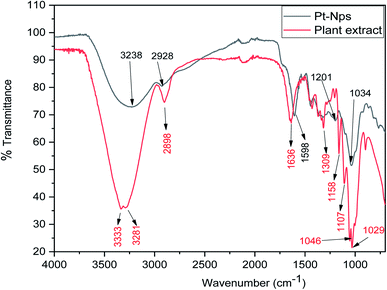
FT-IR analysis was done to determine the formation of platinum nanoparticles with the use of the leaf extract of P. guajava. From the FT-IR analysis of both leaf extract of P. guajava and platinum nanoparticles, it can be concluded that functional groups present in the extract help in the reduction and stabilization of platinum nanoparticles. The (Fig. 6) shows that in the leaf extract of P. guajava, FT-IR peaks at 3333 cm−1 corresponds to [OH– stretching], H-bonded groups in alcohol and phenol, 2898 cm−1 [CHO– stretch], 1636 cm−1 [C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O], 1107 cm−1 [ C–O–C stretch], 1046 cm−1 and 1029 cm−1 [
O], 1107 cm−1 [ C–O–C stretch], 1046 cm−1 and 1029 cm−1 [![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–O–C sym. Stretch].59 The FT-IR spectrum of platinum nanoparticles shows appearance of a broad peak at 3238 cm−1 [OH– stretching] and H-bonded groups, which compared with P. guajava extract shows a decline in the sharpness of the peak and a shift toward lower wavelength. The peak at 2928 cm−1 [CHO– stretch] on comparison with extract shows a decrease in the intensity and shift towards higher wavelength. The peak at 1588 cm−1 [C
C–O–C sym. Stretch].59 The FT-IR spectrum of platinum nanoparticles shows appearance of a broad peak at 3238 cm−1 [OH– stretching] and H-bonded groups, which compared with P. guajava extract shows a decline in the sharpness of the peak and a shift toward lower wavelength. The peak at 2928 cm−1 [CHO– stretch] on comparison with extract shows a decrease in the intensity and shift towards higher wavelength. The peak at 1588 cm−1 [C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O] has shifted to a lower wavelength on comparision with the C
O] has shifted to a lower wavelength on comparision with the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O peak of extract. The results obtained show that these functional groups of different organic compounds of leaf extract of P. guajava are mainly responsible for the bio-reduction and stabilization of platinum nanoparticles.
O peak of extract. The results obtained show that these functional groups of different organic compounds of leaf extract of P. guajava are mainly responsible for the bio-reduction and stabilization of platinum nanoparticles.
Zeta potential and DLS analysis
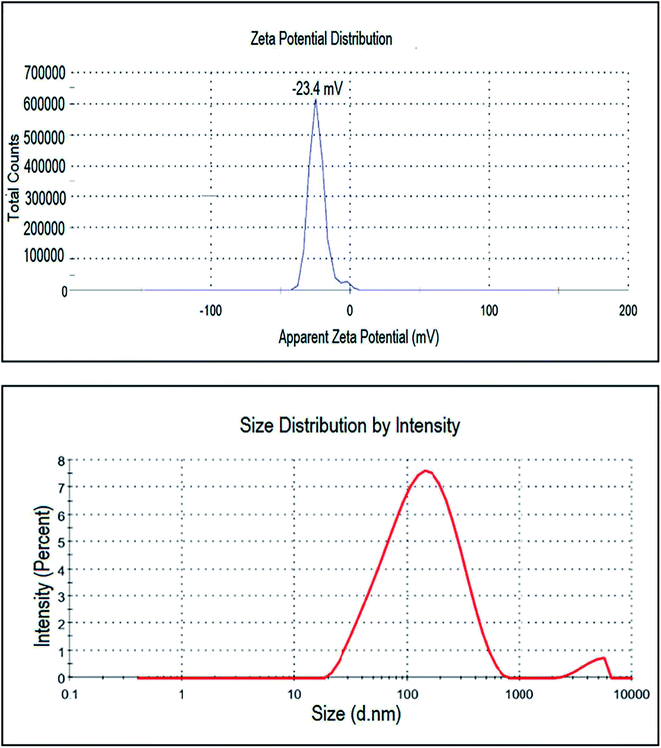
This analysis was used to measure the surface charge of the suspended particles in the solution. It analyses the stability of nanoparticles by the magnitude of charges in which high positive or negative values depict the higher stability of nanoparticles. With larger positive or negative values of zeta potential of the particles, they will exert the force of repulsion on each other and will not come together or agglomerate. In (Fig. 7) sharp peak of the biosynthesized platinum nanoparticles by using P. guajava at −23.4 mV shows that the nanoparticles have a negatively charged surface. Thus, we can conclude that platinum nanoparticles are relatively stable and surrounded by anionic compounds. DLS also known as the photon correlation technique, was used to measure the average size of particles in solution. In this technique, biosynthesized platinum nanoparticles were dissolved in distilled water and the suspension formed was centrifuged at 9000 rpm at 25 °C for 10 min. After that, the supernatant of the solution was separated and sonicated for 5 min. Then, it was analyzed with a computer aided size analyzer of nanoparticles. The size distribution graph showed the average hydrodynamic diameter of platinum nanoparticles to be 108.8 nm with a P.D.I of 0.371 (Fig. 7).HR-TEM and SEM analysis
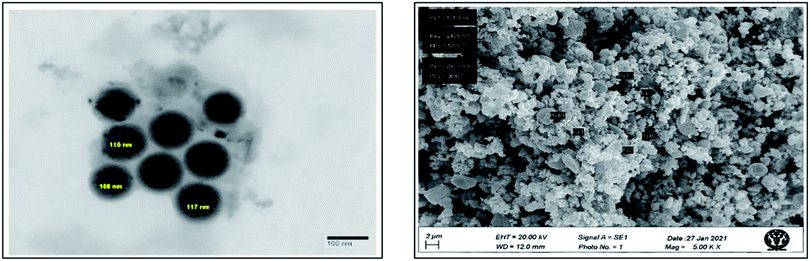
To determine the size and shape of the platinum nanoparticles biosynthesized using leaf extract of P. guajava, TEM analysis was done. In this technique, platinum nanoparticles were dissolved in water in which it will be completely miscible. Then, a drop of this solution was kept on a carbon coated copper grid and then placed in a vacuum desiccator for drying. After complete drying grid was placed on the specimen holder for analysis, the image formed as the electrons interact with the sample and transmit through it. TEM images of synthesized platinum nanoparticles shown in (Fig. 8) reveal that nanoparticles were spherical and the average size of these platinum nanoparticles was found to be around 113.6 nm. To determine the surface morphology of platinum nanoparticles synthesized using the leaf extract of P. guajava, SEM analysis was performed. The (Fig. 8) shows cluster formation of the nanoparticles, which indicates that the nanoparticles agglomerated with each other because of the ample amount of stabilizing agent.XRD analysis
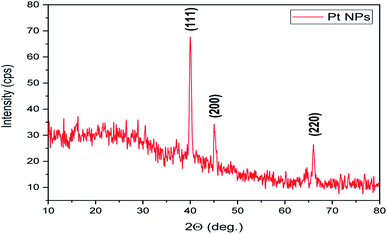
It is an analytical technique used to identify the phase of a crystalline substance and to study the structure of crystals. In this technique, X-rays are produced in a cathode ray tube and get filtered to form a monochromatic radiation that interacts with the sample and forms a constructive interference. Platinum nanoparticles synthesized using the leaf extract of P. guajava confirm the XRD pattern, as shown in (Fig. 9) in which the diffraction peaks are in the 2θ range of 10° to 80° at 40.22°, 46.76° and 67.45° corresponds to the lattice planes of (111), (200) and (220) respectively, which were consistent with face-centered cubic (fcc) structure of platinum.60 Thesize of biosynthesized platinum nanoparticles was calculated using Debye–scherrer equation:where L is the mean particle size, λ is the X-ray wavelength, β is the line broadening at half the maximum intensity and θ is the Bragg angle. The estimated mean crystalline size of the biosynthesized platinum nanoparticles was found to be 113 nm. The result was in close agreement with the result of HR-TEM measurement.
EDAX analysis
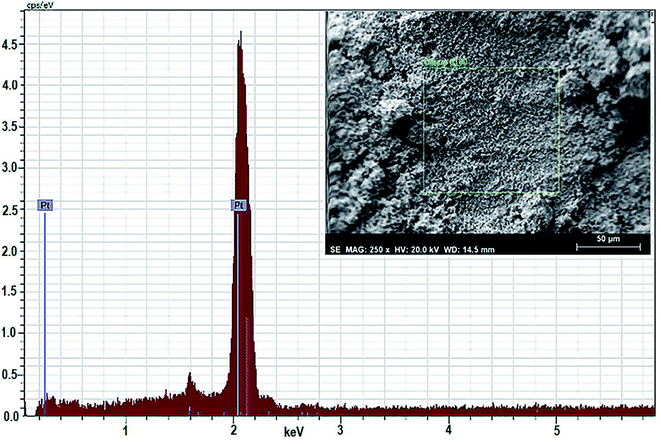
For quantitative estimation of biosynthesized platinum nanoparticles formed using the leaf extract of P. guajava, scientific technique EDAX or EDX is used. In this technique, the sample comes in contact with a source of X-ray excitation. To stimulate the emission of characteristic X-rays from a specimen, a high beam of charged particles such as electron, proton or a beam of X-rays are being studied on the sample. The data obtained in (Fig. 10) clearly show that the content of platinum present in the sample is highest, almost 100% at 2 keV, which confirms the presence of platinum nanoparticles in the sample.Anti-proliferative studies
MTT assay
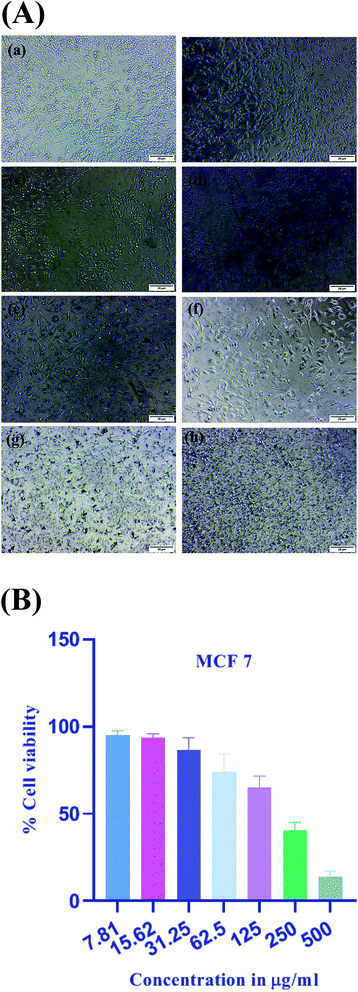
The MTT assay measures cellular metabolism by the use of 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT), which is the tetrazolium salt and it gives the assessment of the number of metabolically active cells.61 This assay depicts the metabolic activity of cell indirectly by the conversion of yellow tetrazolium dye, which is water soluble, into the insoluble crystals of purple formazan by the action of mitochondrial dehydrogenase enzymes. The number of viable cells can be determined by the amount of formazan formed. In case of metabolically active cells, mitochondrial activity remains the same and any change in it can be assessed by the number of active cells.62 It is one of the most widely used in vitro cell viability assays for detecting cytotoxicity of various natural as well as synthetic compounds including biosynthesized nanoparticles. These platinum nanoparticles were tested for in vitro cytotoxic potential against human breast cancer cells MCF-7. The (Fig. 11A) depicts the morphological changes of MCF-7 cells on treatment with platinum nanoparticles. The effectiveness of the platinum nanoparticles was evaluated with a series of dilutions in the range of (7.81–500 μg ml−1). By the use of MTT assay, half-maximal inhibitory concentration (IC50) of the platinum nanoparticles was determined to be 167.2 μg ml−1 at the treatment of 24 h. The (Fig. 11B) depicts the concentration dependent cytotoxic potential of platinum nanoparticles against MCF-7 cells in which 7.81 μg ml−1 concentration showed the percentage of cell viability at 95.16 ± 2.54%. On increasing the concentration to 15.62 μg ml−1, the percentage of cell viability decreased to 93.51 ± 2.23%. The concentration of 31.25 μg ml−1 resulted in further decrease of 86.40 ± 7.23% and the highest concentration of 500 μg ml−1 resulted in 13.91 ± 3.44% of cell viability, which depicts the high cytotoxic inhibition. Thus, it was found that at lower concentrations, cytotoxic inhibition is less and cell viability is significant. On increasing the concentration of platinum nanoparticles, cell viability constantly decreased and cytotoxic inhibition increased, which shows that the activity is dose dependent. Thus, a higher concentration of platinum nanoparticles increases the mortality of breast cancer cells or lowers cell viability.Wound healing assay
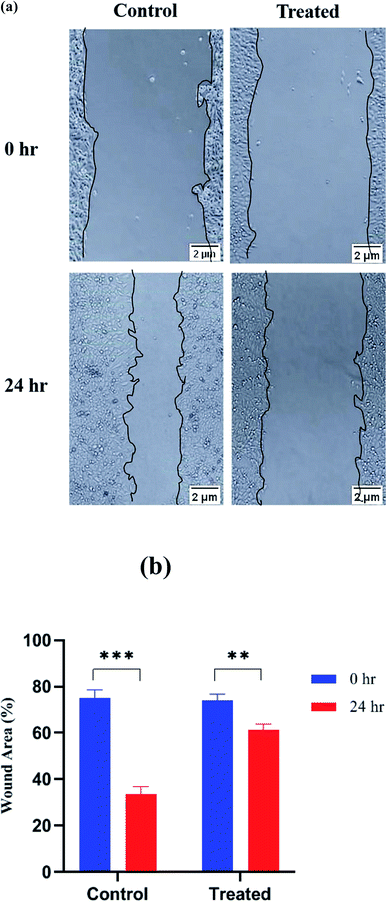
Wound-healing assay or cell scratch assay is used to check the anti-metastatic effect of any drug/agent by evaluating the migration of cancer cells in two dimensions. It is one of the simplest, cost effective and highly reproducible method to evaluate anti-metastatic effect. It is performed by creating a wound in a cell monolayer and then capturing the images at different time intervals. On comparing the images at the beginning and at regular intervals of the cell migration, the rate of migration of the cells can be calculated. It is a process involved in cancer metastasis because after creating an artificial wound, cells migrate to reestablish the interaction. Cancer cells can migrate to the scratched edges and fill the wound area.63 On comparing with the wound area of the formulation-treated cells, migration of the cells was inhibited due to the anti-metastatic potential of the formulation. In the case of the control group, where no treatment was given to the cancer cells, cells migrate to the scratched edges and healed the wound area. This assay examined the impact of platinum nanoparticles on the migratory behavior of MCF-7 cells. In control cells, the area was covered almost entirely between the scratched edges. However, with the platinum nanoparticles, a significant reduction was observed in the ability to migrate between the scratched edge, which produces a noticeable effect on the inhibition of the migratory potential of MCF-7 cells. The percentage of wound healing was calculated and it was found to be 75.24 ± 3.41% for control and 74.05 ± 2.77% for treated cells at time 0 h. The wound area reduced to 33.71 ± 3.18% for control and 61.33 ± 2.45% for treated cells after 24 h. Thus, the (Fig. 12a) microscopic images depict how treated MCF-7 cells show greater inhibition of cell migration than control at 0 h and 24 h and in (Fig. 12b) it is clearly depicted that platinum nanoparticles possess anti-migration potential more than that of platinum nanoparticles free solution.Cell cycle analysis
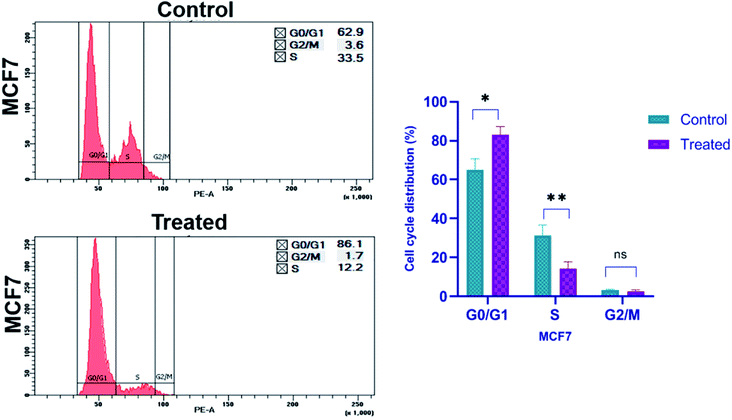
For cell cycle analysis, flow cytometry is a common method used to evaluate the cell cycle by measuring the cellular DNA content. It is a quick and reliable method that determines the chemical as well as physical characteristics of the cell population. During the process a fluorescent dye, propidium iodide is used, which is commonly used to stain cells and it intercalates into the cellular DNA by passing through the permeabilized membrane. PI can also stain RNA, so, after fixing the cells with ethanol, RNase A is also added before staining with PI. This helps to analyze the cell cycle more accurately.64 By measuring the DNA content of cells in different phases of cell cycle, we can assess the effect of any agent on it. The different stages of cell cycle are G0/G1, S, and G2/M, which are compared between the treated and untreated group of cells. To investigate whether platinum nanoparticles inhibit cell proliferation by regulating cell cycle progression, changes to the cell cycle were detected following treatment with nanoparticles for 24 h by propidium iodide staining and flow cytometry analysis. Platinum nanoparticles at 167.2 μg ml−1 induced significant cell cycle arrest at G0/G1 phase (p ≥ 0.05, Fig. 13) and the distribution of cells in G2/M and S phases were notably decreased compared with the control cells. In MCF-7 cells treated with platinum nanoparticles (167.2 μg ml−1), the percentage of cells at G2/M decreased to 1.7%, which was half that of the control group (3.6%; Fig. 13). Compared with the untreated control, in which 62.9% and 33.5% of cells were at G0/G1 and S phase, respectively, MCF-7 cells treated with platinum nanoparticles had 86.1% and 12.2% of cells at the respective phases. The percentage of cells in G0/G1 increases compared to that of the control, whereas the population in the S phase and G2/M decreases. This shows that the platinum nanoparticles were effective in restricting the cells in G0/G1 phase of the cell cycle and also inhibiting the cells to pass on to S phase of cell cycle leading to inhibition of the cell division, and subsequently cell proliferation in breast cancer cells.Anti-bacterial studies
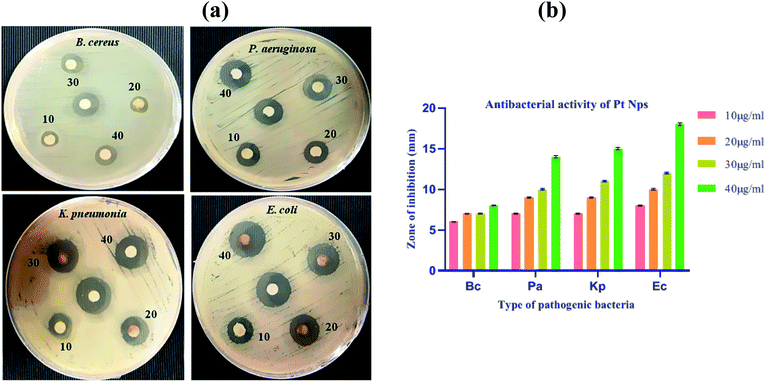
The anti-bacterial property of the biosynthesized platinum nanoparticles was evaluated against Bacillus cereus (B. cereus), Pseudomonas aeruginosa (P. aeruginosa), Klebsiella pneumonia (K. pneumonia) and Escherichia coli (E. coli) pathogenic bacteria. The biosynthesized platinum nanoparticles showed susceptibility in all bacteria, but a significant zone of inhibition was observed in E coli. In all these bacterial strains, a minimum concentration of 10 μg ml−1 also showed the anti-bacterial potential.For all Gram-negative bacteria, synthesized platinum nanoparticles offer consistently superior anti-bacterial effect as compared to Gram-positive bacteria B. cereus. We observed better results in Gram-negative bacteria than Gram positive bacteria. This result might be due to the physical and chemical structural difference in bacteria because the cell wall of Gram-positive is thick compared to Gram-negative bacteria.65 Nanoparticles bind on the surface of the bacterial membrane and rupture the cell wall of the bacteria. The thin cell wall of E. coli ruptures easily than the thick cell wall of the B. cereus and due to this reason, E. coli showed a better result than B. cereus. Platinum nanoparticles showed concentration dependent zone of inhibitions at 10, 20, 30 and 40 μg ml−1, as shown in (Fig. 14a and b) and (Table 2).
| Concentrations of nanoparticles | B. cereus | P. aeruginosa | K. pneumonia | E. coli |
|---|---|---|---|---|
| 10 μg ml−1 | 06 ± 0.02 | 07 ± 0.05 | 07 ± 0.05 | 08 ± 0.06 |
| 20 μg ml−1 | 07 ± 0.04 | 09 ± 0.07 | 09 ± 0.07 | 10 ± 0.09 |
| 30 μg ml−1 | 07 ± 0.05 | 10 ± 0.10 | 11 ± 0.09 | 12 ± 0.11 |
| 40 μg ml−1 | 08 ± 0.06 | 14 ± 0.15 | 15 ± 0.14 | 18 ± 0.18 |
The biosynthesized platinum nanoparticles solution at a maximum concentration of 40 μg ml−1, inhibits the growth of B. cereus with a zone of inhibition (8 ± 0.06 mm), P. aeruginosa with a zone of inhibition (14 ± 0.15 mm), K. pneumonia with a zone of inhibition (15 ± 0.14 mm) and E. coli with a maximum zone of inhibition (18 ± 0.18 mm). However, at the concentration of 10 μg ml−1, the zone of inhibition was found to be (6 ± 0.02 mm) for B. cereus, (7 ± 0.05 mm) for P. aeruginosa, (7 ± 0.05 mm) for K. pneumonia and (08 ± 0.06 mm) for E. coli. Thus, the results clearly show that the anti-bacterial activity increases with the increase in the concentration of platinum nanoparticles, which indicates that the activity is dose dependent.
Several possible mechanisms have been proposed for inhibiting bacteria through metal nanoparticles such as (i) denaturation of the thiol group of the critical enzymes of bacteria by metal nanoparticles, (ii) metal ions interact with bacterial DNA and destroy them, (iii) bacterial species are inactivated through respiratory burst mechanism, (iv) disruption of bacterial cell membrane, (v) disruption of cellular transport system of bacteria, (vi) production of highly reactive oxygen species (ROS) i.e. hydroxyl and superoxide radicals from metal nanoparticles. It has previously been reported that an increase in the concentration of ROS causes damage to the bacterial cells.66
The Xanthium strumarium leaf extract based platinum nanoparticles reported earlier showed antibacterial activity with the maximum zone of inhibition against E. coli (20 ± 0.5 mm), K. pneumonia (19 ± 0.5 mm), P. aeruginosa (18 ± 0.5 mm), S. aureus (22 ± 0.5 mm), and B. subtilis (19 ± 0.5 mm) at the concentration of 100 μg per well.29 While Taraxacum laevigatum extract-based platinum nanoparticles reported antibacterial activity against two bacterial species B. subtilis and P. Aeruginosa, the zone of inhibition of Pt-NPs againstB. subtilis was (18 ± 0.8) mm and P. aeruginosa was (15 ± 0.5) mm.67 Another researcher reported antibacterial activity of Botryococcus braunii algae based platinum nanoparticles against bacterial strains P. aeruginosa, E. coli, K. pneumoniae and S. aureus with a zone of inhibition ranging from 7 to 16 mm.68 Platinum nanoparticles synthesized during the present study using leaf extract of P. guajava showed higher zone of inhibition at lower concentrations (10, 20, 30, 40 μg ml−1) against B. cereus, P. aeruginosa, K. pneumoniae and E. coli respectively. The comparison of the zone of inhibition values with previous studies showed a much better nature of platinum nanoparticles. The anti-bacterial activity of these platinum nanoparticles may also be due to the morphology and size of the nanoparticles.
Conclusion
This investigation involved the use of biological methods for synthesizing metal nanoparticles as this would eliminate any kind of waste generated from chemical and physical methods. This method is cheap, simple and ecofriendly further adds to the advantage. In this work, platinum nanoparticles were synthesized using leaf extract of P. guajava. The characterization of biosynthesized platinum nanoparticles was done using various techniques such as UV-Vis and FT-IR spectroscopy, zeta potential, DLS, TEM, XRD, SEM and EDAX. The biosynthesized platinum nanoparticles have cytotoxic properties that inhibit cell proliferation. Platinum nanoparticles also exhibited anti-metastatic potential on MCF-7 breast cancer cell line as demonstrated by inhibition in migration. In the cell cycle analysis platinum nanoparticles activated cell cycle arrest at the G0/G1 phase. Platinum nanoparticles also displayed antibacterial properties against pathogenic bacteria. These findings asserted that platinum nanoparticles have potential anti-proliferative and anti-bacterial properties and can pave the way for developing future anti-proliferative and anti-bacterial bio-nanomedicine.Abbreviations
| ROS | Reactive oxygen species |
| MTT | 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide |
| DMEM | Dulbecco's modified Eagle medium |
| FBS | Fetal bovine serum |
| PBS | Phosphate buffer saline |
| DMSO | Dimethyl sulphoxide |
| SPR | Surface plasmon resonance |
| FT-IR | Fourier transform infrared spectroscopy |
| DLS | Dynamic light scattering |
| HR-TEM | High resolution transmission electron microscopy |
| XRD | X-ray powder diffraction |
| SEM | Scanning electron microscopy |
| EDAX | Energy-dispersive X-ray spectroscopy |
Funding
This research did not receive any kind of grant from funding agency.Conflicts of interest
The authors declare no competing financial interest.Acknowledgements
We gratefully acknowledge the valuable contribution of SAIF facility at AIIMS, New Delhi, Central Research Facility at IIT Delhi and Central Instrumentation Facility at JMI, New Delhi.References
- X. Gao, Y. Lu, L. Fang, X. Fang, Y. Xing, S. Gou and T. Xi, Eur. J. Med. Chem., 2013, 69, 1–9 CrossRef CAS PubMed
.
- F. Bray, J. Ferlay, I. Soerjomataram, R. Siegel, L. Torre and A. Jemal, Ca-Cancer J. Clin., 2020, 70, 313 Search PubMed
.
- K. J. Wernli, J. M. Hampton, A. Trentham-Dietz and P. A. Newcomb, Pharmacoepidemiol. Drug Saf., 2011, 20, 131–137 CrossRef PubMed
.
- I. Kostova, Recent Pat. Anti-Cancer Drug Discovery, 2006, 1, 1–22 CrossRef CAS PubMed
.
- I. E. Stephens, A. S. Bondarenko, U. Grønbjerg, J. Rossmeisl and I. Chorkendorff, Energy Environ. Sci., 2012, 5, 6744–6762 RSC
.
- E. P. Getaz, S. Beckley, J. Fitzpatrick and A. Dozier, N. Engl. J. Med., 1980, 302, 334–335 CrossRef CAS PubMed
.
- M. Cemazar, R. MilaCciCc, D. MiklavCiC, V. Dolzan and G. Sersa, Anti-Cancer Drugs, 1998, 9, 525–530 CrossRef CAS PubMed
.
- Y. S. Kim, J. S. Kim, H. S. Cho, D. S. Rha, J. M. Kim, J. D. Park, B. S. Choi, R. Lim, H. K. Chang, Y. H. Chung, I. H. Kwon, J. Jeong, B. S. Han and I. J. Yu, Inhalation Toxicol., 2008, 20, 575–583 CrossRef CAS PubMed
.
- R. E. Doherty, I. V. Sazanovich, L. K. McKenzie, A. S. Stasheuski, R. Coyle, E. Baggaley, S. Bottomley, J. A. Weinstein and H. E. Bryant, Sci. Rep., 2016, 6, 22668 CrossRef CAS PubMed
.
- T. C. Johnstone, K. Suntharalingam and S. J. Lippard, Chem. Rev., 2016, 116, 3436–3486 CrossRef CAS PubMed
.
- Z. Wang, L. Chen, C. Huang, Y. Huang and N. Jia, J. Mater. Chem. B, 2017, 5, 3498–3510 RSC
.
- H. Hosaka, R. Haruki, K. Yamada, C. Böttcher and T. Komatsu, PLoS One, 2014, 9, e110541 CrossRef PubMed
.
- S. Li, E. Porcel, H. Remita, S. Marco, M. Réfrégiers, M. Dutertre, F. Confalonieri and S. Lacombe, Cancer Nanotechnol., 2017, 8, 4 CrossRef PubMed
.
- K. McNamara and S. A. M. Tofail, Adv. Phys.: X, 2017, 2, 54–88 CAS
.
- Y. Yoshihisa, A. Honda, Q. L. Zhao, T. Makino, R. Abe, K. Matsui, H. Shimizu, Y. Miyamoto, T. Kondo and T. Shimizu, Exp. Dermatol., 2010, 19, 1000–1006 CrossRef CAS PubMed
.
- M. Jeyaraj, S. Gurunathan, M. Qasim, M.-H. Kang and J.-H. Kim, Nanomaterials, 2019, 9, 1719 CrossRef CAS PubMed
.
- V. Borse, A. Kaler and U. C. Banerjee, Int. J. Emerg. Trends Electr. Electron, 2015, 11 Search PubMed
.
- B. Yogesh, B. Vineeta, N. Rammesh and P. Saili, J. Pharmacopuncture, 2016, 19, 114 CrossRef PubMed
.
- A. Medhat, S. Mansour, S. El-Sonbaty, E. Kandil and M. Mahmoud, Tumor Biol., 2017, 39, 1010428317717259 CAS
.
- J. Pelka, H. Gehrke, M. Esselen, M. Türk, M. Crone, S. Bräse, T. Muller, H. Blank, W. Send and V. Zibat, Chem. Res. Toxicol., 2009, 22, 649–659 Search PubMed
.
- B. Şahin, A. Aygün, H. Gündüz, K. Şahin, E. Demir, S. Akocak and F. Şen, Colloids Surf., B, 2018, 163, 119–124 CrossRef PubMed
.
- S. Diabaté, C. Weiss, S. Mülhopt, H. Paur, V. Niedetzky and M. Seipenbusch, Altern. Lab. Anim., 2003, 36, 285–298 CrossRef PubMed
.
- M. S. Shoshan, T. Vonderach, B. Hattendorf and H. Wennemers, Angew. Chem., Int. Ed., 2019, 58, 4901–4905 CrossRef CAS PubMed
.
- Y. Bendale, V. Bendale and S. Paul, Integr. Med. Res., 2017, 6, 141–148 CrossRef PubMed
.
- N. Beyth, Y. Houri-Haddad, A. Domb, W. Khan and R. Hazan, J. Evidence-Based Complementary Altern. Med., 2015, 2015, 246012 Search PubMed
.
- B. Ramalingam, T. Parandhaman and S. K. Das, ACS Appl. Mater. Interfaces, 2016, 8, 4963–4976 CrossRef CAS PubMed
.
- U. Jeyapaul, M. J. Kala, A. J. Bosco, P. Piruthiviraj and M. Easuraja, Orient. J. Chem., 2018, 34, 783 CrossRef CAS
.
- N. S. Al-Radadi, Arabian J. Chem., 2019, 12, 330–349 CrossRef CAS
.
- P. V. Kumar, S. M. J. Kala and K. Prakash, J. Environ. Chem. Eng., 2019, 7, 103146 CrossRef CAS
.
- C. Dhand, N. Dwivedi, X. J. Loh, A. N. Jie Ying, N. K. Verma, R. W. Beuerman, R. Lakshminarayanan and S. Ramakrishna, RSC Adv., 2015, 5, 105003–105037 RSC
.
- H. Bönnemann and R. M. Richards, Eur. J. Inorg. Chem., 2001, 2001, 2455–2480 CrossRef
.
- Z. Liu, J. Y. Lee, M. Han, W. Chen and L. M. Gan, J. Mater. Chem., 2002, 12, 2453–2458 RSC
.
- K. T. Kim, S.-H. Jin, S.-C. Chang and D.-S. Park, Bull. Korean Chem. Soc., 2013, 34, 3835–3839 CrossRef CAS
.
- J. Zeng, J. Y. Lee and W. Zhou, Appl. Catal., A, 2006, 308, 99–104 CrossRef CAS
.
- S. Abdolhosseinzadeh, S. Sadighikia and S. Alkan Gürsel, ACS Sustainable Chem. Eng., 2018, 6, 3773–3782 CrossRef CAS
.
- S. Meltzer, R. Resch, B. E. Koel, M. E. Thompson, A. Madhukar, A. A. G. Requicha and P. Will, Langmuir, 2001, 17, 1713–1718 CrossRef CAS
.
- M. T. Reetz and M. G. Koch, J. Am. Chem. Soc., 1999, 121, 7933–7934 CrossRef CAS
.
- Y. K. Du, P. Yang, Z. G. Mou, N. P. Hua and L. Jiang, J. Appl. Polym. Sci., 2006, 99, 23–26 CrossRef CAS
.
- K. Okitsu, A. Yue, S. Tanabe and H. Matsumoto, Chem. Mater., 2000, 12, 3006–3011 CrossRef CAS
.
- T. Fujimoto, S.-y. Terauchi, H. Umehara, I. Kojima and W. Henderson, Chem. Mater., 2001, 13, 1057–1060 CrossRef CAS
.
- K. Saminathan, V. Kamavaram, V. Veedu and A. M. Kannan, Int. J. Hydrogen Energy, 2009, 34, 3838–3844 CrossRef CAS
.
- M. Shafiei, A. R. Riahi, F. G. Sen and A. T. Alpas, Surf. Coat. Technol., 2010, 205, 306–311 CrossRef CAS
.
- C. Soundarrajan, A. Sankari, P. Dhandapani, S. Maruthamuthu, S. Ravichandran, G. Sozhan and N. Palaniswamy, Bioprocess Biosyst. Eng., 2012, 35, 827–833 CrossRef CAS PubMed
.
- R. Bali, R. Siegele and A. T. Harris, J. Nanopart. Res., 2010, 12, 3087–3095 CrossRef CAS
.
- J. Y. Song, E.-Y. Kwon and B. S. Kim, Bioprocess Biosyst. Eng., 2010, 33, 159–164 CrossRef CAS PubMed
.
- R. W. Raut, A. S. M. Haroon, Y. S. Malghe, B. T. Nikam and S. B. Kashid, Adv. Mater. Lett., 2013, 4, 650–654 CrossRef CAS
.
- P. Mukherjee, A. Ahmad, D. Mandal, S. Senapati, S. R. Sainkar, M. I. Khan, R. Ramani, R. Parischa, P. Ajayakumar and M. Alam, Angew. Chem., Int. Ed., 2001, 40, 3585–3588 CrossRef CAS PubMed
.
- S. K. Das, T. Parandhaman, N. Pentela, A. Maidul Islam, A. B. Mandal and M. Mukherjee, J. Phys. Chem. C, 2014, 118, 24623–24632 CrossRef CAS
.
- T. Parandhaman, M. D. Dey and S. K. Das, Green Chem., 2019, 21, 5469–5500 RSC
.
- S. Gurunathan, K. Kalishwaralal, R. Vaidyanathan, D. Venkataraman, S. R. Pandian, J. Muniyandi, N. Hariharan and S. H. Eom, Colloids Surf., B, 2009, 74, 328–335 CrossRef CAS PubMed
.
- B. Balraj, N. Senthilkumar, C. Siva, R. Krithikadevi, A. Julie, I. V. Potheher and M. Arulmozhi, Res. Chem. Intermed., 2017, 43, 2367–2376 CrossRef CAS
.
- G. Riva, S. Baronchelli, L. Paoletta, V. Butta, I. Biunno, M. Lavitrano, L. Dalprà and A. Bentivegna, Toxicol. Rep., 2014, 1, 188–199 CrossRef CAS PubMed
.
- M. Imran, M. K. Iqubal, K. Imtiyaz, S. Saleem, S. Mittal, M. M. A. Rizvi, J. Ali and S. Baboota, Int. J. Pharm., 2020, 587, 119705 CrossRef CAS PubMed
.
- J. Liu, Y. He, D. Zhang, Y. Cai, C. Zhang, P. Zhang, H. Zhu, N. Xu and S. Liang, Mol. Med. Rep., 2017, 16, 2595–2603 CrossRef CAS PubMed
.
- I. Ahamad, N. Aziz, A. Zaki and T. Fatma, J. Appl. Phycol., 2021, 1–13 Search PubMed
.
- A. W. Bauer, W. M. Kirby, J. C. Sherris and M. Turck, Am. J. Clin. Pathol., 1966, 45, 493–496 CrossRef CAS PubMed
.
- V. Kandathil, R. B. Dateer, B. Sasidhar, S. A. Patil and S. A. Patil, Catal. Lett., 2018, 148, 1562–1578 CrossRef CAS
.
- M. M. Adina Bragaru, M. Simion, M. D. A. A. Andra Iordanescu, A. R. Ădoi and A. Dinescu, Rom. J. Inf. Sci. Technol., 2010, 13, 350–357 Search PubMed
.
- W. F. Silverstein, M. Robert and D. Kiemle, Spectrometric identification of organic compounds, JohnWiley&Sons, 7th edn, 2005 Search PubMed
.
- R. Venu, T. S. Ramulu, S. Anandakumar, V. S. Rani and C. G. Kim, Colloids Surf., A, 2011, 384, 733–738 CrossRef CAS
.
- C. M. Sayes, K. L. Reed and D. B. Warheit, Toxicol. Sci., 2007, 97, 163–180 CrossRef CAS PubMed
.
- J. C. Stockert, R. W. Horobin, L. L. Colombo and A. Blázquez-Castro, Acta Histochem., 2018, 120, 159–167 CrossRef CAS PubMed
.
- J. E. Jonkman, J. A. Cathcart, F. Xu, M. E. Bartolini, J. E. Amon, K. M. Stevens and P. Colarusso, Cell Adhes. Migr., 2014, 8, 440–451 CrossRef PubMed
.
- R. Nunez, Curr. Issues Mol. Biol., 2001, 3, 67–70 CAS
.
- P. Konieczny, A. G. Goralczyk, R. Szmyd, L. Skalniak, J. Koziel, F. L. Filon, M. Crosera, A. Cierniak, E. K. Zuba-Surma, J. Borowczyk, E. Laczna, J. Drukala, E. Pyza, D. Semik, O. Woznicka, A. Klein and J. Jura, Int. J. Nanomed., 2013, 8, 3963–3975 Search PubMed
.
- K. B. Holt and A. J. Bard, Biochemistry, 2005, 44, 13214–13223 CrossRef CAS PubMed
.
- K. Tahir, S. Nazir, A. Ahmad, B. Li, A. U. Khan, Z. U. H. Khan, F. U. Khan, Q. U. Khan, A. Khan and A. U. Rahman, J. Photochem. Photobiol., B, 2017, 166, 246–251 CrossRef CAS PubMed
.
- A. Anju, K. Gupta and T. S. Chundawat, Turk. J. Pharm. Sci., 2020, 17, 299 CrossRef PubMed
.
| This journal is © The Royal Society of Chemistry 2021 |