Open Access Article
Open Access ArticleTricarbonyl triazolato Re(I) compounds of pyridylbenzimidazole ligands: spectroscopic and antimicrobial activity evaluation†
Ahmed M. Mansour *
*
Department of Chemistry, Faculty of Science, Cairo University, Gamma Street, Giza, Cairo 12613, Egypt. E-mail: mansour@sci.cu.edu.eg; inorganic_am@yahoo.com
First published on 28th June 2021
Abstract
Catalyst-free [3+2] cycloaddition coupling between [Ren(N3)n(CO)3nL] (n = 1, L = 1-ethyl-2-(pyridin-2-yl)benzimidazole (L1) and n = 2, L = 1,1′-(hexane-1,6-diyl)bis[2-(pyridin-2-yl)-1H-benzimidazole] (L2)) and dimethyl acetylene dicarboxylate (DMAD) afforded mono- and binuclear triazolate complexes. Spectroscopic data presented unambiguous evidence for isomerization of the kinetically formed N(1) bound triazolate isomer into the N(2) analogue. The solvatochromism properties were assessed by UV/Vis spectroscopy with the aid of time dependent density functional theory calculations. The free ligands and their tricarbonyl triazolato Re(I) complexes were screened for their potential antimicrobial activity against different bacterial and fungal pathogens.
Tricarbonyl rhenium(I) complexes have received much interest because of their various photophysical and photochemical properties making them useful for diverse applications such as labelling of biomolecules,1 light emitting devices,2 and CO2 reduction.3 Based on the rich photophysical properties of the complexes of Re(I), they were investigated in the context of photoactivated therapy.4 Their mechanism of action against some malignant cells involved the generation of singlet oxygen similar to the conventional porphyrin based photodynamic therapy agents.5–7 Bioconjugation of the Re(CO)3 motif to a variety of biologically active molecules or drugs allows their application as luminescent tags for tracing such bioactive compounds.8
The chemoselective introduction of metal-based compounds into biomolecules is one of the main challenges in the synthesis of metal bioconjugates. A set of chemical reactions, known as biorthogonal reactions, that is orthogonal to most functional groups in the biological systems has so far shown favorable applications in the biological research. Of these reactions, azide–alkyne [3+2] coupling was extensively employed for labelling of biologically active molecules.9 Organic molecules functionalized with azide group reacted with alkynes affording a mixture of 1,4-, and 1,5-disubstituted 1,2,3-triazole derivatives.10 This reaction proceeded regioselectivity to 1,4-isomer by addition of Cu(I) ion,11 and 1,5-isomer by using Ru(II) compounds e.g., pentamethylcyclopentadienyl Ru(II) chloride.12 Metal azide was investigated in the context of click reaction via coupling with alkyne, cyanide, and thiocyanate derivatives to synthesize triazolate and tetrazolate based compounds.13,14 Massi and coworkers examined the photophysical and photochemical properties of [Re(triazolateCOOCH3,COOCH3)(CO)3(NHC)] complexes (where NHC = N-heterocyclic carbene) (Fig. 1), which formed via [3+2] cycloaddition coupling of their azide analogues with DMAD.15 Their crystal structures presented unambiguous evidence for the isomerization of the kinetically formed N(1) triazolate isomer to the N(2) analogue. Their photophysical properties revealed a dependence on both the heterocycle moiety and the auxiliary ligand (triazolate vs. azide). Azide complexes emitted at longer wavelength than the triazolate analogues. A red shift in the emission wavelength was observed on changing the pyrimidine ring with pyridine in the framework of the NHC ligand.15
 | ||
| Fig. 1 Previously reported cases for the inorganic click reactions of azido Re(CO)3I based complexes with electron poor alkynes. | ||
The role of the axial ligand (triazolate vs. Br−) on controlling the lysozyme binding affinity and the solvatochromism properties of terpyridine based Re(CO)3 complexes (Fig. 1) was reported.16 No protein adducts were formed when soaked with triazolate complex, while the bromo analogue reacted with that protein affording four adduct peaks with Re+, Re(CO)(OH2)+, Re(CO)3+, and Re(CO)3(OH2)2+ fragments. It has been concluded that the probability of exchange of Br− with DMSO is the control factor, which probably enables the binding to the model protein.16
The interesting photophysical and photochemical properties of complexes of Re(CO)3 featuring some heterocyclic compounds e.g. 1,10-phenanthroline, and 2,2′-bipyridine derivatives in diverse applications such as luminescent biological molecular probes, and light emitting devices17,18 encouraged me to synthesis new tricarbonyl Re(I) complexes with the formula of [Ren(triazolateCOOCH3,COOCH3)n(CO)3nL] (n = 1 and 2; L = benzimidazole ligands) (Scheme 1). The title complexes were prepared in a catalyst-free cycloaddition coupling of azido Re(I) analogues with DMAD. The effect of exchange of carbene C,N-ligands with N,N-ligands on the nature of the isolated triazolate bound isomer (N1 vs. N2) and the photophysical properties of the complexes were studied. The influence of the axial ligand (triazolate vs. bromide) on the electronic structure of the complexes and the antimicrobial activity were investigated. The electronic absorption transitions of the complexes were examined by executing time-dependent density functional theory (TDDFT) calculations.
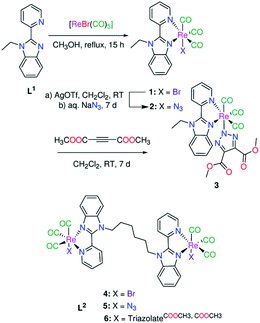 | ||
| Scheme 1 Synthesis of mono- and binuclear Re(I) triazolate complexes under catalyst-free conditions via [3+2] inorganic click reaction. | ||
Results and discussion
Synthesis of the complexes
The pyridylbenzimidazole derivatives, (1-ethyl-2-(pyridin-2-yl) benzimidazole (L1),19 and 1,1′-(hexane-1,6-diyl)bis[2-(pyridin-2-yl)-1H-benzimidazole] (L2)),20 were prepared in two-steps via the deprotonation of 2-(2′-pyridyl)benzimidazole, using DMF and sodium hydride, and reaction with the appropriate alkyl halide. Bromo Re(I)(CO)3 complexes (1 and 4) (Scheme 1) were synthesized in one step literature procedure via reaction of L1,2 with [ReBr(CO)5].21 The azide compounds [Ren(N3)n(CO)3nL] (n = 1, L = L1 and n = 2, L = L2) were prepared from the corresponding bromide complexes by abstraction of the bromo ligand(s) of 1 and 4 with silver triflate and reaction with a 2-fold excess of sodium azide. The IR spectra (Fig. S1†) of 2 and 5 display strong azide bands at 2055 and 2059 cm−1 respectively. The symmetrical and two anti-symmetrical stretching carbon monoxide bands of 2 are found at 2001, 1879, and 1858 cm−1. Azide 5 displays only two bands at 2009 and 1863 cm−1 due to the latter one appearing rather broad. The unique positively charged mass fragments due to [M–N3]+ (m/z 494.0496 (2) and 1054.1222 (5)) were detected in the mass spectra. The 13CO NMR signals are observed at δ = (197.9, 197.4, 191.8) (2) and (198.4, 197.8, 192.3) (5) (Fig. S2 and S3†). In the spectra of the free ligands, the 1H NMR signals of pyridine-H6 are observed at 8.76 (L1), and 8.62 ppm (L2), while those of benzimidazole-H4 are seen at 7.90 (L1) and 7.70 ppm (L2).19,20 Upon the azide formation, the 1H NMR resonances of pyridine-H6 (δ = 9.06 (2) and 9.16 ppm (5)) and benzimidazole-H4 (δ = 7.94 (2) and 8.00 ppm (5)) experience a major down field shift with respect to the free ligands.Complexes 2 and 5 were examined for their efficiency in the catalyst-free click cycloaddition reaction with DMAD. More recently, both NMR and X-crystallographic analysis offered conversant and realistic evidence that although the azide ligand is terminally coordinated to Re(I) ion, the N(2) triazolate bound isomer may be entirely the main isolated product from the azide–alkyne [3+2] cycloaddition reaction.15,16 A non-equivalent mixture of N(1) and N(2) triazolate isomers may be produced from the click reaction.15 A search in the literature showed that the type of the triazolate isomer, synthesized by the inorganic click cycloaddition reaction, is influenced by the bulkiness of the electron poor coupling agent, geometry of the metal-based compounds, synthetic and storing conditions.13,14 Herein, the cycloaddition reaction proceeded at the room temperature in CH2Cl2, from which the triazolate complexes 3 and 6 were easily isolated by precipitation with hexane. The disappearance of ν(N3) mode and appearance of ester ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) modes presented a simple way for following the progress of the click reaction with IR spectroscopy. The ester ν(C
O) modes presented a simple way for following the progress of the click reaction with IR spectroscopy. The ester ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) bands are observed at 1738 (3) and 1725 cm−1 (6) in the IR spectra (Fig. S4†). The CO stretches of 2 and 5 moved to higher wavenumber values upon the formation of 3 and 6. Two triazolate isomers (N(1) and N(2)) are expected to be formed from the reaction of azide compounds 2 and 5 with DMAD. The triazolate coordination mode of 3 and 6 was carefully investigated by NMR analysis (Fig. S5 and S6†). The N(2) coordination mode in 3 is clearly obvious from the presence of only one major ester methyl group signal at δ = 3.60 ppm (6H) and a single methyl ester 13C signal at 52.0 ppm. In the case of N(1) bound triazolate isomer, it is expected that the methyl groups are seen as two separated singlet signals. The 1H NMR spectrum of 6 shows two singlet methyl signals of equal integrations at δ = 3.53 and 3.52 ppm. By inspection of the 13C NMR spectrum of 6 (Fig. S6†), it seems that each signal is a doublet or an overlapped of two resonances (the 13C NMR signals of 6 were given as a median). The NMR spectra of 6 show mainly signals for N2-bound isomer containing two non-isoenergetic Re(CO)3 moieties in agreement with the data of other complexes of the same ligand.19,20,22 Besides, the very small signals in the ester methyl range and in the aromatic region may be attributed to presence of traces of N(1) isomer formed as a side product in the cycloaddition coupling of Re(I) azide with DMAD. These triazolate isomers could not be separated.15
O) bands are observed at 1738 (3) and 1725 cm−1 (6) in the IR spectra (Fig. S4†). The CO stretches of 2 and 5 moved to higher wavenumber values upon the formation of 3 and 6. Two triazolate isomers (N(1) and N(2)) are expected to be formed from the reaction of azide compounds 2 and 5 with DMAD. The triazolate coordination mode of 3 and 6 was carefully investigated by NMR analysis (Fig. S5 and S6†). The N(2) coordination mode in 3 is clearly obvious from the presence of only one major ester methyl group signal at δ = 3.60 ppm (6H) and a single methyl ester 13C signal at 52.0 ppm. In the case of N(1) bound triazolate isomer, it is expected that the methyl groups are seen as two separated singlet signals. The 1H NMR spectrum of 6 shows two singlet methyl signals of equal integrations at δ = 3.53 and 3.52 ppm. By inspection of the 13C NMR spectrum of 6 (Fig. S6†), it seems that each signal is a doublet or an overlapped of two resonances (the 13C NMR signals of 6 were given as a median). The NMR spectra of 6 show mainly signals for N2-bound isomer containing two non-isoenergetic Re(CO)3 moieties in agreement with the data of other complexes of the same ligand.19,20,22 Besides, the very small signals in the ester methyl range and in the aromatic region may be attributed to presence of traces of N(1) isomer formed as a side product in the cycloaddition coupling of Re(I) azide with DMAD. These triazolate isomers could not be separated.15
Electronic structure
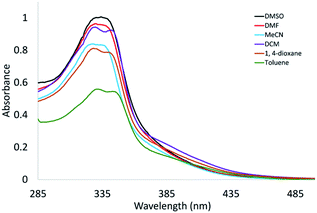
The premise for the modification of the proximity of Re(I) centres in the title complexes was to not affect the electronic structures of the individual Re(CO)3 centres. A search in the literature revealed that presence of a saturated linker in the binuclear complexes will inhibit the substantial communication between the two metal centres.23 Comparing the electronic absorption spectra of triazolate complexes 3 and 6 (in DMSO) showed that λmax is the same at about 336 nm (Fig. S7†). However, the molar absorptivity value of the binuclear complex is higher than that of the mononuclear compound. The λmax of the complexes did not alter on changing the Br− with triazolate group (Fig. S7†). In fact, the biological activities such as permeability, and the energetics of protein insertion are regulated, in part, by variations in the polarity across the phospholipid membrane. Environment polarity can also cause changes in absorption or emission maxima, for a given compound. This is known as solvatochromism.24 Other factors such as H-bond interaction may be contributed to the net effect of the solvent on a given species. The solvatochromic properties of 4 and 6 were examined by UV/Vis using diverse solvents of different polarity and coordinating ability (DMSO, DMF, CH3CN, 1,4-dioxane, CH2Cl2, and toluene). Unfortunately, compound 4 is insoluble in the highly nonpolar tested solvents (toluene and 1,4-dioxane) as well as has poor solubility in CH3CN and CH2Cl2. The influence of the polarity of the solvent on the position of the lowest energy band is not systematic although there is a slight red shift on changing the highly polar solvents (e.g., DMSO and DMF) with CH2Cl2. The exception is the UV/Vis spectrum of 4 in acetonitrile revealing to the probability of exchange of Br− with that solvent.25,26 As shown in Fig. 2, the red shift of the lowest energy transition of 6 on changing the high polar solvents such as DMSO and DMF (336 nm) with the less polar solvents such as 1,4-dioxane (341 nm), and toluene (345 nm) may be attributed to negative solvatochromism,25 which is well recognized for metal carbonyls attributable to reduction of the excited-state electric dipole.27 Based on the obtained data, exchange of Br− with triazolate moiety has no effect on determining the position of the main electronic absorption band of this class of Re(I) complexes.DFT/TDDFT calculations
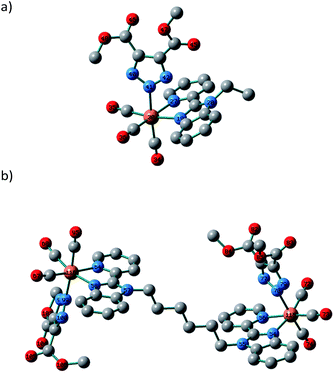
The nature of the observed transitions was examined by attaining first the local minimum structures of the complexes, in the ground-state using Becke 3-parameter (exchange) Lee–Yang–Parr (B3LYP)28 and the effective core potential (ECP) of the Hady and Wadt, LANL2DZ basis set.29,30 The resulting geometries were described as local minima via the harmonic frequency analysis, presenting the absence of the imaginary vibrations. The octahedron sphere around the Re(I) ion of 1 is composed of three CO ligands (1.908–1.925 Å), bidentate pyridylbenzimidazole L1 ligand (Re–N19 = 2.153 and Re–N27 = 2.188 Å) and Br− (2.716 Å).21 In the case of 3 (Fig. 3), the octahedral geometry is formed from three CO ligands [Re–C34 = 1.927, Re–C34 = 1.926, and Re–C34 = 1.922 Å], N,N-bidentate ligand [Re–N19 = 2.162, and Re–N27 = 2.197 Å] and triazolate ligand [Re–N41 = 2.179 Å]. The triazolate ring is positioned approximately perpendicular to the central axis of the pyridyl-benzimidazole system and intersects the C35–Re–C36 angle with a deviation of 3°. It looks like that the bond lengths of the coordination sphere of 1 did not alter significantly on changing Br− with triazolate group except for the bond trans to the axial monodentate ligand. The comparison between the calculated bond lengths of 3 and the only two previously reported crystal structures of a triazolate coordinated to a fac-Re(CO)3 unit, [Re(triazolateCOOCH2CH3,CF3)(CO)3(terpy–k2N1,N2)] (ref. 16) and [Re(triazolateCOOCH3,COOCH3)(CO)3(NHC)] (ref. 15) reveals that the Re–triazolate bond of 3 and the reported values are identical reflecting the suitability of the applied DFT method for such size of the complexes. Similar, the axial C34–Re–N41 angle (175.5°) approaches the linearity in agreement with the values of the previously published structures.15,16 Like 1 and 3, optimization of 6 was done at the same level of theory with RMS Gradient Norm of 0.0056. The root means square difference of the bond lengths between the two coordination spheres of 6 is about 0.01 Å.In the singlet state, the lowest 30 spin-allowed excitation states of 3 and 6 were calculated at CAM-B3LYP (ref. 31)/LANL2DZ level of theory using PCM keyword to introduce the solvent effect. The calculations were done using TDDFT method. The absorption spectra were simulated using GAUSSSUM. Each excited state was included by a Gaussian convolution with the full-width at half-maximum (FWHM) of 3000. The results of the triazolate compounds (3 and 6) were compared with those of the published bromo complexes.21 As reported, the calculated spectra of 1 and 4 show one broad band at around 343 nm allocated for HOMO–1 → LUMO [d(Re)/π(pyridine)/π(Br) → d(Re)/π(CO)/π(Br)].21 The calculated spectrum (Fig. 4) of 3, in the singlet state, displays two main bands at 222 and 294 nm, a shoulder at 251 nm as well as a weak transition at 343 nm allocated for HOMO–5 → LUMO, HOMO–2 → LUMO, HOMO → LUMO+2, and HOMO → LUMO, respectively.
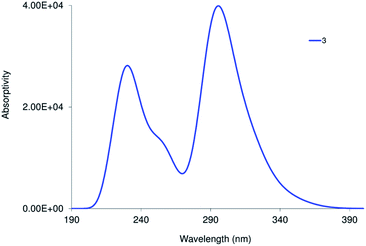 | ||
| Fig. 4 Calculated electronic spectra of 3 at TD/PCM(DMSO)/CAM-B3LYP/LANL2DZ level of theory in the singlet state. | ||
As shown in Fig. 5, the transition at 343 nm has a ground state composed of d(Re)/π(CO) and excited of π*(CO)/π*(L1) forming MLCT. Both 1 and 3 have a transition at 343 nm ruling out the contributing of the orbitals of the axial ligand in determining the wavelength of the lowest energy transition in agreement with the experimental findings. For the lowest energy transition, a discrepancy of 7 nm is observed between the theoretical and experimental finding. Triazolate group contributes only to the highest energy transition at 222 nm.
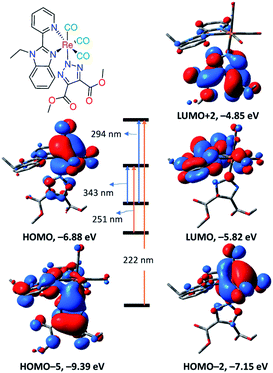 | ||
| Fig. 5 Selected frontier molecular orbitals of 3 obtained at B3LYP/LANL2DZ and main calculated electronic transitions obtained at CAM-B3LYP/LANL2DZ. | ||
The calculated electronic spectrum of 6 (Fig. S8†) is characterized by three main transitions at 255, 295 and 366 nm corresponding to HOMO–2 → LUMO+2, HOMO–5/HOMO–6→LUMO+1 and HOMO → LUMO+1, respectively. The description of Frontier molecular orbitals and the relocation of the electron density of 6 are given in Table S1.† HOMO is composed of d(Re)/π(triazolate)/π(CO) character, and LUMO+1 is of π*(L1) forming LLCT/MLCT.
Antimicrobial activity
The antimicrobial activity of the triazolate compounds (3 and 6) was assessed against two pathogenic fungi (Candida albicans ATCC 90028 and Cryptococcus neoformans var. grubii H99; ATCC 208821), Gram-positive bacterium (Staphylococcus aureus ATCC 43300) and four Gram-negative bacterial strains (Escherichia coli ATCC 25922, Pseudomonas aeruginosa ATCC 27853, Klebsiella pneumoniae ATCC 700603 and Acinetobacter baumannii ATCC 19606) at 32 μg mL−1. The results were compared with those of the free ligands (L1,2), bromo complexes (1 and 4)21 as well as three reference drugs (Fluconazole, Colistin, and Vancomycin). As shown in Fig. 6, the tested ligands and their Re(I) compounds exhibited partially or no inhibitory activity against the tested microbes. Coordination of L1,2 to ReBr(CO)3 reduced the antibacterial activity against Staphylococcus aureus, which further decreased on changing Br− with triazolate moiety. Escherichia coli was the most resistant bacterium to the tested compounds. The difference in the activity may be attributed to the cell wall structure of the bacteria.32 Alternatively, the free ligands and their complexes were inactive against Cryptococcus neoformans. In the case of Candida albicans, coordination of L1,2 to ReBr(CO)3 improved the antifungal activity, which reduced upon the introduction of the triazolate moiety in the complex. In general, the title compounds exhibited weak or no inhibitory activity against the tested microbes compared to the reference drugs.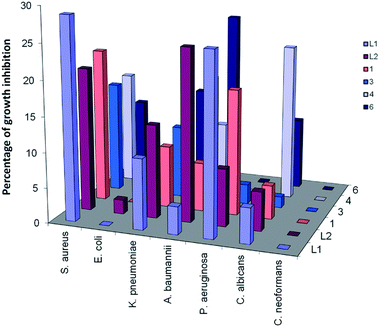 | ||
| Fig. 6 Comparison between the antimicrobial activities of free L1,2 and their Re(I) complexes against some representative microbes. All the experiments were carried out at 32 μg mL−1 in triplicate and the mean results are given using the negative control (media only) and positive control (bacteria without inhibitors) on the same plate as references. | ||
Conclusions
The antimicrobial activity of [Ren(triazolateCOOCH3,COOCH3)n(CO)3nL] (n = 1, L = 1-ethyl-2-(pyridin-2-yl)benzimidazole (L1) and n = 2, L = 1,1′-(hexane-1,6-diyl)bis[2-(pyridin-2-yl)-1H-benzimidazole] (L2)), formed by catalyst free [3+2] cycloaddition coupling of the azide analogues with dimethyl acetylene dicarboxylate, was evaluated using five Gram-positive and Gram-negative bacterial strains as well as two pathogenic fungi, Candida albicans and Cryptococcus neoformans. NMR data confirmed that while the azido ligand is terminally coordinated to fac-Re(CO)3 unit, the kinetically formed N(1) triazolate bound isomer favoured mainly the isomerization to N(2) analogue. The antibacterial activity of L1,2 against Staphylococcus aureus diminished upon the complex formation and reached the minimum values on changing the axial bromo ligand(s) with the triazolate moiety. On the contrast, the antifungal activity of the ligands against Candida albicans improved upon the formation of bromo Re(I) complexes, and then reduced by introduction of the triazolate moiety. With the aid of time dependent density functional theory (TDDFT) calculations, the solvatochromism properties of the bromo and triazolate complexes were investigated. The red shift of the longest wavelength band in the absorption spectrum of the triazolate compound on changing the polar solvents with the less polar solvents may be accounted for the negative solvatochromism because of the reduction of the excited-state electric dipole. The effect of the solvent on the absorption spectrum of the bromo complex was not systematic revealing to the probability of exchange of Br− with the coordinating solvents such acetonitrile and DMSO. Comparison of the maxima of the lower-energy bands in the complexes indicated that exchange of Br− with triazolato ligand has no effect in determining the wavelength of the lowest energy transition as theoretically found. Besides, the λmax of the absorption spectra of the mono- and binuclear complexes are typical. An investigation of the scope of the catalyst free cycloaddition reactions using azido metal based compounds of Mn(I), Re(I), Ru(II), Rh(III), Ir(III), etc. with electron deficient systems is currently underway in our research group.Experimental section
Materials and instruments
The ligands 1-ethyl-2-(pyridin-2-yl)benzimidazole (L1) and 1,1′-(hexane-1,6-diyl)bis[2-(pyridin-2-yl)1H-benzimidazole] (L2) were prepared as reported in the literature.19,20 Bromo Re(I) complexes (1 and 4) were prepared by following the published procedures.21 Bromo pentacarbonyl rhenium(I), DMAD, sodium azide and solvents were obtained from the commercial sources and used as received. Electronic absorption spectra of the azide and triazolate complexes were recorded on Specord 200 Plus spectrophotometer. Infrared spectra were registered on Nicolet 380 FT-IR spectrometer. CHN analyses of the synthesized compounds were carried out by Elementar Vario MICRO cube CHN analyzer or an EA 3000 elemental analyser from HEKtech. The reported error (>0.4%) in the elemental analysis of the synthesized azide and triazolate compounds is expected.33,34 The source of water of hydration that is assumed to be in the molecular formula of 2 and 3, is the aqueous solution of NaN3 though the complexes kept under vacuum for at least one week. The {1H, and 13C} NMR analyses were done using Bruker-Avance 500 (1H, 500.13 MHz; and 13C{1H}, 125.77 MHz) spectrometer. Electrospray ionization mass spectra (ESI MS) were run on ThermoFisher Exactive Plus instrument with an Orbitrap mass analyzer at a resolution of R = 70.000 and a solvent flow rate of 50 μL min.Synthetic procedures
Caution. Azide compounds are subjected to unexpected violent decomposition and thus handling and purification with great care is so critical.
To a round-bottom flask charged with bromide Re(I) complexes (1, 114 mg and 4, 234 mg) and CH2Cl2 (50 mL), Ag(SO3CF3) (0.5 mmol; 128 mg) was added. The reaction mixtures were stirred at the room temperature for two days. Silver bromide was filtered off through Celite. Then aqueous solution (0.5 mL) of NaN3 (1, 40 mg and 5, 80 mg) was added to the reaction mixture. Stirring was continuing for 3 days. Small quantity of AgN3 was cautiously filtered off and then the solvent was removed under vacuum. The resulting orange precipitate was washed with water (3 × 5 mL), hexane (3 × 5 mL) and dried under vacuum.
2. Yield: 81% (87 mg, 0.16 mmol). IR (ATR, diamond): ν = 2927 (w, CH), 2055 (s, N3), 2001 (vs., C
![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) O), 1879 (vs., C
O), 1879 (vs., C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) O), 1858 (vs., C
O), 1858 (vs., C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) O), 1605, 1487, 1439, 1336, 1266, 1135, 746. 1H NMR (DMSO-d6, 500.13 MHz): δ = 9.06 (d, 3JH,H = 5.5 Hz, 1H, py-H6), 8.49 (d, 3JH,H = 8.0 Hz, 1H, py-H3), 8.32 (td, 3JH,H = 7.4 Hz, 4JH,H = 1.5 Hz, 1H, py-H4), 7.94 (m, 1H, bim-H4), 7.76 (m, 2H, bim-H7/py-H5), 7.53 (m, 2H, bim-H5/H6), 4.81 (q, 3JH,H = 7.3 Hz, 2H, CH2), 1.39 (t, 3JH,H = 7.8 Hz, 3H, CH3) ppm. 13CNMR (DMSO-d6, 125.75 MHz): δ = 197.9 (C
O), 1605, 1487, 1439, 1336, 1266, 1135, 746. 1H NMR (DMSO-d6, 500.13 MHz): δ = 9.06 (d, 3JH,H = 5.5 Hz, 1H, py-H6), 8.49 (d, 3JH,H = 8.0 Hz, 1H, py-H3), 8.32 (td, 3JH,H = 7.4 Hz, 4JH,H = 1.5 Hz, 1H, py-H4), 7.94 (m, 1H, bim-H4), 7.76 (m, 2H, bim-H7/py-H5), 7.53 (m, 2H, bim-H5/H6), 4.81 (q, 3JH,H = 7.3 Hz, 2H, CH2), 1.39 (t, 3JH,H = 7.8 Hz, 3H, CH3) ppm. 13CNMR (DMSO-d6, 125.75 MHz): δ = 197.9 (C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) O), 197.4 (C
O), 197.4 (C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) O), 191.8 (C
O), 191.8 (C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) O), 154.6 (py-C6), 151.9 (py-C2), 146.0 (bim-C2), 141.18 (py-C4), 139.4 (bim-C3a), 135.1 (bim-C7a), 128.2 (py-C5), 126.2 (bim-C5), 125.8 (bim-C6), 125.1 (py-C3), 117.8 (bim-C7), 112.4 (bim-C4), 40.8 (CH2), 14.4 (CH3) ppm. ESI-MS (positive mode, methanol): m/z = 494.0496 [M–N3]+. C17H13N6O3Re·2H2O: C 35.72, H 3.00, N 14.70, found C 35.34, H 3.04, N 12.36.
O), 154.6 (py-C6), 151.9 (py-C2), 146.0 (bim-C2), 141.18 (py-C4), 139.4 (bim-C3a), 135.1 (bim-C7a), 128.2 (py-C5), 126.2 (bim-C5), 125.8 (bim-C6), 125.1 (py-C3), 117.8 (bim-C7), 112.4 (bim-C4), 40.8 (CH2), 14.4 (CH3) ppm. ESI-MS (positive mode, methanol): m/z = 494.0496 [M–N3]+. C17H13N6O3Re·2H2O: C 35.72, H 3.00, N 14.70, found C 35.34, H 3.04, N 12.36.
5. Yield: 74% (162 mg, 0.30 mmol). IR (ATR, diamond): ν = 2931 (w, CH), 2059 (s, N3), 2009 (vs., C
![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) O), 1863 (vs., C
O), 1863 (vs., C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) O), 1604, 1439, 1259, 1168, 1030, 746. 1H NMR (DMSO-d6, 500.13 MHz): δ = 9.18 (d, 3JH,H = 5.4 Hz, 4JH,H = 1.2 Hz, 1H, py-H6), 8.53 (d, 3JH,H = 8.0 Hz, 1H, py-H3), 8.40 (td, 3JH,H = 7.8 Hz, 4JH,H = 1.6 Hz, 1H, py-H4), 8.00 (m, 1H, bim-H4), 7.86 (m, 2H, bim-H7/py-H5), 7.63 (m, 2H, bim-H5/H6), 4.84 (m, 2H, NCH2), 1.84 (m, 2H, NCH2CH2), 1.43 (m, 2H, NCH2CH2CH2) ppm. 13C NMR (DMSO-d6, 125.75 MHz): δ = 198.4 (C
O), 1604, 1439, 1259, 1168, 1030, 746. 1H NMR (DMSO-d6, 500.13 MHz): δ = 9.18 (d, 3JH,H = 5.4 Hz, 4JH,H = 1.2 Hz, 1H, py-H6), 8.53 (d, 3JH,H = 8.0 Hz, 1H, py-H3), 8.40 (td, 3JH,H = 7.8 Hz, 4JH,H = 1.6 Hz, 1H, py-H4), 8.00 (m, 1H, bim-H4), 7.86 (m, 2H, bim-H7/py-H5), 7.63 (m, 2H, bim-H5/H6), 4.84 (m, 2H, NCH2), 1.84 (m, 2H, NCH2CH2), 1.43 (m, 2H, NCH2CH2CH2) ppm. 13C NMR (DMSO-d6, 125.75 MHz): δ = 198.4 (C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) O), 197.8 (C
O), 197.8 (C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) O), 192.3 (C
O), 192.3 (C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) O), 155.2 (py-C6), 152.6 (py-C1), 146.6 (bim-C2), 141.0 (py-C4), 139.7 (bim-C3a), 136.2 (bim-C7a), 128.7 (py-C5), 126.7 (bim-C5), 126.3 (bim-C6), 125.6 (py-C3), 118.3 (bim-C7), 113.1 (bim-C4), 45.7 (NCH2), 29.3 (NCH2CH2), 25.9 (NCH2CH2CH2) ppm. ESI-MS (positive mode, acetone): m/z = 1054.1222 [M–N3]+. C36H28N12O6Re2: C 39.41, H 2.57, N 15.32, found C 39.64, H 3.11, N 14.99.
O), 155.2 (py-C6), 152.6 (py-C1), 146.6 (bim-C2), 141.0 (py-C4), 139.7 (bim-C3a), 136.2 (bim-C7a), 128.7 (py-C5), 126.7 (bim-C5), 126.3 (bim-C6), 125.6 (py-C3), 118.3 (bim-C7), 113.1 (bim-C4), 45.7 (NCH2), 29.3 (NCH2CH2), 25.9 (NCH2CH2CH2) ppm. ESI-MS (positive mode, acetone): m/z = 1054.1222 [M–N3]+. C36H28N12O6Re2: C 39.41, H 2.57, N 15.32, found C 39.64, H 3.11, N 14.99.
3. Yield: 58% (39 mg, 0.07 mmol). IR (ATR, diamond): ν = 2956 (w, CH), 2021 (vs., C
![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) O), 1899 (vs., C
O), 1899 (vs., C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) O), 1738 (s, C
O), 1738 (s, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1442, 1202, 1088, 756. 1H NMR (DMSO-d6, 500.13 MHz): δ = 9.19 (d, 3JH,H = 5.0 Hz, 1H, py-H6), 8.62 (d, 3JH,H = 8.8 Hz, 1H, py-H3), 8.41 (t, 3JH,H = 8.4 Hz, 1H, py-H4), 8.03 (m, 1H, bim-H4), 7.85 (m, 1H, bim-H7), 7.78 (m, 1H, py-H5), 7.59 (m, 2H, bim-H5/H6), 4.92 (m, 2H, CH2), 3.60 (s, 6H, CH3COOMe), 1.50 (t, 3JH,H = 7.0 Hz, 3H, CH3) ppm. 13C-NMR (DMSO-d6, 125.75 MHz): δ = 198.1 (C
O), 1442, 1202, 1088, 756. 1H NMR (DMSO-d6, 500.13 MHz): δ = 9.19 (d, 3JH,H = 5.0 Hz, 1H, py-H6), 8.62 (d, 3JH,H = 8.8 Hz, 1H, py-H3), 8.41 (t, 3JH,H = 8.4 Hz, 1H, py-H4), 8.03 (m, 1H, bim-H4), 7.85 (m, 1H, bim-H7), 7.78 (m, 1H, py-H5), 7.59 (m, 2H, bim-H5/H6), 4.92 (m, 2H, CH2), 3.60 (s, 6H, CH3COOMe), 1.50 (t, 3JH,H = 7.0 Hz, 3H, CH3) ppm. 13C-NMR (DMSO-d6, 125.75 MHz): δ = 198.1 (C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) O), 197.7 (C
O), 197.7 (C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) O), 193.9 (C
O), 193.9 (C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) O), 162.3 (C
O), 162.3 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 155.4 (py-C6), 153.8 (py-C2), 147.7 (bim-C2), 141.7 (py-C4), 140.1 (OOC–C=C–COO), 139.1 (bim-C3a), 135.6 (bim-C7a), 128.4 (py-C5), 126.4 (bim-C5), 126.1 (bim-C6), 125.3 (py-C3), 118.3 (bim-C7), 112.8 (bim-C4), 52.0 (OCH3), 41.1 (CH2), 15.0 (CH3) ppm. ESI-MS (positive mode, acetone): m/z = 679.0923 [M + H]+. C26H19N6O5Re·3H2O: C 42.45, H 3.43, N 11.42, found C 42.13, H 3.83, N 10.95.
O), 155.4 (py-C6), 153.8 (py-C2), 147.7 (bim-C2), 141.7 (py-C4), 140.1 (OOC–C=C–COO), 139.1 (bim-C3a), 135.6 (bim-C7a), 128.4 (py-C5), 126.4 (bim-C5), 126.1 (bim-C6), 125.3 (py-C3), 118.3 (bim-C7), 112.8 (bim-C4), 52.0 (OCH3), 41.1 (CH2), 15.0 (CH3) ppm. ESI-MS (positive mode, acetone): m/z = 679.0923 [M + H]+. C26H19N6O5Re·3H2O: C 42.45, H 3.43, N 11.42, found C 42.13, H 3.83, N 10.95.
6. Yield: 64% (81 mg, 0.15 mmol). IR (ATR, diamond): ν = 2949 (w, CH), 2019 (vs., C
![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) O), 1881 (vs., C
O), 1881 (vs., C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) O), 1725 (s, C
O), 1725 (s, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1440, 1226, 1169, 1088, 744. 1H NMR (DMSO-d6, 500.13 MHz): δ = 9.19 (m, 1H, py-H6), 8.52 (m, 1H, py-H3), 8.35 (m, 1H, py-H4), 7.97 (m, 1H, bim-H4), 7.84 (m, 1H, bim-H7), 7.77 (m, 1H, py-H5), 7.56 (m, 2H, bim-H5/H6), 4.83 (m, 2H, NCH2), 3.53 (s, 3H, CH3), 3.52 (s, 3H, CH3), 1.85 (m, 2H, NCH2CH2), 1.49 (m, 2H, NCH2CH2 CH2) ppm. 13C-NMR (DMSO-d6, 125.75 MHz): δ = 198.2 (C
O), 1440, 1226, 1169, 1088, 744. 1H NMR (DMSO-d6, 500.13 MHz): δ = 9.19 (m, 1H, py-H6), 8.52 (m, 1H, py-H3), 8.35 (m, 1H, py-H4), 7.97 (m, 1H, bim-H4), 7.84 (m, 1H, bim-H7), 7.77 (m, 1H, py-H5), 7.56 (m, 2H, bim-H5/H6), 4.83 (m, 2H, NCH2), 3.53 (s, 3H, CH3), 3.52 (s, 3H, CH3), 1.85 (m, 2H, NCH2CH2), 1.49 (m, 2H, NCH2CH2 CH2) ppm. 13C-NMR (DMSO-d6, 125.75 MHz): δ = 198.2 (C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) O), 197.6 (C
O), 197.6 (C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) O), 193.9 (C
O), 193.9 (C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) O), 162.4 (C
O), 162.4 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 155.7 (py-C6), 153.8 (py-C1), 147.6 (bim-C2), 141.6 (py-C4), 139.9 (bim-C3a), 139.1 (OOC–C=C–COO), 136.1 (bim-C7a), 128.5 (py-C5), 126.5 (bim-C5), 126.2 (bim-C6), 125.2 (py-C3), 118.5 (bim-C7), 113.0 (bim-C4), 52.0 (CH3), 52.0 (CH3), 45.6 (NCH2), 29.5 (NCH2CH2), 26.0 (NCH2CH2CH2) ppm. ESI-MS (positive mode, acetone): m/z = 1196.1604 [M–triazolate]+, 1139.1498 [M–2CO–triazolate]+, 1029.1272 [M–6CO–triazolate]+, 928.2286 [M–3CO–Re–triazolate]+. C48H40N12O14Re2: C 41.74, H 2.92, N 12.17, found C 41.71, H 2.86, N 10.26.
O), 155.7 (py-C6), 153.8 (py-C1), 147.6 (bim-C2), 141.6 (py-C4), 139.9 (bim-C3a), 139.1 (OOC–C=C–COO), 136.1 (bim-C7a), 128.5 (py-C5), 126.5 (bim-C5), 126.2 (bim-C6), 125.2 (py-C3), 118.5 (bim-C7), 113.0 (bim-C4), 52.0 (CH3), 52.0 (CH3), 45.6 (NCH2), 29.5 (NCH2CH2), 26.0 (NCH2CH2CH2) ppm. ESI-MS (positive mode, acetone): m/z = 1196.1604 [M–triazolate]+, 1139.1498 [M–2CO–triazolate]+, 1029.1272 [M–6CO–triazolate]+, 928.2286 [M–3CO–Re–triazolate]+. C48H40N12O14Re2: C 41.74, H 2.92, N 12.17, found C 41.71, H 2.86, N 10.26.
Density functional theory calculations
Ground-state geometry optimization and harmonic frequency analysis of the complexes were executed using B3LYP (ref. 28) functional and the effective core potential (ECP) of the Hady and Wadt, LANL2DZ basis set.29,30 In the singlet state, TDDFT calculations were performed at CAM-B3LYP (ref. 31)/LANL2DZ level of theory and the PCM keyword to introduce the effect of the solvent. All the calculations were carried out using Gaussian03.35 Visualization of Frontier molecular orbitals and local minimum structures were done using Gaussview.36Biological activity testing
Antimicrobial screening of the synthesized compounds was performed by CO-ADD (The Community for Antimicrobial Drug Discovery), funded by the Wellcome Trust (UK) and The University of Queensland (Australia). The antimicrobial activity was examined against some representative fungi and bacteria as given within the main text at 32 μg mL−1 according to standard broth microdilution assays.37 For toxic compound, a follow-up hit confirmation is triggered, where the toxicity is established by means of a dose–response assay against the same microbe strain. No animals were used in this study. Cell lines (bacteria, fungi, mammalian) were sourced from the American Type Culture Collection (ATCC). Human blood was sourced from the Australian Red Cross Blood Service with informed consent and its use in haemolysis assays was approved by The University of Queensland Institutional Human Research Ethics Committee, Approval Number 2014000031.Conflicts of interest
There are no conflicts to declare.Acknowledgements
A. Mansour thanks the Alexander von Humboldt Foundation for Georg Forster postdoctoral fellowship, and Cairo University for funding of this work. A. Mansour would like to gratefully acknowledge the Professors at Julius-Maximilians-Universität Würzburg, Germany, that own the specific instrumentation, to facilitate my study. Antimicrobial screening was performed by CO-ADD (The Community for Antimicrobial Drug Discovery), funded by the Wellcome Trust (UK) and The University of Queensland (Australia).References
- K. K.-W. Lo, W.-K. Hui, C.-K. Chung, K. H.-K. Tsang, D. C.-M. Ng, N. Zhu and K.-K. Cheung, Biological labelling reagents and probes derived from luminescent transition metal polypyridine complexes, Coord. Chem. Rev., 2005, 249, 1434 CrossRef CAS.
- T. Yu, D. P. K. Tsang, V. K. M. Au, W. H. Lam, M. Y. Chan and V. W. W. Yam, Deep Red to Near-Infrared Emitting Rhenium (I) Complexes: Synthesis, Characterization, Electrochemistry, Photophysics, and Electroluminescence Studies, Chem.–Eur. J., 2013, 19, 13418 CrossRef CAS.
- C. Sun, S. Prosperini, P. Quagliotto, G. Viscardi, S. S. Yoon, R. Gobetto and C. Nervi, Electrocatalytic reduction of CO 2 by thiophene-substituted rhenium (i) complexes and by their polymerized films, Dalton Trans., 2016, 45, 14678 RSC.
- A. Kumar, S.-S. Sun and A. J. Lees, Photophysics and Photochemistry of Organometallic Rhenium Diimine Complexes, in Photophysics of Organometallics, ed. A. J. Lees, Springer Berlin Heidelberg, Berlin, Heidelberg, 2010, pp. 1–36 Search PubMed.
- K. Wähler, A. Ludewig, P. Szabo, K. Harms and E. Meggers, Rhenium Complexes with Red-Light-Induced Anticancer Activity, Eur. J. Inorg. Chem., 2014, 2014, 807 CrossRef PubMed.
- A. Leonidova, V. Pierroz, R. Rubbiani, J. Heier, S. Ferrari and G. Gasser, Towards Cancer Cell-Specific Phototoxic Organometallic Rhenium(I) Complexes, Dalton Trans., 2014, 43, 4287 RSC.
- A. Leonidova and G. Gasser, Underestimated Potential of Organometallic Rhenium Complexes as Anticancer Agents, ACS Chem. Biol., 2014, 9, 2180 CrossRef CAS PubMed.
- L. Raszeja, A. Maghnouj, S. Hahn and N. Metzler-Nolte, A Novel Organometallic ReI Complex with Favourable Properties for Bioimaging and Applicability in Solid-Phase Peptide Synthesis, ChemBioChem, 2011, 12, 371 CrossRef CAS PubMed.
- P. Thirumurugan, D. Matosiuk and K. Jozwiak, Click chemistry for drug development and diverse chemical biology applications, Chem. Rev., 2013, 113, 4905 CrossRef CAS.
- R. Huisgen, G. Szeimies and L. Mobius, 1.3-Dipolare Cycloadditionen, XXXII. Kinetik der Additionen organischer Azide an CC-Mehrfachbindungen, Chem. Ber., 1967, 100, 2494 CrossRef CAS.
- C. W. Tornoe, C. Christensen and M. Meldal, Peptidotriazoles on solid phase:[1, 2, 3]-triazoles by regiospecific copper (I)-catalyzed 1, 3-dipolar cycloadditions of terminal alkynes to azides, J. Org. Chem., 2002, 67, 3057 CrossRef CAS PubMed.
- L. K. Rasmussen, B. C. Boren and V. V. Fokin, Ruthenium-catalyzed cycloaddition of aryl azides and alkynes, Org. Lett., 2007, 9, 5337 CrossRef CAS PubMed.
- H. W. Frühauf, Organotransition metal [3 + 2] cycloaddition reactions, Coord. Chem. Rev., 2002, 230, 79 CrossRef.
- W. P. Fehlhammer and W. Beck, Azide chemistry-An inorganic perspective, Part II. [3 + 2]-Cycloaddition reactions of metal azides and related systems, Z. Anorg. Allg. Chem., 2015, 641(10), 1599 CrossRef CAS.
- P. V. Simpson, B. W. Skelton, P. Raiteri and M. Massi, Photophysical and photochemical studies of tricarbonyl rhenium (i) N-heterocyclic carbene complexes containing azide and triazolate ligands, New J. Chem., 2016, 40, 5797 RSC.
- A. M. Mansour, K. Radacki and O. R. Shehab, Role of the ancillary ligand in controlling the lysozyme binding affinity and electronic properties of terpyridine fac-Re(CO)3 complexes, Dalton Trans., 2021, 50, 1197 RSC.
- K. K.-W. Lo, M.-W. Louie, K.-S. Sze and J. S.-Y. Lau, Rhenium(I) Polypyridine Biotin Isothiocyanate Complexes as the First Luminescent Biotinylation Reagents:
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Synthesis, Photophysical Properties, Biological Labelling, Cytotoxicity, and Imaging Studies, Inorg. Chem., 2008, 47, 602 CrossRef CAS.
Synthesis, Photophysical Properties, Biological Labelling, Cytotoxicity, and Imaging Studies, Inorg. Chem., 2008, 47, 602 CrossRef CAS. - M. V. Werrett, P. J. Wright, P. V. Simpson, P. Raiteri, B. W. Skelton, S. Stagni, A. G. Buckley, P. J. Rigby and M. Massi, Rhenium tetrazolato complexes coordinated to thioalkyl-functionalised phenanthroline ligands: synthesis, photophysical characterisation, and incubation in live HeLa cells, Dalton Trans., 2015, 44, 20636 RSC.
- A. M. Mansour, O. R. Shehab and K. Radacki, Role of Sulfonate Appendage in the Protein Binding Affinity of Half-Sandwich Ruthenium(II)(η6-p-Cym) Complexes, Eur. J. Inorg. Chem., 2020, 299 CrossRef CAS.
- A. M. Mansour and A. Friedrich, IClick cycloaddition reaction of light-triggered manganese (i) carbonyl complexes, New J. Chem., 2018, 42, 18418 RSC.
- A. M. Mansour, Pyridylbenzimidazole based Re(I)(CO)3 complexes: antimicrobial activity, spectroscopic and density functional theory calculations, RSC Adv., 2019, 9, 15108 RSC.
- A. M. Mansour and K. Radacki, Antimicrobial properties of half-sandwich Ir(III) cyclopentadienyl complexes with pyridylbenzimidazole ligands, Dalton Trans., 2020, 49, 4491 RSC.
- S. Van Wallendael, R. J. Shaver, D. P. Rillema, B. J. Yoblinski, M. Stathis and T. F. Guarr, Ground-state and excited-state properties of monometallic and bimetallic complexes based on rhenium(I) tricarbonyl chloride: effect of an insulating vs. a conducting bridge, Inorg. Chem., 1990, 29, 1761 CrossRef CAS.
- E. G. Randles and P. R. Bergethon, Environment reaction fields for lipophilic fluorphores using solvatochromic shifts, Biophys. J., 2013, 104(2), 83A CrossRef.
- A. M. Mansour, K. Radacki, R. M. Khaled, M. H. Soliman and N. T. Abdel-Ghani, Phototriggered cytotoxic properties of tricarbonyl manganese (I) complexes bearing α-diimine ligands towards HepG2, J. Biol. Inorg Chem., 2021, 26(1), 135 CrossRef CAS PubMed.
- A. M. Mansour and K. Radacki, Terpyridine based ReX(CO)3 compounds (X = Br−, N3− and triazolate): Spectroscopic and DFT studies, Polyhedron, 2021, 194, 114954 CrossRef CAS.
- I. G. Dance and T. R. Miller, Solvatochromic dithiolene α-di-imine nickel complexes, J. Chem. Soc., Chem. Commun., 1973, 433 RSC.
- A. D. Becke, Density-functional thermochemistry. III. The role of exact exchange, J. Chem. Phys., 1993, 98, 5648 CrossRef CAS.
- P. J. Hay and W. R. Wadt, Ab initio effective core potentials for molecular calculations. Potentials for the transition metal atoms Sc to Hg, J. Chem. Phys., 1985, 82, 270 CrossRef CAS.
- P. J. Hay and W. R. Wadt, Ab initio effective core potentials for molecular calculations. Potentials for main group elements Na to Bi, J. Chem. Phys., 1985, 82, 284 CrossRef.
- T. Yanai, D. P. Tew and N. C. Handy, A New Hybrid Exchange-Correlation Functional Using the Coulomb-Attenuating Method (CAM-B3LYP), Chem. Phys. Lett., 2004, 393, 51 CrossRef CAS.
- W. Vollmer, D. Blanot and M. A. De pedro, Peptidoglycan structure and architecture, FEMS Microbiol. Rev., 2008, 32, 149 CrossRef CAS PubMed.
- F. P. Gabbai, P. J. Chirik, D. E. Fogg, K. Meyer, D. J. Mindiola, L. L. Schafer and S. You, An Editorial About Elemental Analysis, Organometallics, 2016, 35, 3255 CrossRef.
- A. M. Mansour and K. Radacki, Spectroscopic and antimicrobial activity of photoactivatable tricarbonyl Mn(I) terpyridine compounds, Inorg. Chim. Acta, 2020, 511, 119806 CrossRef CAS.
- M. J. Frisch, G. W. Trucks, H. B. Schlegel, G. E. Scuseria, M. A. Robb, J. R. Cheeseman, V. G. Zakrzewski, J. A. Montgomery, R. E. Stratmann, J. C. Burant, S. Dapprich, J. M. Millam, A. D. Daniels, K. N. Kudin, M. C. Strain, O. Farkas, J. Tomasi, V. Barone, M. Cossi, R. Cammi, B. Mennucci, C. Pomelli, C. Adamo, S. Clifford, J. Ochterski, G. A. Petersson, P. Y. Ayala, Q. Cui, K. Morokuma, D. K. Malick, A. D. Rabuck, K. Raghavachari, J. B. Foresman, J. Cioslowski, J. V. Ortiz, A. G. Baboul, B. B. Stefanov, G. Liu, A. Liashenko, P. Piskorz, I. Komaromi, R. Gomperts, R. L. Martin, D. J. Fox, T. Keith, M. A. Al-Laham, C. Y. Peng, A. Nanayakkara, C. Gonzalez, M. Challacombe, P. M. W. Gill, B. G. Johnson, W. Chen, M. W. Wong, J. L. Andres, M. Head-Gordon, E. S. Replogle and J. A. Pople, GAUSSIAN 03 (Revision A.9), Gaussian, Inc., Pittsburgh, 2003 Search PubMed.
- A. Frisch, A. B. Nielson and A. J. Holder, Gaussview User Manual, Gaussian, Inc, Pittsburgh, PA 2000 Search PubMed.
- A. Frei, J. Zuegg, A. G. Elliott, M. Baker, S. Braese, C. Brown, F. Chen, C. G. Dowson, G. Dujardin, N. Jung, A. P. King, A. M. Mansour, M. Massi, J. Moat, H. A. Mohamed, A. K. Renfrew, P. J. Rutledge, P. J. Sadler, M. W. Todd, C. E. Willans, J. J. Wilson, M. A. Cooper and M. A. T. Blaskovich, Metal complexes as a promising source for new antibiotics, Chem. Sci., 2020, 11, 2627 RSC.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1ra03063a |
| This journal is © The Royal Society of Chemistry 2021 |