Open Access Article
Open Access ArticleEffective inhibition and eradication of pathogenic biofilms by titanium dioxide nanoparticles synthesized using Carum copticum extract†
Mohammad Altafad,
Mohammad Tarique Zeyad *b,
Md Amiruddin Hashmic,
Salim Manoharadas
*b,
Md Amiruddin Hashmic,
Salim Manoharadas d,
Shaik Althaf Hussaind,
Mohammed Saeed Ali Abuhasile and
Mohammed Abdulaziz M. Almuzainid
d,
Shaik Althaf Hussaind,
Mohammed Saeed Ali Abuhasile and
Mohammed Abdulaziz M. Almuzainid
aDepartment of Chemistry, College of Science, King Saud University, Riyadh, 11451, Saudi Arabia
bDepartment of Agricultural Microbiology, Faculty of Agricultural Sciences, Aligarh Muslim University, Aligarh, UP-202002, India. E-mail: mohd.zeyad@gmail.com
cInterdisciplinary Biotechnology Unit, Aligarh Muslim University, Aligarh, UP-202002, India
dCentral Laboratory, College of Science, King Saud University, Riyadh 11451, Saudi Arabia
eDepartment of Food Science and Nutrition, College of Food & Agriculture Sciences, King Saud University, Riyadh 11451, Saudi Arabia
First published on 27th May 2021
Abstract
Most bacteria exist in nature in the form of biofilms. One of the key survival strategies by bacteria to withstand chemical and physical stresses is by forming biofilms on biotic and abiotic surfaces. A different set of genes are expressed in biofilms compared to the planktonic mode of bacterial growth. According to data from the National Institutes of Health (NIH) and Centers for Disease Control and Prevention (CDC), nearly 80 percent of all human infections are encouraged by biofilms and roughly 65 percent of all hospital-acquired infections are associated with biofilms. Hence, considering the role of biofilms in clinical settings, there is an urgent need for the discovery/development of novel antibiofilm agents. In this study, we have tested the effect of freshly prepared titanium dioxide nanoparticles (TiO2-NPs) synthesized using Carum copticum extract on biofilms, both against Gram +ve and Gram −ve bacteria. Being environment friendly in nature, the green route of nanoparticle synthesis is believed to be advantageous over chemical synthesis of metal nanoparticles. The synthesized nanoparticles were found to be predominantly spherical or spheroidal in shape with an average size of 12.01 ± 5.58 nm. As evident from data, more than 70% inhibition of biofilms of test bacteria was achieved in the presence of TiO2-NPs. Electron microscopic analysis revealed that the adherence and colonization of bacteria on the glass surface were remarkably reduced by the treatment of TiO2-NPs. The EPS secretion of E. coli ATCC 25922 and P. aeruginosa PAO1 were inhibited by 62.08 and 74.94%, respectively. The EPS secretion of S. aureus MTCC 3160 was least inhibited (<55%) compared to other test bacteria. Moreover, TiO2-NPs successfully eradicated the preformed biofilms of E. coli ATCC 25922, P. aeruginosa PAO1, and S. aureus MTCC 3160 by 60.09, 64.14, and 48.30%, respectively. The findings demonstrate the efficacy of green synthesized titanium dioxide nanoparticles in inhibiting and eradicating the biofilms of bacterial pathogens and they may be further exploited for the development of a new alternative antibiofilm agent.
1. Introduction
Earlier, it was thought that bacteria exist and grow in a free-floating form (or in planktonic mode) but later this concept was overturned by the discovery of biofilms.1 Now it is widely accepted that the biofilm mode of bacterial growth is more predominant in nature, industry, and clinical settings. Biofilms are also one of the key survival strategies in which bacteria can resist a number of chemical and physical stresses.2 Briefly, biofilms are bacterial communities attached to abiotic or biotic surfaces that are enclosed in a self-produced polymeric matrix, composed of polysaccharides, proteins, and eDNA, termed as extracellular polymeric substances.3,4 In biofilms mode of bacterial growth, there is an altered expression of certain phenotypes compared to the planktonic counterparts.5 For instance, bacteria in biofilms are more resistant to antibiotics and also to the host's immune response compared to planktonic bacteria.2,6 It has been documented that P. aeruginosa growing as biofilms on urinary catheters were found to be 1000 times more resistant to tobramycin compared to the planktonic growth.7 The enhanced resistance against antibiotics may also lead to the development of chronic infections such as cystic fibrosis, chronic wound infections, chronic-otitis media, etc.8Biofilms have become a serious threat in the treatment of infection in clinical settings.9 Apart from its role in chronic infections, biofilms also develop on medical implants or devices, causing an extra burden on human health.10 According to the National Institutes of Health (NIH) estimates, roughly 80 percent of all known infections in humans are linked to the development of biofilms. Moreover, the Centers for Disease Control and Prevention (CDC) has found that more than 65 percent of hospital-acquired infections are associated with biofilms.11 The biofilms developed in persistent infections are difficult to be overpowered by the immune response of host.12 Moreover, biofilms also induce enhanced overproduction of white blood cells and polymorphonucleocytes, resulting in chronic inflammation and ultimately causes complications in tissue and delay in wound healing.13 Owing to the problems associated with biofilms in human health, the development of novel antibiofilm agents has become need of the hour.
In the last few decades, nanotechnology has received great attention in numerous disciplines of science such as materials science, biomedical engineering, medicine, etc.14 Nanomaterials are typically less than 100 nm but sometimes its size may be more than 100 nm. Inorganic nanomaterials such as silver, gold, platinum, iron, titanium, zinc, etc., are of great choice for biomedical applications in diagnosis, photo-thermal therapy, optical and electrical sensing, etc.15 Furthermore, nanomaterials exhibit great implications for wound dressings, medical devices with anti-microbial properties, drug-eluting stents and, scaffolds to prevent biofilms.15 Due to the ability of nanomaterials to inhibit bacterial growth and biofilms, continuous efforts are being made for the design and synthesis of nanoparticles with promising antibiofilm activities.
The most common procedure of metal nanoparticles is the chemical synthesis. In chemicals synthesis, certain chemicals are used for the reduction process to form metal nanoparticles in which these chemicals or their byproducts pose serious threat to environmental and biological systems.16 The green route of nanoparticle synthesis has numerous advantages over chemical synthesis as it utilizes environment friendly materials (mainly natural extracts) without using harmful chemical entities.17 Moreover, the biodegradability of natural products makes it advantageous and a commonly used material for synthesis of nanoparticles. The biological polymers such as starch, alginate, chitosan, cellulose etc. provide size confined microenvironment in which reduction of metal ions to form nanoparticles can be achieved through adsorption coupled reduction process.18 However, there are certain limitations associated with the green synthesis. The major one is variations in phytochemical profile as it is influenced by seasonal or climate variations that may not remain precise enough to obtain reproducible results.19 Additionally, there is also risk associated that overexploitation of natural resources may cause ecological imbalance of natural bioresources such as microbes and plants.
Titanium nanoparticles have been found to exhibit antimicrobial properties.20 Moreover, there also some findings on the antibiofilm activity of titanium nanoparticles.19,21 To the best of our knowledge, very limited reports are available on the broad-spectrum antibiofilm activity of titanium nanoparticles. Most of the previous findings do not considered the impact of nanoparticles on the biofilm's architecture. Considering the lack of information available in these aspects, titanium dioxide nanoparticles (TiO2-NPs) were synthesized using aqueous extract of Carum copticum and characterized using spectroscopic and microscopic techniques. The nanoparticles were tested for its efficacy to inhibit the biofilms development of both Gram +ve and Gram −ve bacteria. The effect of TiO2-NPs on the biofilm's architecture was studied using light, confocal, and electron microscopy. Moreover, the ability of TiO2-NPs to eradicate the established biofilms of test bacteria was also explored.
2. Materials and methods
2.1. Chemicals and reagents
Titanium(IV) dioxide (GRM3065), acridine orange (MB116), and Luria Bertani Broth were purchased from HiMedia Laboratories, India. Crystal violet (074072) powder was procured from Sisco Research Laboratories (SRL) Pvt. Ltd.2.2. Preparation of plant extract and synthesis of TiO2-NPs
Seeds of Carum copticum were purchased from the local market at Aligarh, UP, India. The seeds were powdered in the blender to prepare fine powder. Aqueous extract of plant material was made by suspending 50 g of the plant powder in 500 ml double distilled water. The mixture was boiled for 30 min in the water bath at 90 °C and then filtered using Whatman filter paper to obtain a clear solution.For the synthesis of titanium dioxide nanoparticles (TiO2-NPs), 20 ml extract was mixed with 300 ml of titanium(IV) dioxide (5 mM in double-distilled water) and mixed using a magnetic stirrer. One millilitre (1 ml) of 1 mM NaOH was added drop-wise in the reaction mixture while continuous stirring. The reaction was allowed to take place for 6 h at 60 °C. The nanoparticles were obtained by centrifugation at 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500 rpm for 30 min. TiO2-NPs were washed three times with distilled water and then ethanol, and finally dried overnight in the oven at 50 °C. TiO2-NPs were stored at room temperature in powdered form for characterization and further studies.
500 rpm for 30 min. TiO2-NPs were washed three times with distilled water and then ethanol, and finally dried overnight in the oven at 50 °C. TiO2-NPs were stored at room temperature in powdered form for characterization and further studies.
2.3. Characterization of TiO2-NPs
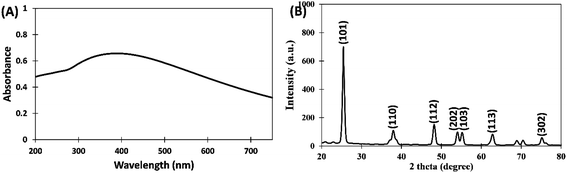
Preliminary characterization of titanium dioxide nanoparticles (TiO2-NPs) synthesized using aqueous extract of C. copticum seeds was performed using UV-vis spectroscopy. TiO2-NPs were suspended in double-distilled water and sonicated for 30 min. The UV-vis absorption spectrum of TiO2-NPs was recorded from 200 to 800 nm using UV-1800 spectrophotometer (Shimadzu, Japan). The baseline was corrected using double distilled water.The diffraction pattern of TiO2-NPs was obtained using an X-ray diffractometer ranging from 20° to 80°. Cu-Kα radiation with the nickel monochromator was used as the light source. The crystalline size of TiO2-NPs was calculated using Scherer's eqn (1):22
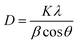 | (1) |
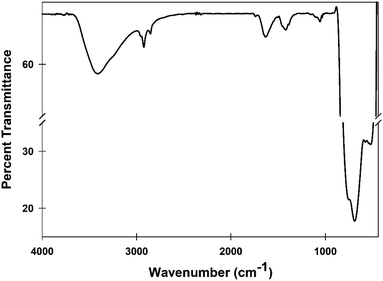
Fourier-transform infrared spectroscopy (FTIR) was used for chemical characterization as this tool deciphers the rotational and vibrational modes of motion of molecules in TiO2-NPs. The nanoparticles were mixed with KBr in ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 to before recording the transmittance spectrum. The FTIR spectrum was recorded using Spectrum Two spectrometer (Perkin Elmer Life and Analytical Sciences, CT, USA) in diffuse reflectance mode at a resolution of 2 cm−1 in the range of 4000 to 400 cm−1.23
100 to before recording the transmittance spectrum. The FTIR spectrum was recorded using Spectrum Two spectrometer (Perkin Elmer Life and Analytical Sciences, CT, USA) in diffuse reflectance mode at a resolution of 2 cm−1 in the range of 4000 to 400 cm−1.23
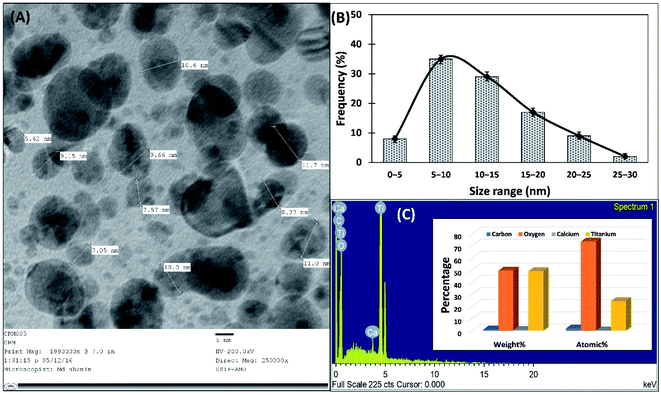
For the analysis of size variation of shape of TiO2-NPs, transmission electron microscopy (TEM) was performed. The aqueous suspension of TiO2-NPs was placed on a TEM grid and air-dried overnight to prepare the grid. The TEM analysis was performed using a JOEL-2100 electron microscope (Tokyo, Japan) and micrographs were captured at 200 kV.24 The elemental analysis of TiO2-NPs was performed by energy-dispersive X-ray (EDX) spectroscopy coupled to JEOL-JSM 6510 LV (Tokyo, Japan) at University Sophisticated Instrumentation Facility (USIF), AMU, Aligarh.25
2.4. Bacterial strains and culture conditions
Staphylococcus aureus MTCC 3160, Pseudomonas aeruginosa PAO1, and Escherichia coli ATCC 25922 were used in this study. All cultures were grown in Luria Bertani (LB) broth at 37 °C otherwise stated. All the experiments were performed in LB broth C otherwise stated.2.5. Quantification of biofilms inhibition
The quantification of biofilm inhibition of test bacteria by TiO2-NPs was performed in 96-well polystyrene plate using standard crystal violet method with slight modifications.26 TiO2-NPs were two-fold serially diluted (8–64 μg ml−1) in autoclaved Luria-Bertani broth. The bacteria were allowed to grow and inoculated in the wells containing varying concentrations of TiO2-NPs. No treatment was given to the control group. The microtitre plate was incubated for 24 h at 37 °C in static incubator. On completion of incubation period, the wells were decanted to remove media and washed with sterile phosphate buffer. The wells were air-dried for 20 min followed by the addition of 200 μl of 0.1% crystal violet to stain the biofilms. After 15 min, the crystal violet was removed and again washed with sterile phosphate buffer to remove unbound crystal violet. The plate was air-dried and finally, biofilms were dissolved in 200 μl ethanol (90% in distilled water). The absorbance of wells of polystyrene plate was recorded at 620 nm using microplate reader. The percent inhibition of biofilms was calculated with respect to the respective controls.2.6. Microscopic analysis of biofilm inhibition on glass surface
The inhibition of biofilms of test bacteria by TiO2-NPs was further studied using an array of microscopic techniques such as light microscopy, confocal laser scanning microscopy, and scanning electron microscopy.2.7. Inhibition of exopolysaccharides
The level of exopolysaccharides (EPS) in control and TiO2-NPs treated cultures were assessed by following the standard procedure with minor modifications.30 Briefly, the test bacteria were grown in presence of different concentrations of TiO2-NPs (8, 16, 32, and 64 μg ml−1) at 37 °C for 24 h. The control group was not given treatment. On completion of incubation period, bacterial cultures were centrifuged and cell-free supernatant was collected. Chilled ethanol was added to the culture supernatant in the ratio of 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and the incubated overnight at 4 °C to precipitate the EPS. The level of EPS was determined by estimating the sugars using the Dubois method.31
1 and the incubated overnight at 4 °C to precipitate the EPS. The level of EPS was determined by estimating the sugars using the Dubois method.31
2.8. Disruption of mature biofilms
The biofilm disruption assay was performed in a 96-well polystyrene plate following the standard procedure with minor modifications.32 The bacteria were grown in microtitre plates for 24 h at 37 °C to form the biofilms in the wells of microtitre pate. The media was then decanted gently and washed using sterile phosphate buffer to remove loosely adhered cells. Freshly prepared media (Luria Bertani broth) was added to the wells of the plate and TiO2-NPs were added, making final concentrations of 8, 16, 32, and 64 μg ml−1. The polystyrene plate was incubated for the next 24 h under static conditions. The wells of plate were washed using sterile phosphate buffer to remove the planktonic cells. The biofilms were stained with crystal violet (0.1% solution in water) for 20 min. The stain was removed and gently washed. Finally, biofilms were dissolved in ethanol (90% in water). The absorbance was recorded using a microplate reader and percent disruption was calculated with respect to the control group.2.9. Statistical analysis
The experiments were performed in three independent replicates and the data is presented as the average with standard deviation (SD). For the analysis of statistical significance, the analysis variance (One-way ANOVA) was performed by Tukey test at the significance level of 0.05. Different letters above each group of the independent variables show that the groups are statistically different from one another at p-value = 0.05.3. Results and discussion
3.1. Synthesis and characterization of TiO2-NPs
The seeds extract of C. copticum was used for green synthesis of titanium dioxide nanoparticles (TiO2-NPs). Being simple, usually one step, and ecofriendly in nature, green synthesis is advantageous over the chemical route of metal nanoparticle synthesis. One of the reasons for selection of this plant was that extracts of Carum copticum has been reported to exhibit antibiofilm properties.33 It is expected that the combination of TiO2-NPs and phytoconstituents of C. copticum will impart enhanced antibiofilm action. Moreover, we have also reported the broad-spectrum antibiofilm and antiquorum sensing activities of silver nanoparticles synthesized using C. copticum exact against Gram −ve bacterial pathogens.27 TiO2-NPs were characterized using a number of spectroscopic and microscopic tools which are described below.3.2. Antibiofilm activity of TiO2-NPs
Before evaluating the antibiofilm activity, test bacteria were grown in presence of TiO2-NPs to assesses its effect on the viable cells. The viability was determined by counting the number of colonies forming units (CFU). The number of viable cells of test bacteria in the absence and presence of 64 μg ml−1 TiO2-NPs is shown in supplementary Fig. S1.† This was the highest tested concentration at which insignificant (p-value > 0.05) difference in the viability of bacterial cell was recorded. Therefore, the antibiofilm activity was assessed at 64 μg ml−1 and lower concentrations of TiO2-NPs. The green synthesized nanoparticles were tested for its efficacy against the biofilms of P. aeruginosa PAO1, E. coli ATCC 25922, and S. aureus MTCC 3160. The findings are discussed below.3.3. Quantification of biofilm inhibition by TiO2-NPs
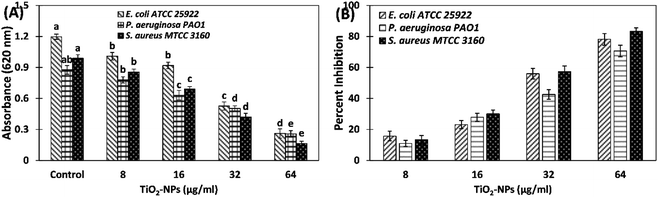
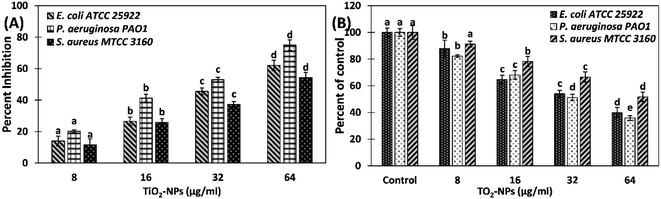
The inhibition of biofilms by TiO2-NPs against S. aureus MTCC 3160, P. aeruginosa PAO1, and E. coli ATCC 25922 was quantified using crystal violet assay in 96-well microtitre plate. The data shows that TiO2-NPs effectively reduced the biofilms of tested bacteria (Fig. 4). Treatment with 8, 16, 32, and 64 μg ml−1 TiO2-NPs inhibited the development of the biofilm of E. coli ATCC 25922 by 15.73, 23.19, 55.97, and 78.19%, respectively. Similarly, there was 11.03, 27.88, 42.60, and 70.67% reduction in biofilms of P. aeruginosa PAO1 in presence of 8, 16, 32, and 64 μg ml−1 TiO2-NPs. The biofilms of tested Gram +ve bacteria (S. aureus MTCC 3160) were maximumly inhibited by TiO2-NPs. The addition of 8, 16, 32, and 64 μg ml−1 TiO2-NPs in culture media decreased the formation of biofilms of S. aureus MTCC 3160 by 13.29, 30.06, and 57.38%, respectively. At the highest tested concentration (64 TiO2-NPs), more than 80% inhibition was recorded. The bacteria growing in biofilms become many folds more resistant to chemotherapeutic agents that pose extra burden for the treatment of bacterial infections. For instance, it has been found that P. aeruginosa growing in biofilms on urinary catheters became up to 1000-folds more resistant to tobramycin compared to the planktonic growth.7 The biofilms are important for clinical point of view and it has been estimated that a vast majority of bacterial infections involve biofilms formation for the establishment of successful infection.41 Moreover, bacteria coordinate the expression of resistance genes and virulent traits in biofilms that makes poses difficulties for chemical as well as physical treatments.42Antimicrobial resistance (AMR) or drug resistance poses a massive threat to public health and as well as the environment, where biofilms are considered as one of the major drivers in the dissemination of drug resistance. The aggregated bacterial communities in biofilms facilitate the transfer of resistant genes and also prevent the entry and diffusion of antibiotics.43 Therefore, inhibition of development of biofilm using nanoparticles is one of the promising approaches to prevent bacterial infection. Previously, bio-fabricated titanium oxide nanoparticles using Ochradenus arabicus have been found to inhibit the pathogenic biofilms of drug resistant P. aeruginosa and S. aureus isolated from diabetic foot infections.19 It has been reported that silver nanoparticles embedding on copper surfaces reduced the viability of E. coli and C. auris by >90% on 7 days of exposure.44 The galinstan (composed of gallium, indium, and tin) liquid metal particles with encapsulated iron were found to successfully reduce both the mono-species and multi-species biofilms.45 It has been reported that extracts of Carum copticum poses good antibiofilm activity antibiotic-resistant bacteria.33 It can be deduced that the antibiofilm activity of TiO2-NPs may be the synergistic or cumulative effects of both titanium nanoparticles and the phytoconstituents of C. copticum. The findings validate the broad-spectrum inhibition of biofilms by green synthesized titanium oxide nanoparticles.
3.4. Microscopic analysis of biofilms inhibition on glass surface using
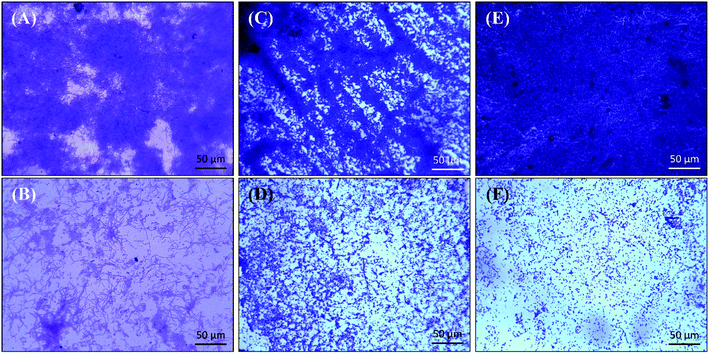
The biofilm inhibition of test bacteria by TiO2-NPs was further studied on a glass surface using an array of microscopic techniques such as using light microscopy, scanning electron microscopy, confocal microscopy, and the findings are discussed below.The detailed analysis of changes in the biofilm's architecture by the treatment of TiO2-NPs was studied by culturing the test bacteria in presence of TiO2-NPs in 24-well tissue culture plates containing glass coverslips. The light microscopic images of control and treated test bacterial biofilms are shown in Fig. 5. The micrographs of untreated biofilms show that bacteria heavily colonized the glass surface and formed thick layer of biofilms. Treatment with TiO2-NPs reduced the bacterial colonization and subsequently a remarkable inhibition in the biofilm development was observed.
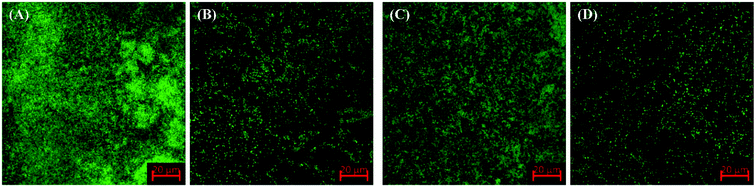
The confocal laser scanning micrographs of untreated and treated bacterial biofilms of P. aeruginosa PAO1 and E. coli ATCC 25922 are shown in Fig. 6. As seen in control images, test bacteria formed very dense biofilms of glass coverslips. Dense clumps of cells in biofilms were found and bacteria were seen to be heavily colonized. However, the presence of TiO2-NPs in culture media substantially reduced the biofilm formation and bacterial cells were found in scattered form. The result in agreement with the previous finding where a confocal microscopic analysis revealed that titanium oxide nanoparticles synthesized using Withania somnifera root extract inhibited the biofilms formation of E. coli, P. aeruginosa, L. monocytogenes, S. marcescens, and methicillin-resistant S. aureus (MRSA) on the glass surface.46
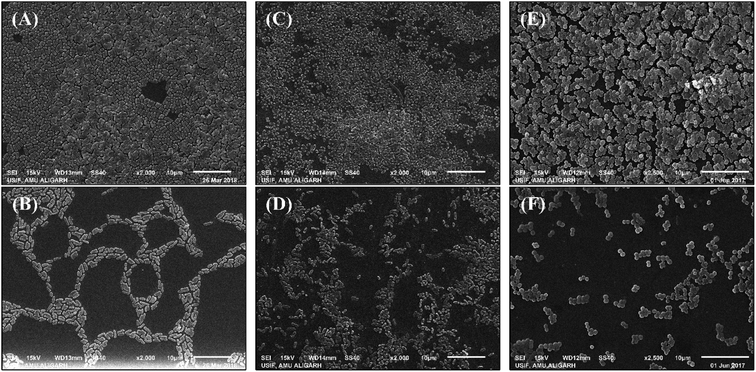
Further analysis of the alterations on the biofilm's architecture by the treatment of TiO2-NPs was studied using scanning electron microscopy. The electron micrographs of control and TiO2-NPs treated biofilms of test bacteria are shown in Fig. 7. The untreated S. aureus MTCC 3160 formed dense biofilms on a glass surface where the morphology of bacterial cells was found to be normal and smooth. The bacterial cells were seen closed in extra polymeric substance. On contrary, treatment with TiO2-NPs reduced the biofilms formation with decreased bacterial colonization. Likewise, the development of the biofilm in control P. aeruginosa PAO1 was more than the treated one. The aggregation of bacteria on the glass surface was reduced in presence of TiO2-NPs that resulted in a fair decrease in the biofilm forming capability of P. aeruginosa PAO1. A similar observation was recorded for E. coli ATCC 25922 biofilms where challenging the bacteria with TiO2-NPs resulted in decreased biofilm formation. It is interesting to note that extra polymeric substances were not seen on the slides treated with TiO2-NPs. Hence, the microscopic analysis confirms the remarkable inhibition of biofilms of S. aureus MTCC 3160, P. aeruginosa PAO1, and P. aeruginosa PAO1.
Most of the bacterial biofilms are pathogenic that cause nosocomial infections. As per National Institutes of Health (NIH) estimates, nearly 65% of all microbial infections and 80% of chronic infections are associated with biofilm development.47 Hence, targeting biofilms, more precisely pathogenic biofilms, are considered as an effective strategy in the management of bacterial infections. Titanium oxide nanoparticles have been found previously to mitigate the biofilms of bacterial pathogens. The nanoparticles were synthesized leaves extract of O. arabicus that inhibited the biofilms of drug-resistant P. aeruginosa and S. aureus isolated from diabetic foot infections of the human subjects with diabetic foot ulcers.19 Similarly, another study has found that biofabricated porous titanium dioxide nanoparticles synthesized using root extract of Withania somnifera exhibited a broad-spectrum antibiofilm activity against P. aeruginosa, E. coli, methicillin-resistant S. aureus, S. marcescens, L. monocytogenes, and C. albicans.46 A study has found that surfaces coated with zinc oxide nanoparticles and ZnO/Ag nanocomposites selectively inhibited the biofilms of E. coli, S. aureus, and C. albicans.48 Moreover, another study has documented that Ag/Cu/GO nanocomposite inhibited the biofilm formation of P. aeruginosa at the concentration which was harmless to human cells.49 Our findings clearly show the ability of TiO2-NPs in inhibiting the biofilms of both Gram +ve and Gram −ve bacteria.
3.5. Inhibition of exopolysaccharides (EPS) production
Extracellular polymeric substances are natural polymers with a high molecular weight that provide structural framework to the biofilms.50 Hence, extracellular polymeric substance is the fundamental component of biofilms and also determines the physicochemical properties. Among different constituents of extracellular polymeric substances, exopolysaccharides (EPS) are the major contributor.51 EPS blocks the entry of chemotherapeutic agents, such as antibiotics, serving as a protective barrier for the bacterial cells.51 The enhanced secretion of EPS alters the biofilm architecture and thereby confers drug resistance against antimicrobial agents.52 There is a positive correlation between EPS secretion of biofilms formation, hence targeting EPS production is also considered as an alternate target for biofilms inhibition.53 TiO2-NPs successfully inhibited the EPS secretion of test bacteria (Fig. 8A). The EPS secretion of S. aureus MTCC 3160 decreased by 11.66, 25.83, 37.20, and 54.27% by the treatment of 8, 16, 32, and 64 μg ml−1 TiO2-NPs, respectively. Similarly, the presence of 8, 16, 32, and 64 μg ml−1 TiO2-NPs reduced the EPS production of E. coli ATCC 25922 by 14.01, 26.56, 45.52, and 62.08%, respectively. The EPS formation of P. aeruginosa PAO1 was maximally inhibited (∼75%) in presence of 64 μg ml−1 TiO2-NPs. EPS is an important constituent of the biofilm's architecture. It provides structural stability to the biofilms and protects bacterial cells from antibiotics and environmental stresses.54 Hence, targeting EPS is considered an alternative strategy to inhibit the biofilms as reduced secretion of EPS is expected to exert adverse effects on the biofilm-forming ability of bacterial pathogens. Porous titanium oxide nanoparticles synthesized using aqueous extract of Withania somnifera roots have been documented to inhibit the EPS production of methicillin-resistant S. aureus, L. monocytogenes, P. aeruginosa, E. coli, and S. marcescens.46 Moreover, green synthesized silver nanoparticles have also been reported to inhibit the EPS secretion in P. aeruginosa, S. aureus, K. pneumoniae, and C. albicans in the presence of their respective inhibitory concentrations.303.6. Eradication of preformed biofilms
Most antibacterial drugs inhibit the planktonic mode of bacterial growth in which few of them are known to inhibit the development of biofilm.55 Inhibition of biofilms is relatively easier compared to the eradication of established biofilms. In most successful infections, the biofilms get established at the site of infection. In this assay, the bacterial strains were allowed to form the biofilms in 96-well microtitre plates followed by treating the established biofilms. The effect of TiO2-NPs on the removal of preformed biofilms is shown in Fig. 8B. The presence of 8, 16, 32, and 64 μg ml−1 TiO2-NPs eradicated the established biofilms of P. aeruginosa PAO1 by 17.77, 31.85, 48.77, and 64.14%, respectively. Similarly, the preformed biofilms of E. coli ATCC 25922 were decreased by 12.11, 35.23, 45.99, and 60.09%, respectively. TiO2-NPs exhibited the least effect on established biofilms of S. aureus MTCC 3160 where the presence of 64 μg ml−1 disrupted the biofilms by 48.30%. The data demonstrate the successful biofilm eradication of the test bacteria. Bacterial biofilms are the clump of bacterial cells embedded in extracellular polymeric substances.56 The major component of extracellular polymeric substances are polysaccharides, nucleic acids, and polypeptides along with some other biochemical components.57 These components act as a barrier for the entry of chemotherapeutic agents.58 Our findings are in agreement with the previous report in which the established biofilms of P. aeruginosa, E. coli, methicillin-resistant S. aureus, and L. monocytogenes were disrupted by 65, 45, 64, and 49%, respectively.46 A study conducted on silver nanoparticles has found that silver nanoparticles can diffuse the biofilms matrix.59 It can be deduced that TiO2-NPs may be able to penetrate the established biofilms that resulted in disruption of biofilms. The extract of C. copticum are documented to disrupt the preformed biofilms of B. cereus, S. aureus, P. aeruginosa, E. coli, A. baumannii, and K. pneumonia.33 The eradication of biofilms may also be attributed to the cumulative effects of both titanium nanoparticles and the phytocompounds of C. copticum. The results show that TiO2-NPs were successful both in the inhibition as well as eradiation of the biofilms of test bacteria.4. Conclusion
Biofilms have great implications both in clinical settings and the environment. The majority of successful bacterial infections are associated and influenced by biofilms development. Here, we have synthesized titanium dioxide nanoparticles (TiO2-NPs) using Carum copticum extract and tested its ability to inhibit the development of biofilm. The nanoparticles were spherical or spheroidal in shape with an average size of 12.01 nm. More than 70% reduction in biofilms formation of the bacteria was found by the treatment of TiO2-NPs. Microscopic analysis of the biofilms deciphered that bacterial adherence and colonization on a solid support (glass surface) was remarkably hampered in presence of TiO2-NPs. The EPS secretion of test bacteria was also inhibited dose-dependently. TiO2-NPs was quite efficient in removing the established biofilms of E. coli ATCC 25922, P. aeruginosa PAO1, and S. aureus MTCC 3160. The data shows the potency of TiO2-NPs against biofilms of bacterial pathogens and it can be exploited further for the development of novel alternative antibiofilm agents.Abbreviation
| TiO2-NPs | Titanium dioxide nanoparticles |
| EPS | Exopolysaccharides |
| FTIR | Fourier transforms infrared |
| TEM | Transmission electron microscopy |
| SEM | Scanning electron microscopy |
| UV-vis | UV-visible |
| XRD | X-ray diffraction |
| EDX | Energy-dispersive X-ray |
Conflicts of interest
The authors declare that they have no conflict of interest.Acknowledgements
The authors would like to extend their sincere appreciation to the Deanship of Scientific Research at King Saud University for funding this work through the research group project number RG-1439-076.References
- J. Costerton, G. Geesey and K. J. Cheng, Sci. Am., 1978, 238, 86–95 CrossRef CAS PubMed.
- C. A. Fux, J. W. Costerton, P. S. Stewart and P. Stoodley, Trends Microbiol., 2005, 13, 34–40 CrossRef CAS PubMed.
- L. Hall-Stoodley and P. Stoodley, Trends Microbiol., 2005, 13, 7–10 CrossRef CAS PubMed.
- R. M. Donlan and J. W. Costerton, Clin. Microbiol. Rev., 2002, 15, 167–193 CrossRef CAS PubMed.
- T. Bjarnsholt, M. Alhede, P. Ø. Jensen, A. K. Nielsen, H. K. Johansen, P. Homøe, N. Høiby, M. Givskov and K. Kirketerp-Møller, Adv. Wound Care, 2015, 4, 363–372 CrossRef PubMed.
- A. Foreman, G. Holtappels, A. J. Psaltis, J. Jervis-Bardy, J. Field, P.-J. Wormald and C. Bachert, Allergy, 2011, 66, 1449–1456 CrossRef CAS PubMed.
- J. C. Nickel, I. Ruseska, J. B. Wright and J. W. Costerton, Antimicrob. Agents Chemother., 1985, 27, 619–624 CrossRef CAS PubMed.
- L. Hall-Stoodley, P. Stoodley, S. Kathju, N. Høiby, C. Moser, J. William Costerton, A. Moter and T. Bjarnsholt, FEMS Immunol. Med. Microbiol., 2012, 65, 127–145 CrossRef CAS PubMed.
- M. K. Yadav, J.-J. Song, B. P. Singh and J. E. Vidal, in New and Future Developments in Microbial Biotechnology and Bioengineering: Microbial Biofilms, Elsevier, 2020, pp. 1–13 Search PubMed.
- S. Veerachamy, T. Yarlagadda, G. Manivasagam and P. K. Yarlagadda, Proc. Inst. Mech. Eng., Part H, 2014, 228, 1083–1099 CrossRef PubMed.
- S. L. Percival, Br. J. Surg., 2017, 104, e85–e94 CrossRef CAS PubMed.
- J. P. S. Woods, E. J. Davis and P. Barnett, in Microbiology of Wounds, CRC Press, London, 2010, pp. 271–92 Search PubMed.
- D. D. Rhoads, R. D. Wolcott and S. L. Percival, Journal of Wound Care, 2008, 17, 502–508 CrossRef CAS PubMed.
- J. V. Rau, R. De Santis and G. Ciofani, Bioact. Mater., 2017, 2, 119–120 CrossRef PubMed.
- V. Bhardwaj and A. Kaushik, Micromachines, 2017, 8, 298 CrossRef PubMed.
- C. K. Tagad, S. R. Dugasani, R. Aiyer, S. Park, A. Kulkarni and S. Sabharwal, Sens. Actuators, B, 2013, 183, 144–149 CrossRef CAS.
- E. S. Abdel-Halim, M. H. El-Rafie and S. S. Al-Deyab, Carbohydr. Polym., 2011, 85, 692–697 CrossRef CAS.
- A. R. Lokanathan, K. M. A. Uddin, O. J. Rojas and J. Laine, Biomacromolecules, 2014, 15, 373–379 CrossRef CAS PubMed.
- M. Zubair, F. M. Husain, F. A. Qais, P. Alam, I. Ahmad, T. Albalawi, N. Ahmad, M. Alam, M. H. Baig, J.-J. Dong, F. Fatima and B. Alsayed, Appl. Nanosci., 2021, 11, 375–387 CrossRef CAS.
- A. J. Haider, Z. N. Jameel and I. H. M. Al-Hussaini, Energy Procedia, 2019, 157, 17–29 CrossRef CAS.
- E. H. Abdulkareem, K. Memarzadeh, R. P. Allaker, J. Huang, J. Pratten and D. Spratt, J. Dent., 2015, 43, 1462–1469 CrossRef CAS PubMed.
- N. A. Al-Shabib, F. M. Husain, I. Hassan, M. S. Khan, F. Ahmed, F. A. Qais, M. Oves, M. Rahman, R. A. Khan, A. Khan, A. Hussain, I. M. Alhazza, S. Aman, S. Noor, H. Ebaid, J. Al-Tamimi, J. M. Khan, A. R. M. Al-Ghadeer, M. K. A. Khan and I. Ahmad, J. Nanomater., 2018, 2018, 1–14 Search PubMed.
- N. A. Al-Shabib, F. M. Husain, N. Ahmad, F. A. Qais, A. Khan, A. Khan, M. S. Khan, J. M. Khan, S. A. Shahzad and I. Ahmad, J. Nanomater., 2018, 2018, 1–11 Search PubMed.
- A. S. Ahmed, A. Iqbal, A. Shafi, F. A. Qais, T. Ahamad and S. Srivastava, J. Cluster Sci., 2020, 77, 1–7 Search PubMed.
- F. M. Husain, I. Hasan, F. A. Qais, R. A. Khan, P. Alam and A. Alsalme, Coatings, 2020, 10, 1190 CrossRef CAS.
- G. A. O'Toole and R. Kolter, Mol. Microbiol., 1998, 30, 295–304 CrossRef PubMed.
- F. A. Qais, A. Shafiq, I. Ahmad, F. M. Husain, R. A. Khan and I. Hassan, Microb. Pathog., 2020, 144, 104172 CrossRef PubMed.
- M. Maheshwari, F. A. Qais, A. S. Althubiani, H. H. Abulreesh and I. Ahmad, Biofouling, 2019, 35, 1026–1039 CrossRef CAS PubMed.
- F. A. Qais, M. S. Khan and I. Ahmad, Microb. Pathog., 2019, 126, 379–392 CrossRef CAS PubMed.
- I. Hasan, F. A. Qais, F. M. Husain, R. A. Khan, A. Alsalme, B. Alenazi, M. Usman, M. H. Jaafar and I. Ahmad, J. Cleaner Prod., 2019, 230, 1148–1155 CrossRef CAS.
- M. DuBois, K. A. Gilles, J. K. Hamilton, P. A. Rebers and F. Smith, Anal. Chem., 1956, 28, 350–356 CrossRef CAS.
- F. A. Qais, Samreen and I. Ahmad, IET Nanobiotechnol., 2018, 12, 325–335 CrossRef.
- M. Mohammadi, F. Masoumipour, M. Hassanshahian and T. Jafarinasab, Microb. Pathog., 2019, 129, 99–105 CrossRef CAS PubMed.
- V. Sivaranjani and P. Philominathan, Wound Medicine, 2016, 12, 1–5 CrossRef.
- T. Santhoshkumar, A. A. Rahuman, C. Jayaseelan, G. Rajakumar, S. Marimuthu, A. V. Kirthi, K. Velayutham, J. Thomas, J. Venkatesan and S.-K. Kim, Asian Pac. J. Trop. Med., 2014, 7, 968–976 CrossRef CAS PubMed.
- S. Subhapriya and P. Gomathipriya, Microb. Pathog., 2018, 116, 215–220 CrossRef CAS PubMed.
- M. Sundrarajan and S. Gowri, Chalcogenide Lett., 2011, 8, 447–451 CAS.
- W. Ahmad, K. K. Jaiswal and S. Soni, Inorg. Nano-Met. Chem., 2020, 50, 1032–1038 CrossRef CAS.
- A. A. Zahir, I. S. Chauhan, A. Bagavan, C. Kamaraj, G. Elango, J. Shankar, N. Arjaria, S. M. Roopan, A. A. Rahuman and N. Singh, Antimicrob. Agents Chemother., 2015, 59, 4782–4799 CrossRef CAS PubMed.
- G. Rajakumar, A. A. Rahuman, B. Priyamvada, V. G. Khanna, D. K. Kumar and P. J. Sujin, Mater. Lett., 2012, 68, 115–117 CrossRef CAS.
- B. Schachter, Nat. Biotechnol., 2003, 21, 361–365 CrossRef CAS PubMed.
- A. Pompilio, V. Crocetta, S. De Nicola, F. Verginelli, E. Fiscarelli and G. Di Bonaventura, Front. Microbiol., 2015, 6, 951 Search PubMed.
- J. L. Balcázar, J. Subirats and C. M. Borrego, Front. Microbiol., 2015, 6, 1216 Search PubMed.
- S. Gangadoo, A. Elbourne, A. E. Medvedev, D. Cozzolino, Y. B. Truong, R. J. Crawford, P.-Y. Wang, V. K. Truong and J. Chapman, Coatings, 2020, 10, 28 CrossRef CAS.
- S. Cheeseman, A. Elbourne, R. Kariuki, A. V. Ramarao, A. Zavabeti, N. Syed, A. J. Christofferson, K. Y. Kwon, W. Jung, M. D. Dickey, K. Kalantar-Zadeh, C. F. McConville, R. J. Crawford, T. Daeneke, J. Chapman and V. K. Truong, J. Mater. Chem. B, 2020, 8, 10776–10787 RSC.
- N. A. Al-Shabib, F. M. Husain, F. A. Qais, N. Ahmad, A. Khan, A. A. Alyousef, M. Arshad, S. Noor, J. M. Khan, P. Alam, T. H. Albalawi and S. A. Shahzad, Front. Microbiol., 2020, 11, 1680 CrossRef PubMed.
- M. Jamal, W. Ahmad, S. Andleeb, F. Jalil, M. Imran, M. A. Nawaz, T. Hussain, M. Ali, M. Rafiq and M. A. Kamil, J. Chin. Med. Assoc., 2018, 81, 7–11 CrossRef PubMed.
- M. Rosenberg, M. Visnapuu, H. Vija, V. Kisand, K. Kasemets, A. Kahru and A. Ivask, Sci. Rep., 2020, 10, 13478 CrossRef CAS PubMed.
- J. Jang, J.-M. Lee, S.-B. Oh, Y. Choi, H.-S. Jung and J. Choi, ACS Appl. Mater. Interfaces, 2020, 12, 35826–35834 CrossRef CAS PubMed.
- E. Maunders and M. Welch, FEMS Microbiol. Lett., 2017, 364 DOI:10.1093/femsle/fnx120.
- K. Sauer and A. K. Camper, J. Bacteriol., 2001, 183, 6579–6589 CrossRef CAS PubMed.
- F. H. Yildiz and G. K. Schoolnik, Proc. Natl. Acad. Sci. U. S. A., 1999, 96, 4028–4033 CrossRef CAS PubMed.
- K. Czaczyk and K. Myszka, Pol. J. Environ. Stud., 2007, 16, 799–806 CAS.
- H.-C. Flemming, T. R. Neu and D. J. Wozniak, J. Bacteriol., 2007, 189, 7945–7947 CrossRef CAS PubMed.
- R. Roy, M. Tiwari, G. Donelli and V. Tiwari, Virulence, 2018, 9, 522–554 CrossRef CAS PubMed.
- Y. Li, P. Xiao, Y. Wang and Y. Hao, ACS Omega, 2020, 5, 22684–22690 CrossRef CAS PubMed.
- L. Karygianni, Z. Ren, H. Koo and T. Thurnheer, Trends Microbiol., 2020, 28, 668–681 CrossRef CAS PubMed.
- N. Høiby, O. Ciofu, H. K. Johansen, Z. Song, C. Moser, P. Ø. Jensen, S. Molin, M. Givskov, T. Tolker-Nielsen and T. Bjarnsholt, Int. J. Oral Sci., 2011, 3, 55–65 CrossRef PubMed.
- T. O. Peulen and K. J. Wilkinson, Environ. Sci. Technol., 2011, 45, 3367–3373 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1ra02876f |
| This journal is © The Royal Society of Chemistry 2021 |