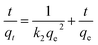Open Access Article
Open Access ArticleBiosorption behavior and mechanism of cadmium from aqueous solutions by Synechocystis sp. PCC6803†
Li Shen abc,
Ran Chena,
Junjun Wangd,
Ling Fana,
Linlin Cuia,
Yejuan Zhanga,
Jinju Chenga,
Xueling Wuab,
Jiaokun Liabd and
Weimin Zeng
abc,
Ran Chena,
Junjun Wangd,
Ling Fana,
Linlin Cuia,
Yejuan Zhanga,
Jinju Chenga,
Xueling Wuab,
Jiaokun Liabd and
Weimin Zeng *ab
*ab
aSchool of Minerals Processing and Bioengineering, Central South University, Changsha 410083, China. E-mail: zengweimin1024@126.com; Fax: +86-731-88879815; Tel: +86-731-88877472
bKey Laboratory of Biometallurgy, Ministry of Education, Changsha 410083, China
cState Key Laboratory of Complex Nonferrous Metal Resources Clean Utilization, Kunming 650093, China
dSchool of Metallurgy and Environment, Central South University, Changsha 410083, China
First published on 25th May 2021
Abstract
Cyanobacteria are promising adsorbents that are widely used for heavy metal removal in aqueous solutions. However, the underlying adsorption mechanism of Synechocystis sp. PCC6803 is currently unclear. In this study, the adsorption behavior and mechanism of cadmium (Cd2+) were investigated. Batch biosorption experiments showed that the optimal adsorption conditions were pH 7.0, 30 °C, 15 min, and an initial ion concentration of 4.0 mg L−1. The adsorption process fitted well with the pseudo-second order kinetic model, mainly based on chemisorption. Complexation of Cd2+ with carboxyl, hydroxyl, carbonyl, and amido groups was demonstrated by Fourier-transform infrared spectroscopy (FTIR), Raman spectroscopy and X-ray photoelectron spectroscopy (XPS) analyses. Scanning electron microscopy (SEM), transmission electron microscopy (TEM), and energy-dispersive X-ray spectrometry (EDX) analyses confirmed the presence of Cd2+ on the cyanobacterial cell surface and intracellularly. Cd2+ could lead to reactive oxygen species (ROS) accumulation and photosynthesis inhibition in cyanobacterial cells, and glutathione (GSH) played an important role in alleviating Cd2+ toxicity. Analyses of three-dimensional fluorescence spectroscopy (3D-EEM) and high performance anion exchange chromatography-pulsed amperometric detection (HPAEC-PAD) revealed the changes of the composition and content of EPS after Cd2+ adsorption, respectively. Real-time quantitative polymerase chain reaction (RT-qPCR) revealed the potential molecular regulatory mechanisms involved in Cd2+ biosorption. These results revealed the adsorption mechanism of Cd2+ by Synechocystis sp. PCC6803 and provided theoretical guidance for insight into the biosorption mechanisms of heavy metals by other strains.
1. Introduction
Environmental pollution caused by heavy metals has become a global issue of great concern due to mineral extraction, water treatment, metal molding, metal coating, and the battery industry.1 Heavy metals are non-biodegradable and highly toxic to the environment and can be enriched through the food chain, posing a serious threat to human health and the ecosystem. Cd2+ is non-essential element and its compounds are considered to be the most toxic substances in the environment. Heavy metals can cause oxidative stress by excessive accumulation of ROS.2,3 In addition, prolonged exposure to Cd2+ can result in dysfunction of the kidneys, liver, and central nervous system.4 Hence, the removal of heavy metals from aqueous environments is necessary for both public and environmental health.Traditional methods for removing heavy metals from water environments include chemical precipitation, ion exchange, and membrane filtration.5 However, most of these methods have a series of disadvantages, such as high operating costs, ineffectiveness, and sludge generation. In recent decades, biosorption has been widely investigated and considered a promising alternative to conventional processes because of its low cost, material regeneration, environmental friendliness, and high performance.6 Various types of biological adsorbents, including fungi, bacteria, yeast, and algae, have been widely applied to remove heavy metals from aqueous solutions.1,5,7
Cyanobacteria are photoautotrophic, prokaryotic, and unicellular microorganisms that usually live in aqueous environments. They are promising biosorbents owing to their high photosynthetic efficiency, simple structure, large surface area, and high uptake capacity.5,8 In addition, cyanobacteria can produce EPS in response to heavy metal toxicity. EPS are heterogeneous macromolecules composed of polysaccharides, proteins, lipids, and nucleic acids.9 EPS have multiple functional groups, including hydroxyl, carboxylic acid, amide, and carbonyl groups, which are likely responsible for the high binding affinity with metal ions.10 EPS can prevent heavy metal ions from entering the cells of microorganisms by electrostatic attraction, complexation, ion exchange, surface precipitation, and other interaction types. In addition, cyanobacteria have developed antioxidant system mechanisms to alleviate oxidative damage caused by heavy metal stress, including enzymatic antioxidants such as superoxide dismutase, catalase, and peroxidase, and non-enzymatic antioxidants such as GSH, ascorbate, and phenolic substances.11 These defense strategies help cyanobacteria reduce oxidative impairment and improve heavy metal tolerance.
Biosorption usually includes passive adsorption on the cell surface and active adsorption within cells (bioaccumulation), passive biosorption is a rapid adsorption process independent of energy. The process of active adsorption is much slower and metabolism-dependent.5,12 However, most adsorption mechanisms are currently unclear. In addition, most related studies have focused on the adsorption behavior and removal efficiency of metal ions, while tolerance and biosorption mechanisms by Synechocystis sp. PCC6803 have not been widely investigated. Hence, this study laid the foundation for in-depth understanding of the adsorption mechanism of Synechocystis sp. PCC6803 or other species in response to heavy metals. In this study, the factors affecting biosorption conditions, such as pH, temperature, contact time, and initial metal concentration were investigated. Adsorption kinetics were employed to explore the adsorption behavior. FTIR, RS, XPS, SEM, and TEM analyses were performed to explore the adsorption mechanism between Synechocystis sp. PCC6803 and cadmium (Cd2+) ions. Moreover, RT-qPCR were used to explore the transcriptional response to Cd2+ stress, which may assist with understanding the molecular mechanisms underlying the Cd2+ resistance and adsorption.
2. Materials and methods
2.1 Cultivation of Synechocystis sp. PCC6803 and effect of Cd2+ stress on cyanobacteria growth
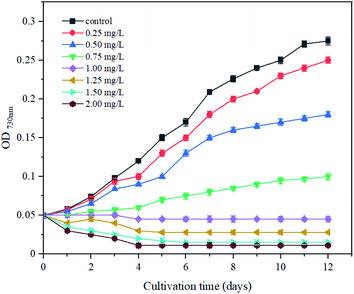
The cyanobacteria strain Synechocystis sp. PCC6803 was originally purchased from the Freshwater Algae Culture Collection at the Institute of Hydrobiology of China (FACHB). Synechocystis sp. PCC6803 cells were grown in conical flasks containing BG-11 medium and placed in a light incubator for static cultivation. Cultivation was performed under a light/dark cycle of 12/12 h with a moderate light intensity of 2000 lx at 25 °C.13 Before the experiment, all instruments and medium were sterilized at 121 °C for 20 min to prevent contamination. During the cultivation period, all flasks were shaken by hand twice daily to avoid agglomeration. In addition, the position of the flasks was changed randomly to reduce the influence of light intensity, as per a previous study.14The Cd2+ solution was prepared by adding CdCl2 to sterile water to form 1 g L−1 mother liquor. Cd2+ solutions were sterilized by filtering them with a 0.22 μm pore size filter.15 Cd2+ solution was separately added to a 250 mL conical flask with 100 mL of culture medium to obtain the final concentrations of 0, 0.25, 0.5, 0.75, 1.0, 1.25, 1.5, and 2.0 mg L−1, and the medium without Cd2+ was defined as the control. Cyanobacteria growth was determined by measurting the optical density at 730 nm (OD730) under different Cd2+ concentrations. The experiments were performed in triplicate. Finally, referred to the chemical-alga growth inhibition test and calculated the 12 day minimal inhibitory concentration (MIC) for Synechocystis sp. PCC6803.
2.2 Determination of non-enzymatic features
After 12 days of cultivation (cells were in the logarithmic phase), the cyanobacteria cells were exposed to Cd2+ for 96 h. Cells were harvested by centrifugation and washed twice with 0.05 M phosphate-buffered saline (pH 7.0) for the following physio-biochemical analysis. Under 1.0 mg L−1 of Cd2+ stress, the contents of chlorophyll and carotenoid, malondialdehyde (MDA), GSH, and ROS were quantified after exposure to Cd2+ for 0 h, 15 min, 24, 48, 72, and 96 h using assay kits (Beijing Solarbio Science &Technology, Beijing; Suzhou Comin Biotechnology, Suzhou, China) according to the manufacturer's instructions. The absorbance of the samples was measured using an ultraviolet spectrophotometer (L5S, Shanghai, China). All sample tests were performed in three biological replicates.2.3 EPS extraction, 3D-EEM, and HPAEC-PAD analyses
When cyanobacteria growth reached the logarithmic phase based on our previous study, Cd2+ was added to the medium, and medium without Cd2+ served as the control. Each treatment was performed in triplicate. The sampling times were 0 h, 15 min, 24, 48, 72, and 96 h. EPS extraction was performed using the methods of Xu et al.,16 all EPS fractions were filtered with 0.45-um membrane filters to remove cell precipitations and other suspended particles.13 The EPS supernatant was stored at −20 °C for further characterization.The 3D-EEM spectra of the EPS solution were recorded using a fluorescence spectrophotometer (F-7000, Hitachi, Japan). The 3D-EEM spectra were collected at 10 nm increments over an excitation range of 200–450 nm with an emission range of 250–550 nm every 5 nm. The excitation and emission slits were set to 2.5 nm of band-pass and the scan speed was set at 1200 nm min−1.17,18 Finally, 3D-EEM data were processed using the Origin 8.0. The measurements were taken in triplicate and the mean values were used for model fitting.
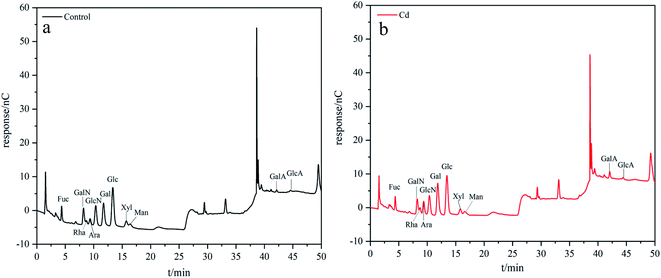
The HPAEC-PAD method was mainly used to analyze the exopolysaccharide components. After EPS extraction, 5 mL of 2 M trifluoroacetic acid was added to the EPS and hydrolyzed at 120 °C for 3 h. Then, the acid hydrolysis solution was accurately absorbed and transferred to a tube for nitrogen blow-drying. Next, 1 mL of water was added, and the solution was vortexed by centrifugation at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 5 min. Finally, remained the supernatant for HPAEC-PAD (ICS5000+ DC, Thermo) analysis.
000 rpm for 5 min. Finally, remained the supernatant for HPAEC-PAD (ICS5000+ DC, Thermo) analysis.
2.4 Batch biosorption experiments
The effects of pH, temperature, initial Cd2+ concentration, and contact time on biosorption efficiency were evaluated.19 The effect of pH was determined at 1.0 mg L−1 Cd2+ and a fixed speed of 180 rpm using solutions with different initial pH values from 2.0 to 8.0 for 3 h. The initial pH was adjusted by adding HCl and NaOH solutions. The effect of temperature was determined at 1.0 mg L−1 Cd2+ and pH 7.0, with temperatures ranging from 20 °C to 40 °C for 3 h of contact time. The initial Cd2+ from 0 to 30 mg L−1 was performed at optimum conditions of pH 7.0 and 30 °C. The effect of contact time was set to 0 h, 5 min, 10 min, 15 min, 30 min, 1 h and 3 h at 1.0 mg L−1 Cd2+ with optimum conditions of pH 7.0 and 30 °C. After biosorption, the cell suspensions were centrifuged at 8000 rpm for 5 min and the supernatant was collected for Cd2+ concentration determination by inductively coupled plasma-optical emission spectrometry (ICP-OES; Optima 5300 DV, PerkinElmer, USA). The biosorption efficiency of Cd2+ was calculated using the following formula:12| Adsorption efficiency (%) = (C0 − Ct)/C0 × 100% |
2.5 Zeta potential measurements
The zero-point charge (pHpzc) was determined by mixing 0.01 g cyanobacteria cells with 0.01 M KCl solution (20 mL), and the pH was adjusted to 1–6 using NaOH or HCl solutions.20 After filtering the adsorbents, the final pH of the solution was determined using a zeta potential analyzer (Zetasizer Nano ZS90, Malvern).2.6 Adsorption kinetics and desorption experiments
The kinetics of adsorption is needed to determine the adsorption velocity.4 Two kinetic models including pseudo-first and -second order kinetic models, were tested to fit the kinetics experimental data. The pseudo-first and -second order kinetic models and formulas are as follows:21| ln(qe − qt) = lnqe − k1t |
where t is the adsorption time (min), qt and qe correspond to the adsorption amount of metal ions (mg g−1) at time t and adsorption equilibrium, and k1 (min−1) and k2 (g mg−1 min−1) correspond to the pseudo-first and -second order adsorption rate constants, respectively.
The desorption study of Cd2+ was performed by treating biosorbents with different desorbents such as 0.1 M EDTA-Na2, 0.1 M HCl, 0.1 M HNO3, 0.1 M NaCl, and distilled water.4,19 The cyanobacteria were first centrifuged at 8000 rpm for 5 min after Cd2+ biosorption (pH 7.0, 30 °C, 180 rpm for 3 h) and the supernatants were retained for Cd2+ concentration detection. Then, the Cd-adsorbed cyanobacterial precipitations were added to 100 mL of the desorbent in 250 mL conical flasks and shaken at 180 rpm, 30 °C for 3 h. Supernatants were detected as described above by ICP-OES after centrifugation.
2.7 FTIR, Raman spectrum, and XPS analyses
The surface functional groups of Synechocystis sp. PCC6803 before and after Cd2+ biosorption were evaluated by FTIR spectroscopy (IS5, Thermo Fisher Scientific, USA) and Raman spectroscopy (HORIBA, LabRAM HR Evolution, France). Cyanobacterial precipitations were obtained by centrifugation at 8000 rpm for 5 min, washing twice with deionized water, freeze-drying, and grinding into powder. Samples (30 mg) were pressed with KBr at a ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 for FTIR analysis, and 50 mg samples were used for Raman spectrum analysis.
100 for FTIR analysis, and 50 mg samples were used for Raman spectrum analysis.
XPS (K-Alpha+, Thermo Fisher Scientific, USA) was used to characterize the oxygen, carbon, and nitrogen valence-electron changes of cyanobacteria before and after Cd2+ adsorption. The binding energies were calibrated using the C1s peak with a fixed binding energy of 284.8 eV to compensate for the surface charging effects.22 Avantage software was used to fit the XPS spectra peaks.
2.8 SEM and TEM-EDX analyses
After 15 min, 24, 48, 72, and 96 h of adsorption with or without 1.0 mg L−1 Cd2+, the cyanobacteria cells were used for SEM analysis, cells were fixed with 2.5% glutaraldehyde at 4 °C for 12 h, washed with phosphate buffer (0.05 M, pH 7.0) then dehydrated with 30%, 50%, 70%, 90% and 100% ethanol, finally, freeze-dried samples were coated with gold as reported by Zhang et al.23 The morphology of the cells was observed using scanning electron microscope (MIRA3, Tescan) coupled with EDX analysis (Oxford X-Max 20, UK).Cyanobacteria cells before and after 1.0 mg L−1 Cd2+ adsorption were used for TEM analysis, sample preparation was performed according to standard procedures,24 including cell fixation, dehydration, embedding, ultrathin sectioning, and dyeing. The distribution and morphology of Cd2+ on the surface and interior of the cells before and after adsorption were characterized using transmission electron microscopy (H-7650, Hitachi).
2.9 Real-time quantitative PCR
Three genes related to Cd2+ adsorption and one gene involved in exopolysaccharides synthesis were selected to calculate expression levels by RT-qPCR. After 15 min, 24, 48, 72, and 96 h of stress with or without 1.0 mg L−1 Cd2+, the cyanobacterial suspensions were used to extract total RNA. Total RNA was isolated from the control and stress groups using a plant RNA extraction kit (Tiangen, China). Specific primers for RT-qPCR were designed using Primer Premier 5.0, synthesized by Tsingke Biological Technology (Changsha, China), and the gap1 gene was used as an internal control gene14 (Table 1). The cDNA synthesized by HiScript II Q RT SuperMix for qPCR containing a gDNA wiper (Vazyme, China) was used as a template for RT-qPCR. RT-qPCR was carried out on a Real-Time PCR Detection Systerm (Gentier 48R, China) with ChamQ Universal SYBR Master Mix (Vazyme, China) according to the manufacturer's instructions. Three experiments were conducted to ensure the accuracy of the results. Relative expression levels were calculated using the 2−ΔΔCT method.25| Gene ID | Annotation name | Forward primer (5′ to 3′) | Reverse primer (5′ to 3′) | Product (bp) |
|---|---|---|---|---|
| MYO_18125 | CusA/CzcA family heavy metal efflux RND transporter | TGGTGACGGAACGGCTAC | TGATGGGAGAAGATACAGGAG | 87 |
| MYO_11210 | TrkA family potassium uptake protein | CAGGAGGATGCCCTAAAT | TCTGTGGTGCTGATAATGTG | 140 |
| MYO_14270 | Cadmium-translocating P-type ATPase | CGTGCCTTGGTTCTGTTG | GCCGTGAGGGAGTCTAAAA | 140 |
| MYO_05725 | Exopolysaccharide biosynthesis protein | TTCTTTCCCTCCCTTCGG | AATGCCTTGGGCCTTGC | 167 |
| gap1 | Reference gene | GCTACCGCAGTTAAAAGGTA | GTAACTCGTGGCTTTTTCC | 106 |
2.10 Statistical analysis
All experiments were performed in triplicate, and the data are expressed as the mean ± standard error for each group. Statistical analyses were performed with one-way analysis of variance (ANOVA), and differences were considered significant at p-value <0.05 (SPSS 20.0 version).3. Results and discussion
3.1 Effect of Cd2+ stress on the growth of Synechocystis sp. PCC6803
To investigate the tolerance of Synechocystis sp. PCC6803 to different Cd2+ concentrations, the growth performance of cyanobacteria under Cd2+ stress was investigated (Fig. 1). The cells grew well in the control group, while the growth curve lagged and the growth rate decreased gradually with increasing Cd2+ concentrations during the 12 day culture period. When the Cd2+ concentration was 1.0 mg L−1, cell biomass did not increase with the extension of cultivation time. The highest biomass yield (OD730) substantially declined to 0.011 when exposed to 2.0 mg L−1 Cd2+, indicating that the inhibitory effect of Cd2+ on cyanobacteria growth was dose-dependent. According to chemical-algae growth inhibition tests,26 the minimal inhibitory concentration (MIC) calculated was 1.18 mg L−1 for Synechocystis sp. PCC6803. The subsequent experiments were all carried out at 1.0 mg L−1 Cd2+ concentration.3.2 Effects of Cd2+ stress on the physiology and biochemistry of Synechocystis sp. PCC6803
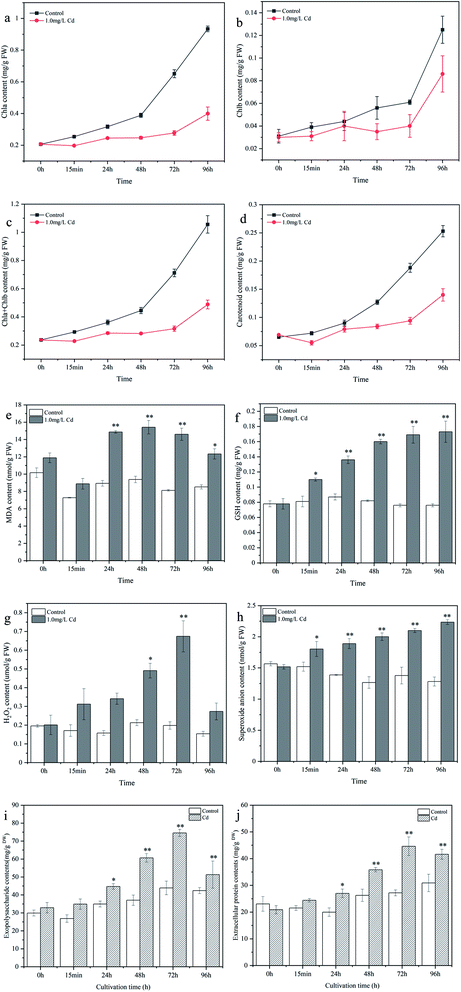
Physiological and biochemical analyses were performed to elucidate the effect of Cd2+ stress on cyanobacterial pigments and oxidative stress tolerance, as shown in Fig. 2. In general, the change trends of chlorophyll a, chlorophyll b, and carotenoid were similar. As the culture time increased, the chlorophyll and carotenoid contents increased in the control, while pigment synthesis was significantly inhibited after Cd2+ stress. The chlorophyll a, chlorophyll b, and chlorophyll a+b contents of Cd2+ stressed samples were 57.28%, 38.13% and 53.79% lower than those in the control at 96 h, respectively (Fig. 2a–c). The carotenoid content in the Cd2+ stressed samples decreased by 44.66% compared to that of the control at 96 h (Fig. 2d). Compared with the control, MDA content was the highest at 48 h (64.41% increase) and decreased thereafter (Fig. 2e). GSH content increased gradually and the highest content was observed at 96 h (1.277-fold increase) compared to that of the control (Fig. 2f). H2O2 content peaked at 72 h (2.404-fold increase) and then induced (Fig. 2g). The O2− content increased gradually and peaked at 96 h (74.34% increase) (Fig. 2h).Cd2+ stress can damage the membrane lipids, proteins and nucleic acids of cyanobacteria cell and disturb the homeostasis of the organism, especially the accumulation of ROS. The increase in H2O2 and O2− levels may be attributed to the excessive accumulation of ROS.27 MDA is the end product of lipid peroxidation, and an increase in MDA content could reflect the degree of lipid peroxidation.28 As a product of sulfur metabolism, GSH is the precursor of phytochelatins, which can chelate with heavy metal ions to reduce Cd2+ toxicity.29 The increase in GSH was responsible for cyanobacteria resisting heavy metal stress. Chlorophyll is closely related to the photosynthetic reaction, and the decrease in chlorophyll content may be due to the distortion of the chlorophyll ultrastructure, inhibition of the synthesis of photosynthetic pigments and enzymes.30 These results revealed that Cd2+ can inhibit cyanobacteria photosynthesis and cause cell oxidative damage, and that GSH played an important role in interacting with Cd2+ and relieving metal toxicity. Consequently, the determination results revealed the toxic effects of Cd2+ on cyanobacteria and the physiological response mechanism of cyanobacteria in response to Cd2+ stress, providing a reference basis for the better application of cyanobacteria in heavy metal wastewater treatment.
3.3 Content changes of EPS before and after Cd2+ adsorption
The changes in EPS contents under Cd2+ stress are depicted in Fig. 2i and j. Extracellular proteins and exopolysaccharides increased by 13.432% and 29.862% (15 min), 35.105% and 28.013% (24 h), 36.459% and 63.688% (48 h), 64.107% and 69.703% (72 h), and 34.711% and 20.997% (96 h) compared to control group, respectively. Besides, the contents of exopolysaccharides were higher than that of extracellular proteins during the cultivation period, indicating that the EPS of Synechocystis sp. PCC6803 was mainly composed of exopolysaccharides, Teng et al.31 also found that the concentrations of protein in EPS were basically less than that of polysaccharides after Pb(II) stress, indicating that polysaccharides was main components of EPS in Leclercia adecarboxylata.Extracellular protein and exopolysaccharide contents showed an obvious increase in the first 72 h, and then decreased slowly at 96 h (still higher than the control) compared to 72 h. To resist the cellular damage caused by Cd2+, microorganisms tend to secrete EPS as a self-protection strategy against abiotic stress.31,32 Li et al.33 found that the adsorption of Ni(II) by EPS extracted from AGS and AnGS were 65.77 and 54.18 mg g−1, respectively, suggesting that EPS can make a certain contribution for heavy metal removal from aqueous environment in sludge biosorption process. Ozturk et al.15 also observed a significant and regular increase in EPS production of Synechocystis sp. BASO67l by Cr(VI), Cd(II) and Cr(VI) + Cd(II) stress at a concentration of 35 ppm. Extracellular protein and exopolysaccharides were rich in many functional groups, which provide numerous binding sites for metal ions, thus reducing the toxicity to the cells. In addition, as the concentration of free Cd2+ in the culture medium decreases, Cd2+ stress become weaker, the secretion of EPS is reduced slowly. Therefore, the results obtained in this investigation also showed that the secretion of EPS in Synechocystis sp. PCC6803 was regulated by the pressure of heavy metal ions.
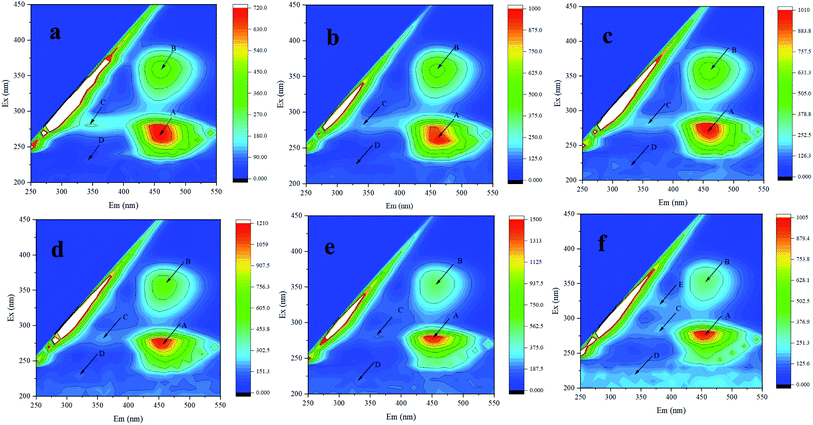 | ||
| Fig. 3 3D-EEM of Synechocystis sp. PCC6803 exposed to Cd2+. (a–f) represent control, 15 min, 24, 48, 72, and 96 h treatment, respectively. | ||
Previous study have found that fluorescence of humic-like and protein-like fluorophores in Synechocystis sp. could be statically quenched by Cu(II), indicating formation of Cu(II)–EPS complexes, confirmed by 3D-EEM.37 Therefore, it was speculated that biochemical reaction happened between Cd2+ and EPS and the increase of extracellular protein was related to heavy metal resistance. In addition, after Cd2+ adsorption, various fluorescence peaks have different degrees of red shift (longer wavelengths) or blue shift (shorter wavelengths), which may be related to the increase or reduction of functional groups, such as hydroxyl, carboxyl, carbonyl, and amino groups.38
The contents of exopolysaccharides changed after Cd2+ adsorption, which could be explained by Cd2+ stress potentially effect the expression of exopolysaccharides genes. Therefore, we used RT-qPCR to futher verify the function of exopolysaccharides during Cd2+ adsorption process. The expression level of the gene related to exopolysaccharides synthesis was evaluated by RT-qPCR (Fig. 9d). The results of RT-qPCR indicated that the expression level of the exopolysaccharides synthesis gene was up-regulated and most significant at 72 h compared to control, which suggested that the exopolysaccharide gene may participate in regulating the adsorption of heavy metals. Shen et al.14 also found that the gene expression of exoD, which is found to be related to EPS production, rose 9.31 times after Cd2+ exposure for 3 days compared with the initial inoculation in Synechocystis sp. PCC6803. These experimental results demonstrated that exopolysaccharides secreted by cyanobacteria participated in the adsorption and resistance of Cd2+ and alleviated toxicity caused by heavy metals.
3.5 Effect of pH, temperature, initial Cd2+ concentration, and contact time on adsorption
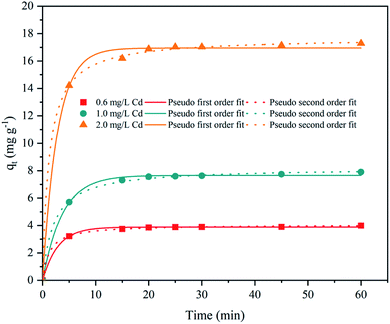
Those findings implied that the optimal adsorption conditions for cyanobacteria were pH 7.0, 30 °C, contact time of 15 min, and an initial Cd2+ concentration of 4.0 mg L−1. The biosorption of Cd2+ by Synechocystis sp. PCC6803 is a complicated process controlled by many environmental variables such as pH, temperature, contact time, and initial Cd2+ concentration. Through the adsorption experiment, we can further understand the effect of heavy metals on the adsorption behavior of Synechocystis sp. PCC6803.
| Adsorbent | Initial ion (mg L−1) | First order kinetic model | Second order kinetic model | ||||
|---|---|---|---|---|---|---|---|
| qe | K1 | R2 | qe | K2 | R2 | ||
| Synechocystis sp. PCC6803 | 0.6 | 3.884 | 0.351 | 0.9977 | 4.066 | 0.190 | 0.9996 |
| 1.0 | 7.660 | 0.269 | 0.9973 | 8.213 | 0.058 | 0.9989 | |
| 2.0 | 16.950 | 0.362 | 0.9970 | 17.712 | 0.046 | 0.9995 | |
Cd2+ released by NaCl can be regarded as the fraction adsorbed by ion exchange, while the fraction released by EDTA-Na2 was considered to be adsorbed by complexation.44 Cd2+ can also bind weakly by physical trapping, which can be easily desorbed by H2O.45 For HCl and HNO3, H+ can compete with Cd2+ for the binding sites on the adsorbent surface, resulting in the release of Cd2+. Suitable desorbents can make the adsorbents efficiently reused during the metal treatment process. Ghoneim et al.46 also observed that HCl has the highest recovery efficiency for cadmium in Ulva lactuca. The desorption experiments suggested that the regeneration of the biosorbents was possible for repeated use with regard to Cd2+.
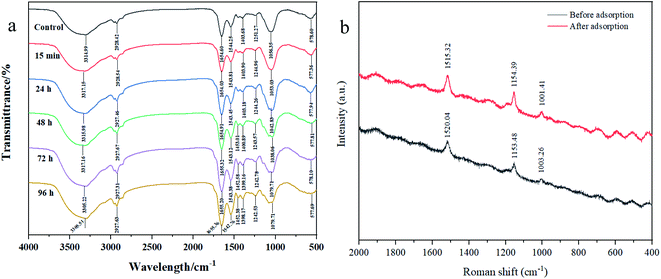
3.7 FTIR, Raman spectrum, and XPS analyses
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O in the carboxyl group.22,23 The peak at 1242.53–1251.27 cm−1 was associated with the stretching vibration of P
O in the carboxyl group.22,23 The peak at 1242.53–1251.27 cm−1 was associated with the stretching vibration of P![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O and the band at 1038.06–1079.71 cm−1 was assigned to the stretching vibration of C–O–C in uronic acid.22,23 Thereafter, these functional groups, such as hydroxyl, carboxyl, methyl, and aldehyde groups on cell wall or EPS that involved in the removal of cadmium by complexation with them.
O and the band at 1038.06–1079.71 cm−1 was assigned to the stretching vibration of C–O–C in uronic acid.22,23 Thereafter, these functional groups, such as hydroxyl, carboxyl, methyl, and aldehyde groups on cell wall or EPS that involved in the removal of cadmium by complexation with them.
Zhang et al.23 also used FTIR and found the adsorption mechanism of Cd2+ by Burkholderia cepacia GYP1 was mainly through complexation of functional groups with Cd2+. It was also reported that hydroxyl, carbonyl, carboxyl, and amino groups in Cupriavidus pauculus 1490 that contributed a major role under the simulation of Cd(II), Ni(II), Cu(II) and Co(II).50 Therefore, FTIR results demonstrated that these functional groups may participate in the Cd2+ adsorption process or play a major role in cyanobacteria resistance to heavy metals.
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C, C–C, and phenylalanine.51,52 In addition, the peak intensities after adsorption were significantly higher than those of the control. This indicated that C
C, C–C, and phenylalanine.51,52 In addition, the peak intensities after adsorption were significantly higher than those of the control. This indicated that C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C, C–C, and amino acid side chain (phenylalanine) exerted a prominent function in the Cd2+ adsorption process. Similar results were found in Synechococcus PCC7002, Yu et al.53 found the C–C stretching and C–H2 deformation after Cs(I) adsorption by Raman spectrum.
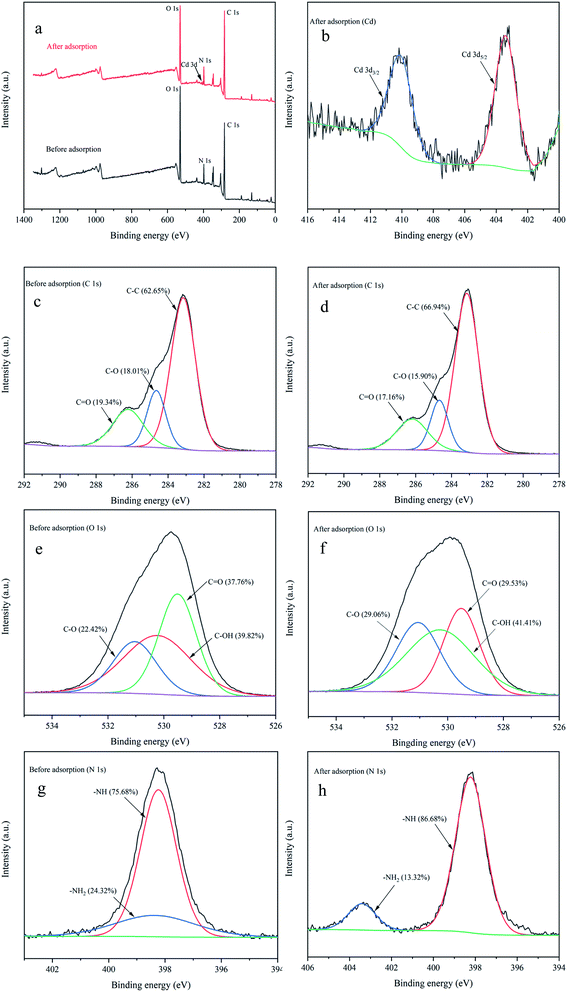
C, C–C, and amino acid side chain (phenylalanine) exerted a prominent function in the Cd2+ adsorption process. Similar results were found in Synechococcus PCC7002, Yu et al.53 found the C–C stretching and C–H2 deformation after Cs(I) adsorption by Raman spectrum.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O.54 After adsorption, the atomic content of C–C increased from 62.63% to 66.94%, and C–O (18.01%) and C
O.54 After adsorption, the atomic content of C–C increased from 62.63% to 66.94%, and C–O (18.01%) and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O (19.34%) decreased to 15.90% and 17.16%, respectively. Fig. 7e and f show the high-resolution O 1s spectrum. Three peaks detected (529.51, 530.23, and 531.04 eV) were assigned to C
O (19.34%) decreased to 15.90% and 17.16%, respectively. Fig. 7e and f show the high-resolution O 1s spectrum. Three peaks detected (529.51, 530.23, and 531.04 eV) were assigned to C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, C–OH, and C–O.54,55 After adsorption, the atomic content of C
O, C–OH, and C–O.54,55 After adsorption, the atomic content of C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O decreased from 37.76% to 29.53%, and C–OH (39.82%) and C–O (22.42%) increased to 41.41% and 29.06%, respectively. The high-resolution N 1s spectrum (Fig. 7g and h) could be divided into two peaks including –NH (398.23 eV) and –NH2 (398.35 eV),56,57 and the atomic content of –NH increased from 75.68% to 86.68%, while –NH2 decreased from 24.32% to 13.32% after adsorption. Thus the positions and atomic contents of these functional groups that transformed after Cd2+ adsorption, may be attributed to the complexation between metal ions and functional groups.
O decreased from 37.76% to 29.53%, and C–OH (39.82%) and C–O (22.42%) increased to 41.41% and 29.06%, respectively. The high-resolution N 1s spectrum (Fig. 7g and h) could be divided into two peaks including –NH (398.23 eV) and –NH2 (398.35 eV),56,57 and the atomic content of –NH increased from 75.68% to 86.68%, while –NH2 decreased from 24.32% to 13.32% after adsorption. Thus the positions and atomic contents of these functional groups that transformed after Cd2+ adsorption, may be attributed to the complexation between metal ions and functional groups.
The results were consistent with previous reports, Xie et al.22 used XPS and found that oxygen atom in protein carboxyl C–O might be the key site responsible for the EPS–Cd(II) complexation in Chlorella vulgaris. Based on XPS spectra analysis, Song et al.57 also reported that the high adsorbability of Aspergillus niger was greatly due to a large number of nitrogen- and oxygen-containing functional groups on the surface of mycelia, which can easily form complexes with Co(II) and Eu(III). Thus XPS results in this research indicated that C–C, C–O, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, C–OH, –NH, and –NH2 contributed major sites responsible for heavy metal adsorption.
O, C–OH, –NH, and –NH2 contributed major sites responsible for heavy metal adsorption.
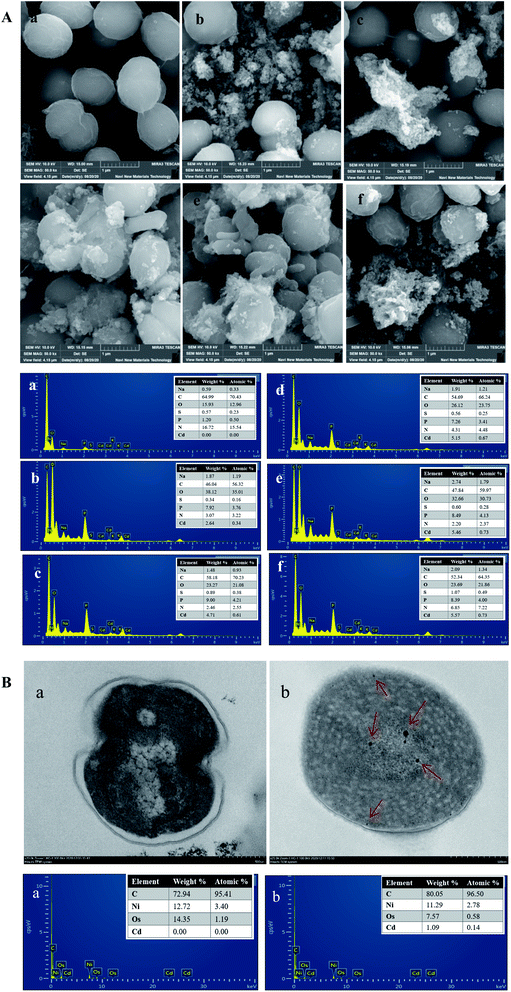
3.8 SEM and TEM-EDX analyses
The surface morphology of cyanobacteria before and after Cd2+ adsorption was observed using SEM (Fig. 8A). Before Cd2+ adsorption, the cells exhibited integrated spherical structures, surface smoothness, and have specific dimensions. After adsorption, the surfaces of the cells were rough with some attachments aggregated on the surface. This could be due to Cd2+ aggregation around the cell surface and linked to the functional groups of EPS secreted by cyanobacteria cells, which was consistent with the results of a previous study. Ghoneim et al.46 studied the surface morphology of Ulva lactuca before and after Cd2+ adsorption by SEM and found that Cd2+ precipitated around the cell surface. In this study, EDX analysis confirmed that Cd2+ accumulated on the cell surface.TEM was mainly conducted to observe the spatial location of heavy metals adsorbed on the surface or interior of the cells. Significant changes were observed after Cd2+ adsorption, as shown in Fig. 8B. The cell walls of cyanobacteria in the control group were smooth and complete, and the cytoplasm was relatively uniform, while the cell walls under Cd2+ stress were irregular and the various positions, sizes, and shapes of the metal particle aggregations were observed both the cell surface and intracellularly, indicating that Cd2+ could cross the membrane by other mechanisms and was not merely fixed on the cell surface, which was similar to the results of previous studies.19,23 Therefore, it is possible that two mechanisms existed in the Cd2+ adsorption process. Cd2+ was rapidly adsorbed on the cell surface by EPS and prevented it from entering the cells. In addition, Cd2+ may enter the cytoplasm through the cytomembrane by transport systems. The EDX energy spectrum showed that Cd2+ entered the intracellular region.
3.9 The expression of genes involved in Cd2+ adsorption
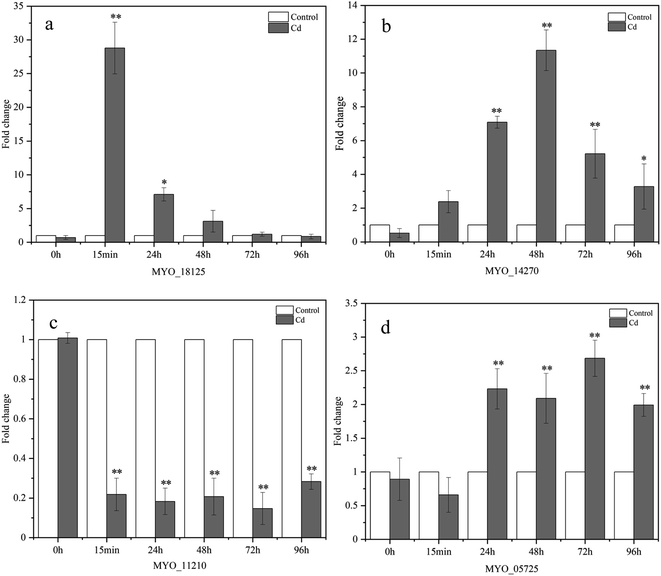
In this research, RT-qPCR was performed to further investigate the expression profiles of genes involved in Cd2+ adsorption at the genetic level. The expression levels of these four genes are shown in Fig. 9. The expression of the MYO_18125 gene was up-regulated by 27.779, 6.098, 2.112, and 0.189 times at 15 min, 24, 48, and 72 h, respectively, and the most significant increase occurred at 15 min compared with the control. MYO_18125 gene encodes the CzcA family heavy metal efflux RND transporter, the high expression of the gene is possibly because of transporting Cd2+ to extracellular space to reduce toxicity.58 The MYO_14270 gene was up-regulated separately by 1.379, 6.086, 10.342, 4.218, and 2.275 times at five time points, and the most significant up-regulation appeared at 48 h compared to the control. MYO_14270 gene encodes cadmium-translocating P-type ATPase (CadA), which could prevent Cd2+ from re-entering the cells once metal efflux is complete, and high expression of this gene may be in response to its detoxification of Cd2+. Furthermore, the gene expression levels of MYO_18125 and MYO_14270 increased initially and then decreased, up-regulation may be because the sites on the adsorbent's surface were sufficient and the Cd2+ concentration was high. The decrease in expression levels may be attributed to the low free Cd2+ concentration in the medium, heavy metal stress became weaker, and saturation of adsorption sites. In addition, The up-regulation of MYO_18125 gene was faster than that of MYO_14270 gene, and the maximum up-regulation multiple of MYO_18125 gene was 2.686-fold that of MYO_14270 gene, it was speculated that CzcA efflux system might play a major role in metal detoxification in Synechocystis sp. PCC6803. Nongkhlaw et al.59 found that CzcA expression in Chryseobacterium sp. PMSZPI was 2-fold higher compared to CadA expression after 0.5 mM Cd2+ stress for 3 h, indicating that CzcA efflux system might play a major role in Cd2+ tolerance. The MYO_11210 gene encodes TrkA potassium uptake protein and the expression of the MYO_11210 gene was down-regulated at all time points, which was contrary to the previous studies, Chi et al.12 found that the expression of the TrkA gene after cadmium adsorption was up-regulated compared to that of the control in Bacillus paranthracis NT1. In this study, down-regulation can be speculated that the presence of Cd2+ may have inhibited potassium uptake protein activity. The expression of the MYO_05725 gene was up-regulated separately by 1.232, 1.091, 1.684, and 0.993 times at 24, 48, 72, and 96 h compared to the control, and the most significant increase occurred at 72 h. The MYO_05725 gene encodes the exopolysaccharide synthesis protein. The results of the gene expression levels were consistent with those of the content changes of exopolysaccharides. The high expression of this gene was probably because cyanobacteria secreted more exopolysaccharides to absorb heavy metals.The results of RT-qPCR reflected that MYO_18125, MYO_14270, and MYO_05725 gene may participate in the regulation process of heavy metal adsorption, which preliminarily revealed the molecular regulation mechanism of Synechocystis sp. PCC6803 in response to Cd2+ adsorption. While the adsorption mechanism of heavy metals by cyanobacteria is a complex process, it may be regulated by a series of genes involved in growth and metabolism, heavy metal resistance, and transmembrane transport. Therefore, there are still many genes involved in Cd2+ adsorption that need to further identify. In addition, based on the results of RT-qPCR, genetic engineering technology can be applied to the treatment of heavy metals by cyanobacteria. Constructing mutants by silencing or over-expressing some genes related to Cd2+ adsorption and resistance, thus improving the adsorption efficiency of cyanobacteria to heavy metals.
4. Conclusion
In this study, Synechocystis sp. PCC6803 was used to evaluate the Cd2+ biosorption behavior and mechanism. The adsorption process was affected by many factors, including pH, temperature, initial Cd2+ concentration, and contact time. The adsorption process was fast and followed the pseudo-second order kinetic model, mainly based on chemical adsorption. The functional groups complexed with Cd2+ were mainly hydroxyl, amino, carbonyl, and carboxyl groups. GSH played an important role in alleviating Cd2+ toxicity as a component of the antioxidant system. In addition, EPS produced by Synechocystis sp. PCC6803 had significant impacts on Cd2+ adsorption by combining with heavy metals. SEM and TEM confirmed that Cd2+ can enter the cell, not only enriched on the cell surface. Moreover, RT-qPCR revealed the potential molecular regulatory mechanisms of Synechocystis sp. PCC6803 in the Cd2+ biosorption process. These results could provide a foundation for further investigation of the adsorption mechanism of heavy metals by other strains and provide theoretical guidance for the development of late-model biosorbents.Conflicts of interest
The authors declare no actual or potential conflict of interest.Acknowledgements
This work was supported by the National Natural Science Foundation of China (No. 52074353), South China Key Laboratory of Applied Microbiology Open Fund Project (SKLAM006-2018), GDAS′ Special Project of Science and Technology Development (2020GDASYL-20200402001), Fundamental Research Funds for the Central Universities of Central South University (2020zzts725), Hunan Provincial Innovation Foundation for Postgraduate (CX20200323). State Key Laboratory of Complex Nonferrous Metal Resources Clean Utilization, Kunming University of Science and Technology (CNMRCUKF2109).References
- W. Huang and Z. M. Liu, Colloids Surf., B, 2013, 105, 113–119 CrossRef CAS PubMed.
- T. Jiao, X. H. Chu, Z. Q. Gao, T. T. Yang, Y. Liu, L. Yang, D. Z. Zhang, J. L. Wang, B. P. Tang, K. Wu, Q. N. Liu and L. S. Dai, Ecotoxicol. Environ. Saf., 2019, 182, 109388 CrossRef CAS PubMed.
- Z. Xu, M. Dong, X. Peng, W. Ku, Y. Zhao and G. Yang, Ecotoxicol. Environ. Saf., 2019, 171, 301–312 CrossRef CAS PubMed.
- I. Vishan, B. Saha, S. Sivaprakasam and A. Kalamdhad, Environ. Technol. Innovation, 2019, 14, 100323 CrossRef.
- Y. K. Leong and J. S. Chang, Bioresour. Technol., 2020, 303, 122886 CrossRef CAS PubMed.
- J. He and J. P. Chen, Bioresour. Technol., 2014, 160, 67–78 CrossRef CAS PubMed.
- M. Jena, D. Pradhan and T. Das, Int. J. Environ. Waste Manage., 2012, 9, 221–231 CrossRef CAS.
- E. N. Bakatula, E. M. Cukrowska, I. M. Weiersbye, L. Mihaly-Cozmuta, A. Peter and H. Tutu, J. Geochem. Explor., 2014, 144, 492–503 CrossRef CAS.
- D. Zhang, X. Pan, K. M. Mostofa, X. Chen, G. Mu, F. Wu, J. Liu, W. Song, J. Yang, Y. Liu and Q. Fu, J. Hazard. Mater., 2010, 175, 359–365 CrossRef CAS PubMed.
- J. N. Nkoh, R. K. Xu, J. Yan, J. Jiang, J. Y. Li and M. A. Kamran, Environ. Pollut., 2019, 247, 136–145 CrossRef CAS PubMed.
- Y. Zhang, X. Wang, T. Shan, S. Pang and N. Xu, Marine Genomics, 2019, 47, 100671 CrossRef PubMed.
- Y. Chi, Y. Huang, J. Wang, X. Chen, S. Chu, K. Hayat, Z. Xu, H. Xu, P. Zhou and D. Zhang, Sci. Total Environ., 2020, 741, 140422 CrossRef CAS PubMed.
- S. Naveed, Q. Yu, C. Zhang and Y. Ge, Environ. Pollut., 2020, 261, 114233 CrossRef CAS PubMed.
- L. Shen, Z. Li, J. Wang, A. Liu, Z. Li, R. Yu, X. Wu, Y. Liu, J. Li and W. Zeng, Environ. Sci. Pollut. Res., 2018, 25, 20713–20722 CrossRef CAS PubMed.
- S. Ozturk, B. Aslim, Z. Suludere and S. Tan, Carbohydr. Polym., 2014, 101, 265–271 CrossRef CAS PubMed.
- H. Xu, H. Cai, G. Yu and H. Jiang, Water Res., 2013, 47, 2005–2014 CrossRef CAS PubMed.
- L. Zhu, J. Zhou, M. Lv, H. Yu, H. Zhao and X. Xu, Chemosphere, 2015, 121, 26–32 CrossRef CAS PubMed.
- X. Pan, J. Liu and D. Zhang, Colloids Surf., B, 2010, 80, 103–106 CrossRef CAS PubMed.
- T. Lan, Y. Feng, J. Liao, X. Li, C. Ding, D. Zhang, J. Yang, J. Zeng, Y. Yang, J. Tang and N. Liu, J. Environ. Radioact., 2014, 134, 6–13 CrossRef CAS PubMed.
- L. Fang, C. Zhou, P. Cai, W. Chen, X. Rong, K. Dai, W. Liang, J. D. Gu and Q. Huang, J. Hazard. Mater., 2011, 190, 810–815 CrossRef CAS PubMed.
- Y. G. Bermúdez, I. L. R. Rico, O. G. Bermúdez and E. Guibal, Chem. Eng. J., 2011, 166, 122–131 CrossRef.
- Q. Xie, N. Liu, D. Lin, R. Qu, Q. Zhou and F. Ge, Environ. Pollut., 2020, 263, 114102 CrossRef CAS PubMed.
- J. Zhang, Q. Li, Y. Zeng, J. Zhang, G. Lu, Z. Dang and C. Guo, Ecotoxicol. Environ. Saf., 2019, 176, 162–169 CrossRef CAS PubMed.
- W. Shi, G. Zhang, F. Li, J. Feng and X. Chen, Sci. Total Environ., 2020, 742, 140606 CrossRef CAS PubMed.
- T. D. Schmittgen and K. J. Livak, Nat. Protoc., 2008, 3, 1101–1108 CrossRef CAS PubMed.
- C. General, OECD Guidelines for the Testing of Chemicals, 2006, 1, 1 Search PubMed.
- Q. L. Zhu, S. N. Guo, F. Wen, X. L. Zhang, C. C. Wang, L. F. Si, J. L. Zheng and J. Liu, Aquat. Toxicol., 2019, 207, 153–162 CrossRef CAS PubMed.
- H. X. Li, Y. Xiao, L. L. Cao, X. Yan, C. Li, H. Y. Shi, J. W. Wang and Y. H. Ye, PLoS One, 2013, 8, e73380 CrossRef CAS PubMed.
- D. Laporte, N. Valdes, A. Gonzalez, C. A. Saez, A. Zuniga, A. Navarrete, C. Meneses and A. Moenne, Aquat. Toxicol., 2016, 177, 433–440 CrossRef CAS PubMed.
- C. S. Seth, P. Kumar Chaturvedi and V. Misra, Ecotoxicol. Environ. Saf., 2008, 71, 76–85 CrossRef CAS PubMed.
- Z. Teng, W. Shao, K. Zhang, Y. Huo, J. Zhu and M. Li, Chem. Eng. J., 2019, 375, 122113 CrossRef CAS.
- B. Ma, Z. Li, S. Wang, Z. Liu, S. Li, Z. She, N. Yu, C. Zhao, C. Jin, Y. Zhao, L. Guo and M. Gao, Environ. Pollut., 2019, 251, 81–89 CrossRef CAS PubMed.
- N. Li, D. Wei, S. Wang, L. Hu, W. Xu, B. Du and Q. Wei, J. Colloid Interface Sci., 2017, 490, 754–761 CrossRef CAS PubMed.
- J. Hou, Y. Yang, P. Wang, C. Wang, L. Miao, X. Wang, B. Lv, G. You and Z. Liu, Environ. Sci. Pollut. Res., 2017, 24, 226–235 CrossRef CAS PubMed.
- G. P. Sheng and H. Q. Yu, Water Res., 2006, 40, 1233–1239 CrossRef CAS PubMed.
- W. Chen, P. Westerhoff, J. A. Leenheer and K. Booksh, Environ. Sci. Technol., 2003, 37, 5701–5710 CrossRef CAS PubMed.
- X. Pan, J. Liu, W. Song and D. Zhang, Front. Environ. Sci. Eng., 2012, 6, 493–497 CrossRef CAS.
- L. Zhu, H. Y. Qi, M. L. Lv, Y. Kong, Y. W. Yu and X. Y. Xu, Bioresour. Technol., 2012, 124, 455–459 CrossRef CAS PubMed.
- M. Alyassin, G. M. Campbell, H. Masey O'Neill and M. R. Bedford, Food Chem., 2020, 315, 126221 CrossRef CAS PubMed.
- F. Çolak, N. Atar, D. Yazıcıoğlu and A. Olgun, Chem. Eng. J., 2011, 173, 422–428 CrossRef.
- M. Aryal and M. Liakopoulou-Kyriakides, Environ. Monit. Assess., 2015, 187, 4173 CrossRef PubMed.
- S. E. Oh, S. H. A. Hassan and J. H. Joo, World J. Microbiol. Biotechnol., 2009, 25, 1771–1778 CrossRef CAS.
- X. Ma, W. Cui, L. Yang, Y. Yang, H. Chen and K. Wang, Bioresour. Technol., 2015, 185, 70–78 CrossRef CAS PubMed.
- J. Bai, X. Yang, R. Du, Y. Chen, S. Wang and R. Qiu, J. Environ. Sci., 2014, 26, 2056–2064 CrossRef PubMed.
- X. Li, D. Li, Z. Yan and Y. Ao, RSC Adv., 2018, 8, 30902–30911 RSC.
- M. M. Ghoneim, H. S. El-Desoky, K. M. El-Moselhy, A. Amer, E. H. Abou El-Naga, L. I. Mohamedein and A. E. Al-Prol, Egypt. J. Aquat. Res., 2014, 40, 235–242 CrossRef.
- P. B. Vilela, C. A. Matias, A. Dalalibera, V. A. Becegato and A. T. Paulino, J. Environ. Chem. Eng., 2019, 7, 103327 CrossRef.
- P. M. Thabede, N. D. Shooto, T. Xaba and E. B. Naidoo, J. Environ. Chem. Eng., 2020, 8, 104045 CrossRef CAS.
- Z. Zhang, K. Yan, L. Zhang, Q. Wang, R. Guo, Z. Yan and J. Chen, J. Hazard. Mater., 2019, 374, 420–427 CrossRef CAS PubMed.
- W. Zeng, S. Zhang, M. Xia, X. Wu, G. Qiu and L. Shen, RSC Adv., 2020, 10, 20385–20394 RSC.
- C. E. E. Grace, P. K. Lakshmi, S. Meenakshi, S. Vaidyanathan, S. Srisudha and M. B. Mary, Spectrochim. Acta, Part A, 2020, 224, 117382 CrossRef CAS PubMed.
- M. Tang, G. D. McEwen, Y. Wu, C. D. Miller and A. Zhou, Anal. Bioanal. Chem., 2013, 405, 1577–1591 CrossRef CAS PubMed.
- R. Yu, H. Chai, Z. Yu, X. Wu, Y. Liu, L. Shen, J. Li, J. Ye, D. Liu, T. Ma, F. Gao and W. Zeng, Microorganisms, 2020, 8, 491 CrossRef CAS PubMed.
- X. Xie, X. Zhao, X. Luo, T. Su, Y. Zhang, Z. Qin and H. Ji, Chin. J. Chem. Eng., 2020 DOI:10.1016/j.cjche.2020.06.003.
- J. J. Ojeda, M. E. Romero-González, R. T. Bachmann, R. G. Edyvean and S. A. Banwart, Langmuir, 2008, 24, 4032–4040 CrossRef CAS PubMed.
- S. J. Yuan, M. Sun, G. P. Sheng, Y. Li, W. W. Li, R. S. Yao and H. Q. Yu, Environ. Sci. Technol., 2011, 45, 1152–1157 CrossRef CAS PubMed.
- W. Song, J. Liang, T. Wen, X. Wang, J. Hu, T. Hayat, A. Alsaedi and X. Wang, Chem. Eng. J., 2016, 304, 186–193 CrossRef CAS.
- X. Xia, S. Wu, Z. Zhou and G. Wang, J. Hazard. Mater., 2021, 401, 123685 CrossRef CAS PubMed.
- M. Nongkhlaw and S. R. Joshi, PLoS One, 2019, 14, e0216995 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1ra02366g |
| This journal is © The Royal Society of Chemistry 2021 |