Open Access Article
Open Access ArticleNetwork of gold conjugates for enhanced sensitive immunochromatographic assays of troponins†
Nadezhda A. Taranova,
Vladislav D. Slobodenuyk,
Anatoly V. Zherdev and
Boris B. Dzantiev
and
Boris B. Dzantiev *
*
A.N. Bach Institute of Biochemistry, Research Center of Biotechnology of the Russian Academy of Sciences, Leninsky Prospect 33, 119071 Moscow, Russia. E-mail: dzantiev@inbi.ras.ru
First published on 4th May 2021
Abstract
Highly sensitive detection of cardiac troponins I and T (cTnI and cTnT) was completed by immunochromatography with double amplification, through the binding of functionalized gold nanoparticles (GNPs). The robust nature of the approach, based on the formation of nanoparticle networks through the biotin–streptavidin interaction, was confirmed; the choice of the best assay parameters for maximal increase in ICA sensitivity was demonstrated. A bifunctional conjugate of GNPs with biotinylated specific IgG and two auxiliary conjugates, GNP–biotin and GNP–streptavidin, form three-component aggregates in the analytical zone of the test strip. The inclusion of abundant gold labels in the resulting immune complex leads to an amplified colorimetric signal. The limits of detection (LoDs) of cTnI and cTnT were 0.9 and 0.4 ng mL−1, respectively, which is 3 times lower than the LoDs of more commonly used systems. Visual LoDs were 10-fold lower in concentration. The enhancement has been realized both in single and double assay formats; analysis of cTnI and cTnT presented the same characteristics.
1. Introduction
Among modern analytical methods, immunochromatographic analysis (ICA) is widely used due to its efficiency, ease-of-use outside of a laboratory and capacity to allow for visual assessment of results.1 ICA begins with the pre-application of reagents at several zones on multimembrane test strips, followed by the movement of the liquid along the membranes after contact with the sample, resulting in all analyte-revealing reactions.1 The presence of an analyte in a sample is characterized by coloration in the analytical zone of the test strip. The result of the analysis may be estimated visually (qualitatively) or by a portable detector (quantitatively).2 While ICA allows for a simple implementation of a rapid assay, it is less sensitive than other, instrumental methods of analysis (such as immunoenzyme assay and chromatography), and therefore is limited in its capacity to detect extremely low concentrations of analytes.3–5Various approaches have been proposed to increase ICA sensitivity.6–8 One approach is to replace the most commonly used label, gold nanoparticles (GNPs), with alternatives (fluorophores, magnetic nanoparticles, etc.).1,9,10 Another approach is to use the post-assay growth of GNPs bound in the analytical zone.11,12 However, such methods require the inclusion of additional solutions in assay kits. The required manipulations with these reagents complicate and lengthen the analytical process.
A promising alternative method is to increase the number of GNPs in the analytical zone, resulting in more intense coloration. The use of small-sized functionalized nanoparticles and their aggregation to the assay will limit the constraints for lateral flow and nonspecific binding.13,14 For the formation of aggregates, intermolecular interactions such as biotin–streptavidin, antibody–anti-species-antibody and antibody–antigen have been considered.15,16 The ease of implementation and the fact that additional steps or washing are not required are advantages of this approach. Our prior study successfully implemented this enhancement based on the mixture of the three types of functionalized nanoparticles for the detection of procalcitonin.17 However, its applicability for other antigens remains unstudied and the mechanisms that provide increased sensitivity were not determined.
This study demonstrates the robust nature of the proposed method through its application in the determination of cardiac troponins I (cTnI) and T (cTnT). The immunochemical measurement of biomarkers released to the blood is the ‘gold standard’ for diagnosing myocardial infarction.18 Cardiac troponins are efficient biomarkers for this purpose.19 In this study, we propose a method of choosing the optimal ratio of functionalized GNP conjugates in the test system for effective signal amplification in ICA. We also demonstrate the potential to apply this enhancement method in a multiplex test system for the simultaneous determination of cTnI and cTnT.
2. Experimental procedure
2.1. Chemicals and materials
Mouse monoclonal antibodies of troponin I (Ab1/cTnI and Ab2/cTnI) and troponin T (Ab3/cTnT and Ab4/cTnT), recombinant troponin I, and troponin T were purchased from Bialexa (Moscow, Russia). The specificity of the antibodies was confirmed by the absence of cross-reactions with other blood components.20 Goat anti-mouse IgG antibodies were purchased from Arista Biologicals (Allentown, PA, USA). Bovine serum albumin (BSA), streptavidin (Stp), biotin-NHS ester (biotin), biotinamidohexanoyl-6-aminohexanoic acid N-hydroxysuccinimide ester (AH-biotin), NHS-d-(polyethylene glycol)4-biotin (PEG-biotin), Tween-20 and sodium azide were purchased from Sigma (St. Louis, MO, USA). Gold(III) chloride was purchased from Fluka (Buchs, Switzerland).Other chemicals (such as solvents and analytically pure salts) were purchased from Chimmed (Moscow, Russia). All solutions for synthesis and assay were prepared using water which was purified by the Sartorius arium® pro system (Sartorius, Germany).
The immunochromatographic test system was made using Mdi Easypack (Advanced Microdevices, India) membrane kits, which include a working CNPC nitrocellulose membrane with a 15 μm pore size, PT-R7 glass fibre membrane, GFB-R4 separation membrane and an AP045 adsorption membrane.
2.2. Abs and BSA biotinylation
The Abs and BSA were biotinylated as described by Hermanson.21 During the synthesis, the biotin![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Abs ratio was 15
Abs ratio was 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, and the biotin
1, and the biotin![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) BSA ratio was 8
BSA ratio was 8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. The synthesized preparations were stored at −20 °C.
1. The synthesized preparations were stored at −20 °C.
2.3. Preparation of GNPs and their conjugates with proteins
GNPs were prepared by citrate reduction of HAuCl4 to Au0, as described by Hermanson.21 The GNP conjugates with Stp, biotinylated BSA, and native and biotinylated Abs2 were obtained through physical adsorption (see Section 3.1).21 After 0.5 h of incubation at room temperature, 10% BSA or BSA–biotin water solution was added (v![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) v = 40
v = 40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1). The reaction mixture was centrifuged, the supernatant was discarded and the precipitate (GNPs with adsorbed molecules) was dissolved and stored at 4 °C.
1). The reaction mixture was centrifuged, the supernatant was discarded and the precipitate (GNPs with adsorbed molecules) was dissolved and stored at 4 °C.
2.4. Spectral characteristics of conjugates
To measure optical absorption of the ternary complexes, solutions of GNP conjugates were first mixed in the appropriate ratio. Absorption spectra were recorded on an EnSpire Multimode Plate Reader (PerkinElmer, Boston, Massachusetts, USA) and the wavelengths of the absorption maximum were then observed. Cuvette path length was 1 cm. The molar extinction coefficient equal to (2.93 ± 0.02) × 109 M−1 cm−1 was earlier determined for spherical nanoparticles of comparable size.222.5. Fabrication of immunochromatographic tests
The glass fibre was treated with the following: for the common test system, membranes were coated with the GNPs conjugates with A520(cTnI) = 5.0 and A520(cTnT) = 2.0; for the triple-test system, one membrane was coated with GNP–Abs–biotin (A520(cTnI) = 5.0 and A520(cTnT) = 2.0) and GNP–BSA–biotin conjugates (A520 = 5.0 for cTnI and A520 = 4.0 for cTnT) and a second membrane was coated with the GNP–Stp conjugate (A520 = 1.0 for cTnI and A520 = 0.5 for cTnT). The load of each conjugate solution was 32 μL per cm of membrane length. After dispensing, the membranes were left to dry at room temperature for at least one day.Using an IsoFlow dispenser (Imagene Technology, Hanover, NH, USA), Abs1/cTnI or Abs3/cTnT antibodies (2 mg mL−1, forming the analytical zone) and goat anti-mouse IgG (1 mg mL−1, forming the control zone) were applied in phosphate-buffered solution (PBS) onto the nitrocellulose membrane. The load was 1.2 μL of both solutions per cm of membrane length. The nitrocellulose membrane was then dried at room temperature for at least one day.
The resulting membranes were assembled, and the sheets were cut into 3.5 mm wide strips. The sheets were split and packed in plastic and laminated aluminium foil at 20–22 °C, with relative humidity under 30%.
2.6. Immunochromatographic assay and data processing
The commercial troponin-free serum TruLab N (DiaSys Diagnostic Systems GmbH, Germany) was used. Stock solutions of antigens were prepared using recombinant troponins from Bialexa (Moscow, Russia), and working solutions were prepared by diluting the stock solutions in the TruLab N serum.The test strip was vertically submerged in a sample for 15 min at room temperature. The binding of the label in the analytical zone was then recorded using the Epson Perfection V600 Photo scanner (Epson, Suwa, Japan) and processed using TotalLAB software (Nonlinear Dynamics, Newcastle, UK).
The visual limit of detection (vLoD) was defined as the minimum analyte concentration at which the coloured line in the analytical zone could be observed visually. Instrumental LoD was defined as the analyte concentration at which the analytical zone coloration intensity exceeds the standard deviation for background coloration of the analytical zone by three times (i.e., for samples without cTns).
3. Results and discussion
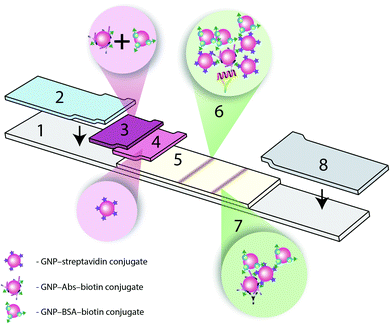
The amplification of colorimetric signal in ICA via biotin–Stp interaction of GNP–Abs–biotin and GNPs–Stp conjugates was completed. As a result of the interaction, the aggregates with the composition GNP–Abs–biotin–GNP–Stp–GNP–BSA–biotin were formed. Three variants of immunochromatographic test systems were created:(a) A common test system using only GNP–IgG conjugate,20
(b) A triple test system (Fig. 1) with a detecting GNP–IgG–biotin conjugate and two amplifying GNP–Stp and GNP–biotin conjugates, and
(c) A triple test system for simultaneous detection of two troponins (duplex assay), described above.
3.1. Choosing and obtaining reactants for immunochromatography
Anti-cTn antibodies were characterized by ELISA (Fig. S1 and S2; Tables S1 and S2†). Combinations of antibodies on the microplate surface and labelled antibodies with the lowest LoD were chosen, namely the IC19 (immobilized)–IC4 (detected) pair with an LoD value of cTnI of 0.66 ng mL−1, and the 7G7 (immobilised)–1F11 (detected) pair with an LoD value of cTnT of 1.7 ng mL−1.The synthesized GNPs had a diameter of 29.3 ± 0.9 nm, according to transmission electron microscopy data (Fig. S3†). Using Abs/cTnI IC4, Abs/cTnT 1F11, BSA–biotin and Stp, the GNP–Abs–biotin, GNP–BSA–biotin, GNP–Stp and GNP–Abs conjugates were prepared by physical adsorption. This method is characterized by easy manipulation and high efficiency. During the syntheses, the concentration of GNPs matched with their optical density at 520 nm (A520) of 1.0. The flocculation curves were used to select protein concentrations.21 (Data on the effect of the antibody concentration on the optical properties of the conjugates are given in Fig. S4†). The optimal concentrations of all antibodies and Stp were 10 μg mL−1. The concentration of the BSA–biotin conjugate was 50 μg mL−1, with a large excess. The size characteristics for all synthesized conjugates are recorded in the ESI (Fig. S3†).
3.2. Common immunochromatographic test system
Test system development included choosing concentrations of immunoreagents and conjugates and the mediums for their interactions. The main criterion was ensuring minimum LoDs for cTnT and cTnI. The optical density of the GNP–Ab2/cTnI and GNP–Ab4/cTnT conjugates varied from 1 to 5 optical units. The chosen optimal values were A520 = 5.0 for the GNP–Ab2/cTnI conjugate and A520 = 2.0 for the GNP–Ab4/cTnT conjugate (Table S3†).The evaluation of the effect of the concentration of antibodies immobilized in the analytical zone showed an optimal concentration of 2 mg mL−1 for both systems (see Table S4† as an example for cTnI).
To ensure complete elution of the GNP conjugates applied to the glass fibre membrane, uniform movement of the serum along the test strip and the absence of non-specific interactions, the Tween-20 detergent was added to the pre-processed sample. Within the tested range of 0.1%–2.0% (v/v), its optimum concentration was 1.0%. Higher concentrations of detergent caused coloration with the absence of antigens (Table S5†).20
The common test system with the only GNP–Abs/cTnI conjugate reached visual and photometric LoDs of 11 ng mL−1 and 3.7 ng mL−1, respectively (Fig. 2A and B). In the case of cTnT ICA, the visual and photometric LoDs reached were 11 ng mL−1 and 3.7 ng mL−1, respectively (Fig. 2C).
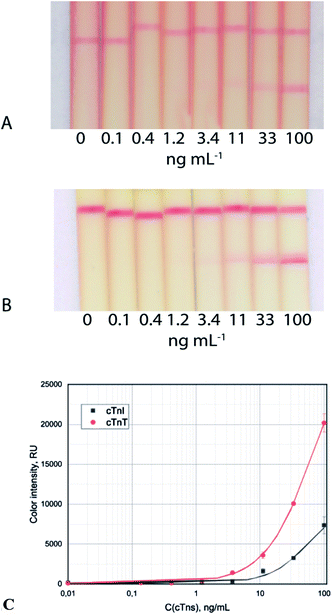 | ||
| Fig. 2 Test strip images of cTnI (A) and cTnT (B) detection in serum and calibration curves (C) for the common immunochromatographic test system. | ||
3.3. Triple immunochromatographic test system
The formation of the GNP aggregates in the analytical zone, resulting from the biotin–Stp reaction, was used for amplification. The conjugate of GNP–biotin was an additional amplifying reagent.17 For the GNPs to precipitate, the particles must have multiple binding sites.23,24 However, it is very difficult to determine the best ratio for the formation of a ternary complex and a subsequent increase in the analytical signal.Optimizing the triple immunochromatographic test system required:
• Studying the influence of the length of the spacer between protein and biotin
• Assessing the effect of the concentration of GNP–Stp on the formation of the GNP–Abs–biotin complex
• Choosing the GNP–Stp![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) GNP–Abs–biotin
GNP–Abs–biotin![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) GNP–BSA–biotin ratio using spectral data
GNP–BSA–biotin ratio using spectral data
The first parameter affecting aggregate formation is the length of the spacer between protein and biotin that influences the availability for biotin's interaction with Stp. Abs/cTnT 1F11 and BSA were biotinylated with biotin, AH-biotin or PEG4-biotin with a succinimide reaction.
Table 1 shows the relationship between LoDs and the length of the spacer in the biotin ester molecule. The minimum LoD for cTnT was achieved using a spacer of maximum length.
| Conjugate | Biotin | AH-biotin | PEG4-biotin |
| Spacer length, Å | 13.5 | 30.5 | 29 |
| M, g mol−1 | 341.4 | 567.7 | 588.7 |
| LoDs, ng mL−1 | 1.2 | 0.4 | 3.9 |
Fewer charged groups in the AH-biotin spacer (generally a neutral charge) allow biotin to remain mobile and available for binding to streptavidin. PEG4-biotin contains a large number of negatively charged oxy groups that can interact with positively charged amino groups of the protein and thereby reduce the distance of biotin from the protein molecule. The use of a spacer of a different nature or greater length seems promising for further study of the process of aggregation of functionalized nanoparticles.
The potential for GNP–Stp and GNP–Abs/cTnT conjugates to aggregate in a two-component system was assessed using a spectral analysis. With varying concentrations of GNP–Stp and GNP–Abs/cTnT–biotin conjugates, an increase in optical density occurred without a shift in the absorption peak. Fig. S5† shows the absence of this shift at different concentrations of the GNP–Stp conjugate, which indicates that there is an absence of a double complex (aggregation). In all cases, the absorption peak was recorded at 535 nm. For further analysis, the optical density of the GNP–Stp conjugate was selected at 0.5.
To achieve a spectral shift, it is necessary to increase the amount of biotin. The GNP–BSA–biotin conjugate was therefore introduced. The maximal displacement of the absorption peak (i.e. the formation of aggregates)25,26 and the maximum optical density was observed for the following concentrations of conjugates: A(GNP–Abs–biotin) = 2.0 and A(GNP–BSA–biotin) = 4.0 (Fig. 3, arrow), which corresponds to a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 molar ratio of the conjugates. Neither lower nor higher concentrations of the GNP–BSA–biotin conjugate led to the formation of a comparable number of aggregates, thus reducing the desired enhancement (see Table 2).
2 molar ratio of the conjugates. Neither lower nor higher concentrations of the GNP–BSA–biotin conjugate led to the formation of a comparable number of aggregates, thus reducing the desired enhancement (see Table 2).
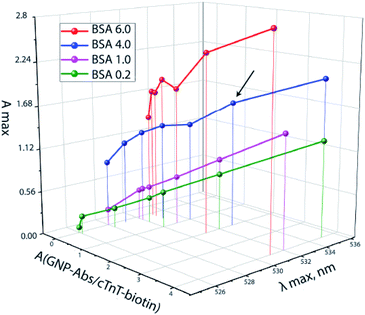 | ||
| Fig. 3 The ratio of GNP–Abs/cTnT–biotin and GNP–BSA–biotin at A(GNP–Stp) = 0.5 and the resulting absorption maxima of a mixture of conjugates. | ||
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) GNP–BSA–biotin
GNP–BSA–biotin![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) GNP–Abs/cTnT–biotin conjugates and shift of the adsorption peak and the resulting LoDs
GNP–Abs/cTnT–biotin conjugates and shift of the adsorption peak and the resulting LoDs
Molar ratios GNP–Stp![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) GNP–BSA–biotin GNP–BSA–biotin![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) GNP–Abs/cTnT–biotin GNP–Abs/cTnT–biotin |
Peak, nm | LoDs (cTnT), ng mL−1 |
|---|---|---|
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.4 0.4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 4 |
526 | 7.5 |
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 2 |
530 | 3.9 |
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 8 8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 4 |
529 | 0.4 |
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 8 8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 8 8 |
528 | 1.4 |
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 16 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 4 |
531 | 4.2 |
According to the resulting data, the optimal molar ratio of the GNP–Stp![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) GNP–BSA–biotin
GNP–BSA–biotin![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) GNP–Abs/cTnT–biotin conjugates for the formation of ternary aggregates was 1
GNP–Abs/cTnT–biotin conjugates for the formation of ternary aggregates was 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 8
8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4. In the next stage, we chose the optimal concentration of the GNP–Str conjugate in the three-component mixture. Based on the size of the proteins and GNPs,27,28 the minimum and maximum amounts of GNP–Stp conjugate needed for complete binding of all available biotin groups were calculated (see Table S6†).29 The following assumptions were made for the calculations: (a) 90% of the protein is adsorbed on the GNP surface,30,31 and (b) after the adsorption of Stp, each Stp molecule binds only 3 biotin molecules because 1 binding site is blocked by the surface of the nanoparticle. The minimum optical density of the GNP–Stp conjugate (proteins form a monolayer on the surface of nanoparticles upon adsorption) was 0.25, and the maximum (with the adsorption of the entire amount of protein used for the synthesis) – 0.9. The concentration of GNP–Stp varied, ranging from 0 to 1.1 optical units (Fig. S6†).
4. In the next stage, we chose the optimal concentration of the GNP–Str conjugate in the three-component mixture. Based on the size of the proteins and GNPs,27,28 the minimum and maximum amounts of GNP–Stp conjugate needed for complete binding of all available biotin groups were calculated (see Table S6†).29 The following assumptions were made for the calculations: (a) 90% of the protein is adsorbed on the GNP surface,30,31 and (b) after the adsorption of Stp, each Stp molecule binds only 3 biotin molecules because 1 binding site is blocked by the surface of the nanoparticle. The minimum optical density of the GNP–Stp conjugate (proteins form a monolayer on the surface of nanoparticles upon adsorption) was 0.25, and the maximum (with the adsorption of the entire amount of protein used for the synthesis) – 0.9. The concentration of GNP–Stp varied, ranging from 0 to 1.1 optical units (Fig. S6†).
The data indicate that the maximum coloration of the analytical zone is achieved at A(GNP–Stp) of 0.5. Lower amounts of GNP–Stp do not allow the binding of a sufficient number of particles containing biotin molecules. Therefore, no aggregates, or only a small number of double aggregates, are formed. An excess of GNP–Str leads to a complete overlap of both GNP–BSA–biotin and GNP–Abs/cTnT–biotin, which leads to the formation of double aggregates, but not triple aggregates.
Table 3 presents the ratio of components in the three-component system, GNP–Str![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) GNP–BSA–biotin
GNP–BSA–biotin![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) GNP–Abs/cTnT–biotin, and the resulting LoDs. The results show that the minimum detection limit with minimal background coloration is observed when the molar ratio of the components of the detecting mixture is 1
GNP–Abs/cTnT–biotin, and the resulting LoDs. The results show that the minimum detection limit with minimal background coloration is observed when the molar ratio of the components of the detecting mixture is 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 8
8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4. Similar data for cTnI are presented in Table 4. The results also show that the minimum detection limit with minimal background coloration is observed when the molar ratio of the components of the detecting mixture is 1
4. Similar data for cTnI are presented in Table 4. The results also show that the minimum detection limit with minimal background coloration is observed when the molar ratio of the components of the detecting mixture is 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10
10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10. Concentrations of GNP conjugates with specific antibodies were previously selected (common systems) and are constant. Additional increase in GNP–BSA–biotin leads to an increase in the background signal, arising from the unreacted components.
10. Concentrations of GNP conjugates with specific antibodies were previously selected (common systems) and are constant. Additional increase in GNP–BSA–biotin leads to an increase in the background signal, arising from the unreacted components.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) GNP–BSA–biotin
GNP–BSA–biotin![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) GNP–Abs/cTnT–biotin and the resulting LoDs
GNP–Abs/cTnT–biotin and the resulting LoDs
| Ratios | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 2 |
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 8 8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 4 |
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 8 8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 8 8 |
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 16 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 4 |
| LoD, ng mL−1 | 3.9 | 0.4 | 1.4 | 4.2 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) GNP–BSA–biotin
GNP–BSA–biotin![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) GNP–Abs/cTnI–biotin and the resulting LoD
GNP–Abs/cTnI–biotin and the resulting LoD
| Ratios | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 10 |
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 10 |
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 7 7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 10 |
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 10 |
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5 5 |
| LoD, ng mL−1 | 1.5 | 1.1 | 1.0 | 0.9 | 4.1 |
The optimal ratios of GNP–Stp![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) GNP–BSA–biotin
GNP–BSA–biotin![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) GNP–Abs–biotin conjugates for cTnT and cTnI is different because the optimal concentrations of the specific conjugates (GNP–Abs–biotin) is initially different. A significant increase in the concentration of the conjugate leads to an increase in the background signal.
GNP–Abs–biotin conjugates for cTnT and cTnI is different because the optimal concentrations of the specific conjugates (GNP–Abs–biotin) is initially different. A significant increase in the concentration of the conjugate leads to an increase in the background signal.
No difference was observed in the detection time between the common and triple systems (Fig. S7†). The analysis duration was 15 min. The sharp increase in the intensity of staining of the analytical zone 30 s after the introduction of the sample (2nd point in Fig. S7†) is explained by the passage of the liquid front, which does not carry the coloured marker.
The proposed triple-test system allows for the detection of cTnI with visual and photometric LoDs of 1.2 ng mL−1 and 0.9 ng mL−1, respectively (Fig. 4A). The triple-test system for cTnT detection is characterised by visual and photometric LoDs of 1.2 ng mL−1 and 0.4 ng mL−1, respectively (Fig. 4B). The stability of the developed test systems was shown during long-term storage (within 6 months) at 20–22 °C. The coefficient of variability did not exceed 15%. Evaluation of the specificity of the test systems showed the absence of false-positive results in the presence of 2000 and 5000-fold excess of human serum albumin and human immunoglobulins G, the main protein compounds of serum samples (Table S7†).
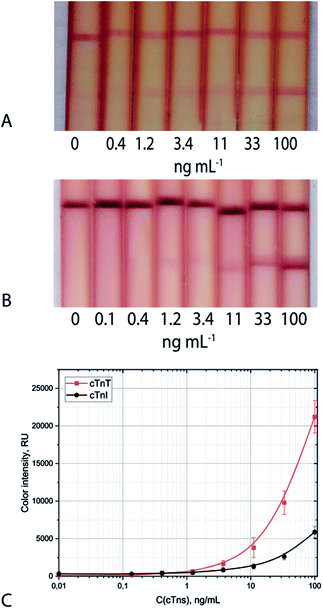 | ||
| Fig. 4 Appearance of test strips and calibration curves (C) for cTnI (A) and cTnT (B) detection in serum: triple system. | ||
Because cTnI and cTnT are parts of the triple troponin complex with troponin C, detecting them separately is complicated.32 This is the reason for the excessively low concentration of troponins in the blood of healthy people (5–6 pg mL−1).33 With myocardial infarction, there is a sharp increase in the concentration of troponins in the blood. Depending on the antigen, a 3–10-fold decrease in the LoDs of troponins was observed using the formation of aggregates in the analytical zone of the test strip. The prevalence of heart attacks resulting in no troponin concentration in the blood and the consequent necessity of adequate treatment for these patients justifies the need for a highly sensitive analytical method of detecting troponins.34,35
An increase in clinical sensitivity in the determination of troponins is achieved due to a decrease in assay specificity, which presents an additional diagnostic problem for clinicians, though LoDs of cTnI could be decreased to 0.05 ng mL−1 (ref. 36) and lower.37 The rapid and accurate detection of low troponin concentrations will allow for efficient diagnoses that may prevent deaths.36
3.4. Triple immunochromatographic system for simultaneous detection of two troponins
This study is the first to present the potential to implement a duplex test for the simultaneous determination of cTnI and cTnT in blood serum (Fig. 5). vLoDs were 1.2 ng mL−1 for cTnI and cTnT for both mixture and individual presence.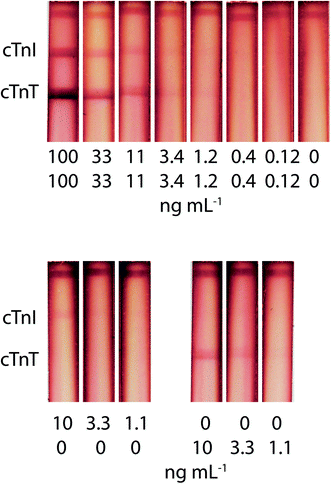 | ||
| Fig. 5 Appearance of test strips and calibration curves for simultaneous cTnI and cTnT detection in serum, using the triple immunochromatographic system. | ||
The so-called troponin-free infarction underscores the need for a multiplex test system for the simultaneous determination of troponins.38 Rapid point-of-care control of troponins will allow for faster and more accurate diagnoses.
The risks of creating an enhanced double test were as follows: an insufficient amount of the GNP–Stp conjugate, the appearance of a high background signal due to an excess of the stained label, and different degrees of amplification when forming a zone closer and further away from the start line (due to different times in the zone flow). Despite the competition for reagents in the aggregate formation, the double test system retains the analytical characteristics of the single test system and presents a 10-fold decrease in LoDs. The absence of interference of antigens in the other analytical zone was experimentally confirmed (Fig. 5). Also, the advantage of this amplification method is the simplicity of its implementation (stored one-stage protocol).
3.5. Utilization of the proposed approach among other aggregation-based enhanced ICAs
Other methods for increasing the sensitivity of ICA using aggregates were discussed in the Introduction. In the ICA of other analytes, increased marker size is commonly used to amplify the signal and to increase sensitivity of the approach. A more advanced approach is combining conventional nanoparticle conjugates and additional reagents, which provide formation of complex aggregates directly during the assay (Table 5).39,40| Interacting compounds | Nanoparticle quantity | Multiassay | Effect | Ref. |
|---|---|---|---|---|
| BSA–Abs/BSA | 2 | No | 10–25-Fold | 16 |
| Stp–biotin | 3 | No | 10-Fold | 17 |
| 3 | Yes | 10-Fold | This work | |
| 2 | No | 30-Fold | 41 | |
| DNA | 2 | No | 10–15-Fold | 42 |
| 2 | No | 30-Fold | 43 | |
| Dendrimer | 2 | No | 20-Fold | 44 |
| Biotin–Abs/biotin | 2 | No | 27-Fold | 45 |
| Anti-species antibodies | 2 | No | 5-Fold | 46 |
The studies discussed in previous sections of this article use mainly two types of nanoparticles to form aggregates, which lead to a decrease in LoDs by a factor of 5–30. In all works, except for the author's, a combination of two types of particles is used. Direct sorption of biotin onto the surface of nanoparticles reduced the LoD by a factor of 30.41 In the ELISA format,15 it was possible to further reduce the sensitivity, but this required the formation of a network of nanoparticles labelled with peroxidase, and the subsequent registration of their enzymatic activity. Due to the strength of the connection, the Stp–biotin pair appears to be a promising tool to enhance the ICA method.41,47 In addition, this study is the first to describe a multiplex test system with amplification of an analytical signal.
4. Conclusions
This study demonstrated the process of increasing the sensitivity of ICA, through the aggregation of three conjugates of GNPs, in order to detect cTnI and cTnT in blood serum. A spectral method for assessing the optimal Stp–biotin ratio in a three-component system is proposed. LoDs were 0.9 and 0.4 ng mL−1, which is 3 times lower in concentration than LoDs of common systems. Visual LoDs were 10-fold lower in concentration. The robustness of the proposed technique for increasing the sensitivity of ICA in a “sandwich” format was shown, and the efficiency of the given amplification in the implementation of multiplex test systems that preserve analytical characteristics was demonstrated. Unlike other amplifying methods, the proposed method does not complicate ICA and does not lengthen the time required for analysis; it is also characterized by high sensitivity.Author contributions
Experimental design: NAT, AVZ and BBD. Experiment performed by: NAT and VDS. Data analysis: NAT and AVZ. Contributed reagents/materials/analysis tools: AVZ. Paper (initial draft) written by: NAT. Paper (final version) written by: NAT, AVZ and BBD.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was financially supported by the Russian Science Foundation (grant no. 20-73-00325).References
- Y. Huang, T. Xu, W. Wang, Y. Wen, K. Li, L. Qian, X. Zhang and G. Liu, Microchim. Acta, 2020, 187, 1–25 CrossRef PubMed.
- W. Xiao, C. Huang, F. Xu, J. Yan, H. Bian, Q. Fu, K. Xie, L. Wang and Y. Tang, Sens. Actuators, B, 2018, 266, 63–70 CrossRef CAS PubMed.
- B. B. Dzantiev, N. A. Byzova, A. E. Urusov and A. V. Zherdev, Trends Anal. Chem., 2014, 55, 81–93 CrossRef CAS.
- S. K. Fischer, K. Williams, L. Wang, E. Capio and M. Briman, Bioanalysis, 2019, 11, 1777–1785 CrossRef CAS PubMed.
- V. Yelleswarapu, J. R. Buser, M. Haber, J. Baron, E. Inapuri and D. Issadore, Proc. Natl. Acad. Sci. U. S. A., 2019, 116, 4489–4495 CrossRef CAS PubMed.
- L. Liu, D. Yang and G. Liu, Biosens. Bioelectron., 2019, 136, 60–75 CrossRef CAS PubMed.
- H. Ye and X. Xia, J. Mater. Chem. B, 2018, 6, 7102–7111 RSC.
- A. V. Zherdev and B. B. Dzantiev, Rapid Test—Advances in Design, Format and Diagnostic Applications, ed. L. Anfossi, 2018, pp. 9–43 Search PubMed.
- R. Banerjee and A. Jaiswal, Analyst, 2018, 143, 1970–1996 RSC.
- X. Gong, J. Cai, B. Zhang, Q. Zhao, J. Piao, W. Peng, W. Gao, D. Zhou, M. Zhao and J. Chang, J. Mater. Chem. B, 2017, 5, 5079–5091 RSC.
- M. O. Rodriguez, L. B. Covian, A. C. Garcia and M. C. Blanco-Lopez, Talanta, 2016, 148, 272–278 CrossRef CAS PubMed.
- V.-T. Nguyen, S. Song, S. Park and C. Joo, Biosens. Bioelectron., 2020, 152, 112015 CrossRef CAS.
- J. D. Bishop, H. V. Hsieh, D. J. Gasperino and B. H. Weigl, Lab Chip, 2019, 19, 2486–2499 RSC.
- V. B. Borse, A. N. Konwar, R. D. Jayant and P. O. Patil, Drug Delivery Transl. Res., 2020, 10, 878–902 CrossRef CAS PubMed.
- I.-H. Cho and J. Irudayaraj, Int. J. Food Microbiol., 2013, 164, 70–75 CrossRef CAS PubMed.
- Y. Zhong, Y. Chen, L. Yao, D. Zhao, L. Zheng, G. Liu, Y. Ye and W. Chen, Microchim. Acta, 2016, 183, 1989–1994 CrossRef CAS.
- N. A. Taranova, A. E. Urusov, E. G. Sadykhov, A. V. Zherdev and B. B. Dzantiev, Microchim. Acta, 2017, 184, 4189–4195 CrossRef CAS.
- K. Thygesen, J. S. Alpert, A. S. Jaffe, B. R. Chaitman, J. J. Bax, D. A. Morrow and H. D. White, Circulation, 2018, 138, e618–e651 CrossRef PubMed.
- D. S. Herman, P. A. Kavsak and D. N. Greene, Am. J. Clin. Pathol., 2017, 148, 281–295 CrossRef CAS PubMed.
- N. A. Byzova, A. V. Zherdev, Y. Y. Vengerov, T. A. Starovoitova and B. B. Dzantiev, Microchim. Acta, 2017, 184, 463–471 CrossRef CAS.
- G. T. Hermanson, Bioconjugate Techniques, Academic Press, 2008 Search PubMed.
- X. Liu, M. Atwater, J. Wang and Q. Huo, Colloids Surf., B, 2007, 58, 3–7 CrossRef CAS PubMed.
- K. J. Levinson, M. De Jesus and N. J. Mantis, Infect. Immun., 2015, 83, 1674–1683 CrossRef CAS PubMed.
- C. M. Dundas, D. Demonte and S. Park, Appl. Microbiol. Biotechnol., 2013, 97, 9343–9353 CrossRef CAS PubMed.
- A. Gole and C. J. Murphy, Langmuir, 2005, 21, 10756–10762 CrossRef CAS PubMed.
- N. G. Khlebtsov, L. A. Trachuk and A. G. Mel'nikov, Opt. Spectrosc., 2005, 98, 77–83 CrossRef CAS.
- A. Kuzuya, K. Numajiri, M. Kimura and M. Komiyama, Nucleic Acids Symp. Ser., 2008, 52, 681–682 CrossRef CAS PubMed.
- B. Jachimska and A. Pajor, Bioelectrochemistry, 2012, 87, 138–146 CrossRef CAS PubMed.
- K. Aslan, C. C. Luhrs and V. H. Pérez-Luna, J. Phys. Chem. B, 2004, 108, 15631–15639 CrossRef CAS.
- D. V. Sotnikov, A. N. Berlina, V. S. Ivanov, A. V. Zherdev and B. B. Dzantiev, Colloids Surf., B, 2019, 173, 557–563 CrossRef CAS PubMed.
- D. V. Sotnikov, A. V. Zherdev and B. B. Dzantiev, Int. J. Mol. Sci., 2015, 16, 907–923 CrossRef CAS PubMed.
- M. S. Parmacek and R. J. Solaro, Prog. Cardiovasc. Dis., 2004, 47, 159–176 CrossRef CAS PubMed.
- V. S. Mahajan and P. Jarolim, Circulation, 2011, 124, 2350–2354 CrossRef PubMed.
- D. M. Kimenai, E. B. Janssen, K. M. Eggers, B. Lindahl, H. M. den Ruijter, O. Bekers, Y. Appelman and S. J. Meex, Clin. Chem., 2018, 64, 1034–1043 CrossRef CAS PubMed.
- A. R. Chapman, A. Bularga and N. L. Mills, Circulation, 2020, 141, 1733–1735 CrossRef CAS PubMed.
- D. Lou, L. Fan, Y. Ji, N. Gu and Y. Zhang, Anal. Methods, 2019, 11, 3506–3513 RSC.
- D. H. Choi, S. K. Lee, Y. K. Oh, B. W. Bae, S. D. Lee, S. Kim, Y.-B. Shin and M.-G. Kim, Biosens. Bioelectron., 2010, 25, 1999–2002 CrossRef CAS PubMed.
- D. A. Morrow, Circulation, 2017, 135, 1586–1597 CrossRef PubMed.
- D. Saerens, F. Frederix, G. Reekmans, K. Conrath, K. Jans, L. Brys, L. Huang, E. Bosmans, G. Maes and G. Borghs, Anal. Chem., 2005, 77, 7547–7555 CrossRef CAS PubMed.
- Y. Berdichevsky, R. Lamed, D. Frenkel, U. Gophna, E. A. Bayer, S. Yaron, Y. Shoham and I. Benhar, Protein Expression Purif., 1999, 17, 249–259 CrossRef CAS PubMed.
- Y. Shen and G. Shen, ACS Omega, 2019, 4, 5083–5087 CrossRef CAS.
- C. Ge, L. Yu, Z. Fang and L. Zeng, Anal. Chem., 2013, 85, 9343–9349 CrossRef CAS PubMed.
- G. Y. Shen, S. B. Zhang and X. Hu, Clin. Biochem., 2013, 46, 1734–1738 CrossRef CAS.
- G. Y. Shen, H. Xu, A. S. Gurung, Y. H. Yang and G. D. Liu, Anal. Sci., 2013, 29, 799–804 CrossRef CAS PubMed.
- J. Si, J. Li, L. Zhang, W. Zhang, J. Yao, T. Li, W. Wang, W. Zhu, J. P. Allain and Y. Fu, J. Med. Virol., 2019, 91, 1301–1306 CrossRef CAS PubMed.
- L. Dou, B. Zhao, T. Bu, W. Zhang, Q. Huang, L. Yan, L. Huang, Y. Wang, J. Wang and D. Zhang, Anal. Bioanal. Chem., 2018, 410, 3161–3170 CrossRef CAS PubMed.
- G. U. Lee, D. A. Kidwell and R. J. Colton, Langmuir, 1994, 10, 354–357 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1ra02212a |
| This journal is © The Royal Society of Chemistry 2021 |