Open Access Article
Open Access ArticleStochastic biosensors based on N- and S-doped graphene for the enantioanalysis of aspartic acid in biological samples†
Raluca-Ioana Stefan-van Staden *ab,
Damaris-Cristina Gheorgheab,
Ruxandra-Maria Ilie-Mihai*a,
Lucian-Barbu Tudoranc and
Stela Maria Pruneanuc
*ab,
Damaris-Cristina Gheorgheab,
Ruxandra-Maria Ilie-Mihai*a,
Lucian-Barbu Tudoranc and
Stela Maria Pruneanuc
aLaboratory of Electrochemistry and PATLAB, National Institute of Research for Electrochemistry and Condensed Matter, 202 Splaiul Independentei Str., 060021, Bucharest-6, Romania. E-mail: ralucavanstaden@gmail.com; i.ruxandra04@yahoo.com
bFaculty of Applied Chemistry and Material Science, Politehnica University of Bucharest, Bucharest, Romania
cNational Institute for Research and Development of Isotopic and Molecular Technologies, Donat Street, 67-103, Cluj-Napoca, Romania
First published on 1st July 2021
Abstract
Metabolomics plays a very important role in cancer mechanism and also in the early diagnosis of cancer. The enantioanalysis of aminoacids found in biological samples may favor the development of new screening tests for the early diagnosis of cancer. Two stochastic biosensors, based on the immobilization of α-hemolysin and hemin in exfoliated reduced graphene modified with nitrogen and sulphur, were proposed for the enantioanalysis of aspartic acid in biological samples (whole blood, urine, saliva, and tissue). The proposed biosensors showed low limits of quantification, high sensitivity, and large linear concentration ranges. The recoveries of D- and L-aspartic acid in biological samples were higher than 95.00%, with a relative standard deviation lower than 1.00%.
1. Introduction
Metabolic and metabolomics in gastric cancer play an important role in identifying new biomarkers such as amino acids1–5 Cancer metabolism is based on central carbon metabolism, including glycolysis and the tricarboxylic acid cycle (the citric acid cycle, TCA cycle). Be that as it may, new surveys have revealed the significant part of amino acids in cancer metabolism. While the L-enantiomer of the chiral amino acids is usually found in the human body, the D-enantiomer may occur while the racemization process takes place or by the twisting of the DNA.Enantioanalysis is essential when the determination of aspartic acid (Asp) is requested by the medical practitioner, because the presence of both L-Asp and D-Asp, or the absence of D-Asp may indicate different diseases. While the concentration of L-Asp is rising to μmol L−1 magnitude order for the patients with gastric cancer, the concentration of D-Asp is also rising to the same level, or higher, especially for patients having diseases like neuroendocrine-related diseases.4 High plasma levels of aspartic acid were seen in old patients or patients with neurodegenerative disorders.3,6 The changes in the serum levels of amino acids seen in patients with gastric cancer may be firmly connected with the dietary requests of tumor cells; i.e., tumor movement requires a high supplement take-up.7
To date, different methods were proposed for the analysis of Asp.8 These methods include: thin layer chromatography,9 gas chromatography,10 high performance liquid chromatography,11 chiral ligand-exchange capillary electrophoresis,12 spectrophotometry,13 fluorimetry,14 and electrochemistry.15 Due to the simplicity and low cost of an electrochemical method, it is necessary to develop an electrochemical system that is both highly selective and sensitive for the aspartic acid investigation. The simplicity of electrochemical techniques mixed with the small size of the apparatus and cheap instrumentation enhanced procedures, such as voltammetry, potentiometry and electrochemical impedance spectroscopy (EIS) are profoundly appealing for the location of bioactive substances.16 In comparison with these mentioned methods, electrochemical sensors are more commonly used, due to the fact that their cost is low and they can be used with ease.17
To date, stochastic sensors have been used for the early diagnosis of different types of diseases, such as diabetes.18–20 The response of the stochastic sensors is based on the channel conductivity: the first stage is the one in which the molecule enters the channel, and the current drops to zero (qualitative analysis is performed during this stage, the measured toff value being known as the signature of the analyte); during the second stage, redox and binding processes take place inside the channel (the measured value of ton is correlated with the concentration of the analyte). The advantages of using the stochastic sensors versus other electrochemical sensors are: they can perform a reliable qualitative analysis by being able to identify specific analytes, and even enantiomers; the quantitative analysis is also reliable, the quantification being performed inside the channel found in the powders used for the design of the sensor.18–20
In this paper, two stochastic biosensors based on α-hemolysin and hemin, immobilized in an exfoliated graphene doped with nitrogen and sulphur were proposed for the enantioanalysis of aspartic acid in biological samples: whole blood, urine, saliva, and tissue. The selection of α-hemolysin and hemin for the design of the stochastic sensors is due to the structures of α-hemolysin and hemin; α-hemolysin and hemin present the necessary pores needed for obtaining the stochastic sensing. The conductivity of the material used as matrix (graphene material): selection of N- and S-dopped graphene increased the conductivity of graphene, and also the signal/noise ratio for the stochastic sensors; no stochastic signal was obtained for the enantioanalysis of Asp when we modified plane graphene with the same biomolecules. Going from graphite material – which proved in the laboratory to be the most difficult matrix for stochastic sensors due to the instability of channels (due to the van der Waals forces taking place between the graphene layers of graphite), the graphene material was far more easy to use because of high stability of the signal, and higher reproducibility of the measurements compared with those obtained using the graphite pastes.
2. Materials and methods
2.1. Materials and reagents
D- and L-aspartic acid, ammonium thiocyanate (NH4SCN), and ammonia were bought from Sigma Aldrich (Milwaukee, USA). Paraffin oil was bought from Fluka (Buchs, Switzerland).Standard solutions of aspartic acid 10−3 mol L−1, buffered with phosphate buffer, pH = 7.50 were prepared. The solutions were diluted to 10−16 mol L−1 using a serial dilution method.
At the end of the exfoliation process, the material collected in the reaction vessel was washed with large amounts of water (10 L), filtered with paper to remove the large particles and finally dried by lyophilization. According to the elemental analysis, the sample had 3.99 wt% nitrogen and 1.06 wt% sulphur, and was denoted E-NSGr.
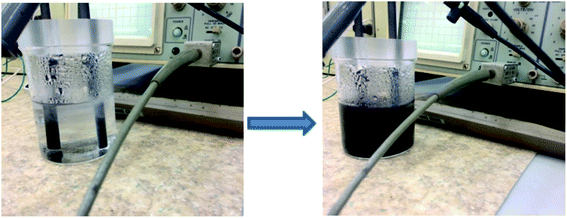
The successful integration of hetero atoms into the graphene generally requires high temperature, therefore it is of paramount importance to develop facile approaches towards the synthesis of doped graphene materials. In our case, the electrolyte based on NH4SCN and ammonia will generate ions by dissociation, such as NH4+ and SCN−. The ions will be electrochemically attracted towards the cathode respectively the anode, leading to the exfoliation of graphite. We suggest that the network defects formed during exfoliation may keep the ions trapped within the graphene layers, forming the NS-doped graphene.
2.2. Apparatus and methods
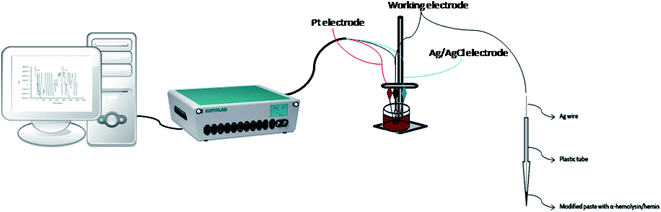
All measurements were performed using an AUTOLAB/PGSTAT 302 (Metrohm, Utrecht, The Netherlands), connected to a computer on which the GPES software was installed; the computer was used to record all the measurements. In order to obtain the electrochemical set-up, three electrodes were used: the counter electrode, represented by a platinum wire, the working electrode, which in our case was represented by the stochastic biosensor and the Ag/AgCl wire, which represents the reference electrode (0.1 mol L−1 KCl). For this method, the values of toff and ton are required, therefore, a chronoamperometric method was used; moreover, a constant potential of 125 mV was applied.The morphology of the graphene sample was investigated using the Scanning Electron Microscopy (SEM) technique (SU-8230 STEM system, Hitachi, Japan). The UV-Vis spectrum of graphene sample ultrasonically dispersed in distilled water (0.05 mg mL−1) was recorded with a SPECORD 250 PLUS, Analytic Jena (Germany). The FTIR spectrum of graphene embedded in the KBr pellet was recorded with a Bruker Tensor II spectrometer (Germany) while the Raman spectrum with a SOLAR TII spectrophotometer coupled with an Olympus IX71 microscope. The elemental analysis of the E-NSGr sample was obtained with an Elemental Analyzer—FLASH EA 1112 (Italy).
A Christ alpha 1-4 LSC freeze dryer was used for the lyophilization of the sample.
2.3. Design of the stochastic biosensors
The two stochastic biosensors were constructed as follows: the exfoliated graphene doped with N and S was mixed with paraffin oil in order to obtain a homogeneous paste. The homogeneous paste was split in half; one half was modified with α-hemolysin and the other half was modified with hemin, by physically mixing the graphene paste and the solution of α-hemolysin and hemin (10−3 mol L−1), respectively. Practically, 100 mg of graphene powder was mixed with 100 μL of α-hemolysin or hemin solution (10−3 mol L−1). α-Hemolysin and hemin are biomolecules able to provide the pores/channels needed for the stochastic sensing. Each modified paste was placed in a non-conducting plastic tube (i.d. of 100 μm). In order to create the connection between the pastes and the external circuit, silver wires were placed inside the plastic tubes containing the modified pastes (Fig. 2). When not in use, the biosensors were kept in the fridge, at a temperature between 2–8 °C.2.4. Stochastic mode
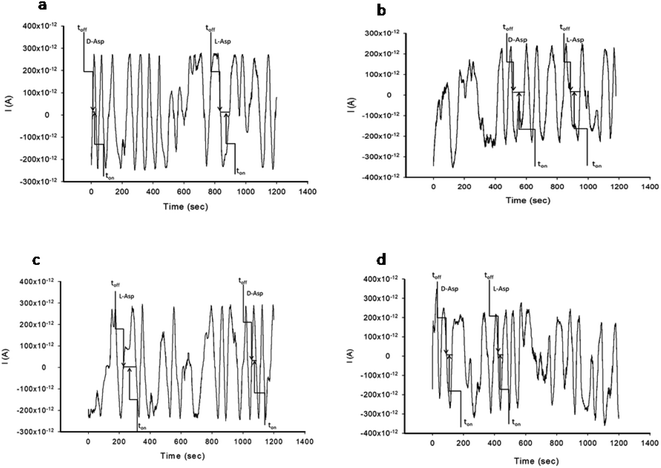
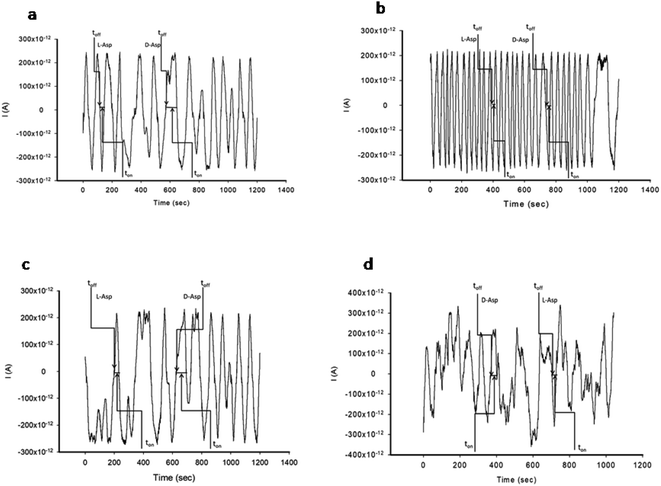
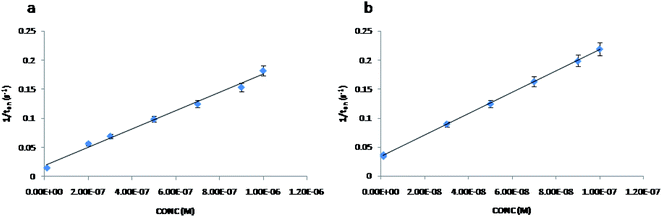
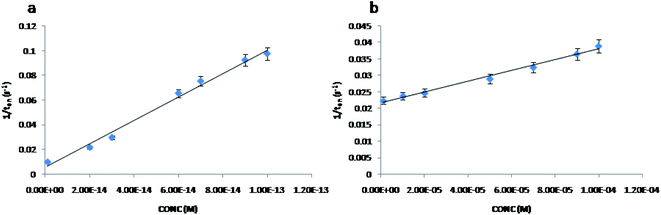
The stochastic mode was employed for all measurements. All measurements were done at 125 mV vs. Ag/AgCl. The functioning of the stochastic biosensors is based on the channel conductivity.Qualitative and quantitative enantioanalysis of Asp from whole blood, tissue, urine and saliva samples can be performed using the proposed biosensors. The parameters of interest were: toff and ton, respectively (Fig. 3 and 4). toff is known as the signature of the analyte and is used for the qualitative analysis. ton is known as the necessary time of equilibrium for the interaction of the analyte with the wall channel; this parameter is used in quantitative enantioanalysis. The equations of calibration (1/ton = a + bxConcenantiomer) and all parameters of the sensors were obtained using the linear regression method. The unknown concentration of the enantiomer was determined by using the equation of calibration (Table 2, Fig. 5 and 6). The time needed for the calibration measurements was 360 s, while the biological samples were assayed for 1200 s.
 | ||
| Fig. 5 The calibration curves for the biosensor modified with α-hemolysin for L-Asp (a) and D-Asp (b). | ||
2.5. Samples
County Emergency Hospital from Targu-Mures provided samples of whole blood, tissue, urine and saliva from confirmed patients with gastric cancer, but also from healthy patients (Ethics committee approval number 32647/2018 awarded by the County Emergency Hospital from Targu-Mures). Written consent was obtained from all patients. No sample pre-treatment was performed for the samples analyzed using the proposed biosensors.3. Results and discussions
3.1. Morphological and structural characterization of the E-NSGr sample
In Fig. 7a and b are shown representative SEM images of graphene flakes obtained after the electrochemical exfoliation of graphite rods in a solution containing 0.1 mol L−1 NH4SCN and 0.2 mol L−1 ammonia.Both images reveal that the sample is composed of flakes with large and smooth regions that correspond to the basal plane of graphene as well as by narrow and white regions, where the edges of graphene roll up. Overall, the sample morphology is a porous one, therefore it has a large surface area, which brings many advantages when the material is employed in the electrochemical sensing of various molecules.
The structural characterization of E-NSGr was next performed by UV-Vis, FTIR, and Raman spectroscopy. In Fig. S1† one can see the UV-Vis spectrum of graphene sample previously sonicated in water for 3 minutes. The absorbance peak of graphene can be generally found at wavelengths between 250 and 300 nm, in contrast with that corresponding to graphene oxide which appears at around 230 nm. In the case of the E-NSGr sample, a strong absorption peak occurs at 275 nm which is due to the π → π* transitions within the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C bonds. The small shoulder around 315 nm originates from the n → π* transitions within the oxygen-containing functional groups. This indicates that the sample contains a small amount of flakes bearing oxygen groups.
C bonds. The small shoulder around 315 nm originates from the n → π* transitions within the oxygen-containing functional groups. This indicates that the sample contains a small amount of flakes bearing oxygen groups.
The FTIR spectrum of E-NSGr (Fig. S2a and b†) further indicates the presence of vibrations corresponding to oxygen-containing groups attached to the graphene surface, such as: the strong and broad O–H stretching vibration at 3432 cm−1 due to the adsorbed water molecules, the O–H deformation vibration at 1386 cm−1, the C–O (alcoxy) stretching vibration at 1072 cm−1. The intense bands at 1400 cm−1 may be attributed to NH4+ vibration while the weaker band at 1165 cm−1 indicates the existence of C–N/C–S vibrations.22
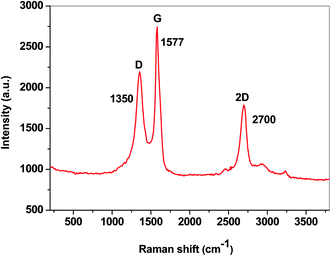
In order to evaluate the degree of the structural disorder within the E-NSGr sample, the Raman spectrum was also recorded (Fig. 8). Three important bands can be seen in this spectrum: the defect (D) band at 1350 cm−1, the graphite (G) band located at 1577 cm−1 and the 2D band at 2700 cm−1.23
According to eqn (1), the ID/IG ratio may be related to the in-plane crystallite size (La), giving an indication of the defect-free domains within the graphene sample:24
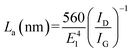 | (1) |
| Sample | G (cm−1) | IG (a.u.) | D (cm−1) | ID (a.u.) | ID/IG | La (nm) |
|---|---|---|---|---|---|---|
| E-NSGr | 1577 | 1794 | 1350 | 1249 | 0.7 | 27.2 |
3.2. Response characteristics of the stochastic biosensors
The response of the stochastic biosensors is based on the channel conductivity: when a constant potential of 125 mV is applied, the current is modified when the molecules of D- or L-aspartic acid enter the channel. In order for the enantiomers to be recognized, two phases need to be completed. During the pattern recognition phase, also known as the first phase, the enantiomers are extracted into the channel, blocking it, the intensity of the current drops to zero until the whole molecules enter the channel; the time spent at this phase is known as toff and represents the signature of the analyte; its value is used for the qualitative analysis of the enantiomers. The binding phase, also known as the second phase,18–20 takes place when the analytes of interest interact with the wall channel and redox processes take place.The response characteristics of the proposed stochastic biosensors used for the enantioanalysis of D- and L-aspartic are shown in Table 2. The signatures obtained for the enantiomers of Asp (the toff values) had different values (for the same biosensors) showing that the biosensors are enantioselective. Accordingly, the proposed biosensors are enantioselective, and can be used for the enantioanalysis of aspartic acid.
| Biosensor based on E-NSGr and | Calibration equation and correlation coefficient (r)a | Linear concentration range (mol L−1) | toff (s) | Sensitivity (s−1/mol L−1) | Limit of determination (mol L−1) |
|---|---|---|---|---|---|
| a 1/ton = s−1, C = mol L−1. | |||||
| α-Hemolysin | |||||
| L-Asp | 1/ton = 0.01 + (7.58 ± 0.03)×1011 × C, r = 0.9999 | 1.0 × 10−15 to 1.0 × 10−3 | 0.7 ± 0.01 | 7.58 × 1011 | 1.0 × 10−15 |
| D-Asp | 1/ton = 0.02 + (1.16 ± 0.03)×102 × C, r = 0.9999 | 1.0 × 10−6 to 1.0 × 10−2 | 2.0 ± 0.01 | 1.16 × 102 | 1.0 × 10−6 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||||
| Hemin | |||||
| L-Asp | 1/ton = 0.06 + (3.75 ± 0.02)×104 × C, r = 0.9999 | 1.0 × 10−8 to 1.0 × 10−3 | 0.6 ± 0.01 | 3.75 × 104 | 1.0 × 10−8 |
| D-Asp | 1/ton = 0.03 + (3.04 ± 0.02)×105 × C, r = 0.9991 | 1.0 × 10−9 to 1.0 × 10−3 | 1.8 ± 0.02 | 3.04 × 105 | 1.0 × 10−13 |
The linear concentration ranges recorded using the biosensors covered the healthy patients, the asymptomatic patients, as well as patients in different stages of gastric cancer. Accordingly, they can be used for the screening tests of healthy, asymptomatic and clinically ill patients.
The lowest limit of determination for L-Asp as well as the highest sensitivity were obtained for the biosensor based on α-hemolysin. The lowest limit of determination for D-Asp as well as the highest sensitivity were obtained for the biosensor based on hemin.
The proposed biosensors were tested daily for a period of 6 months. During this period of time, their sensitivities did not change with more than 1.00% from the values measured in the first day of utilization for screening tests of biological samples.
3.3. Selectivity and enantioselectivity of the stochastic biosensors
The selectivity of the stochastic biosensors is given by the signatures obtained for different biomarkers or substances determined with the same biosensor; differences in the values recorded for the main analyte and other substances means selectivity. Different signature values for D- and L-aspartic acid were obtained for the proposed stochastic biosensor proving their enantioselectivity (Table 2). The selectivity of the biosensors were checked versus other aminoacids: histidine, lysine, glutamine, serine, triptophane, arginine, and alanine. For the signature of these aminoacids, values higher than 2.0 were recorded (histidine 2.3, lysine 2.8, glutamine 2.1, serine 2.5, triptophane 3.2, arginine 3.5, and alanine 3.0), proving that the biosensors are selective.3.4. Analytical applications
The determination of the enantiomers in biological samples (whole blood, urine, saliva, and tissue) was done in accordance with the procedure described in the stochastic mode: the signatures of the two enantiomers were identified in the diagrams (Fig. 2 and 3), and the ton values were read and used for the calculation of the concentrations of the enantiomers.The validation of the proposed stochastic biosensors was done in two steps. During the first step, recovery tests were performed. Whole blood samples from healthy patients were spiked with L- and D-Asp, after an initial screening test, when the amounts of L- and D-Asp were determined. The results shown in Table 3 proved that the recovery of the enantiomers is higher than 95.00% in the biological samples (whole blood, urine, saliva, tissue), with RSD values lower than 1.00%.
L![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) D (mol:mol) Stochastic biosensor based on E-NSGr and D (mol:mol) Stochastic biosensor based on E-NSGr and |
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 99 99 |
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
99![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
|---|---|---|---|
| %, Recovery | |||
| a All results are expressed in μmol L−1. | |||
| Whole blood | |||
| α-Hemolysin | 99.83 ± 0.03 | 99.54 ± 0.05 | 99.90 ± 0.03 |
| Hemin | 99.64 ± 0.05 | 99.70 ± 0.07 | 99.81 ± 0.08 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||
| Saliva | |||
| α-Hemolysin | 99.76 ± 0.04 | 99.81 ± 0.03 | 99.79 ± 0.05 |
| Hemin | 99.80 ± 0.08 | 99.83 ± 0.07 | 99.93 ± 0.05 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||
| Urine | |||
| α-Hemolysin | 99.84 ± 0.04 | 99.97 ± 0.02 | 99.95 ± 0.03 |
| Hemin | 99.81 ± 0.09 | 99.80 ± 0.05 | 99.76 ± 0.06 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||
| Tissue | |||
| α-Hemolysin | 99.57 ± 0.02 | 99.76 ± 0.04 | 99.89 ± 0.03 |
| Hemin | 99.53 ± 0.05 | 99.85 ± 0.06 | 99.85 ± 0.03 |
The second stage of validation was done using real samples of whole blood from healthy patients, and of whole blood, urine, saliva, and tissue samples from confirmed patients with gastric cancer.
The results shown in Tables 4 and 5 proved a good correlation between the concentrations obtained using the two biosensors. While in the case of the confirmed patients with gastric cancer, both enantiomers L- and D- of Asp were identified and quantified (Table 3), for healthy patients, only L-Asp was identified and quantified (Table 5). Accordingly, this test may be used for fast screening of biological samples, contributing to a fast and in some cases early diagnosis of gastric cancer and neuroendocrine or neurodegenerative diseases.
| Sample no. | Enantiomer of aspartic acida | Stochastic biosensor based on E-NSGr and | |
|---|---|---|---|
| α-Hemolysin | Hemin | ||
| a All results are expressed in μmol L−1. | |||
| Whole blood | |||
| 1 | L | 20.74 ± 0.05 | 20.26 ± 0.08 |
| D | 108.77 ± 0.02 | 109.81 ± 0.07 | |
| 2 | L | 25.27 ± 0.03 | 26.95 ± 0.07 |
| D | 87.76 ± 0.03 | 86.96 ± 0.09 | |
| 3 | L | 92.34 ± 0.04 | 92.37 ± 0.07 |
| D | 225.40 ± 0.02 | 224.97 ± 0.08 | |
| 4 | L | 39.54 ± 0.03 | 38.24 ± 0.07 |
| D | 157.78 ± 0.05 | 153.67 ± 0.09 | |
| 5 | L | 22.37 ± 0.05 | 20.26 ± 0.08 |
| D | 40.74 ± 0.08 | 42.62 ± 0.05 | |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||
| Saliva | |||
| 1 | L | 53.84 ± 0.02 | 52.12 ± 0.08 |
| D | 41.40 ± 0.03 | 43.35 ± 0.06 | |
| 2 | L | 39.54 ± 0.02 | 39.25 ± 0.04 |
| D | 18.69 ± 0.04 | 19.28 ± 0.07 | |
| 3 | D | 16.74 ± 0.03 | 14.21 ± 0.09 |
| D | 33.12 ± 0.02 | 33.54 ± 0.08 | |
| 4 | L | 12.29 ± 0.03 | 11.41 ± 0.07 |
| D | 40.74 ± 0.03 | 42.65 ± 0.08 | |
| 5 | L | 35.84 ± 0.04 | 35.48 ± 0.07 |
| D | 22.52 ± 0.03 | 21.80 ± 0.07 | |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||
| Urine | |||
| 1 | L | 22.37 ± 0.04 | 21.25 ± 0.08 |
| D | 55.34 ± 0.03 | 54.01 ± 0.05 | |
| 2 | L | 23.16 ± 0.05 | 23.81 ± 0.07 |
| D | 18.55 ± 0.03 | 18.39 ± 0.09 | |
| 3 | L | 9.80 ± 0.04 | 10.84 ± 0.08 |
| D | 18.04 ± 0.02 | 18.50 ± 0.07 | |
| 4 | L | 10.26 ± 0.03 | 10.98 ± 0.05 |
| D | 33.11 ± 0.02 | 32.78 ± 0.09 | |
| 5 | L | 21.59 ± 0.02 | 20.63 ± 0.07 |
| D | 10.71 ± 0.02 | 10.21 ± 0.08 | |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||
| Tissue | |||
| 1 | L | 15.64 ± 0.03 | 16.71 ± 0.07 |
| D | 28.15 ± 0.05 | 28.79 ± 0.08 | |
| 2 | L | 19.55 ± 0.02 | 18.80 ± 0.09 |
| D | 13.16 ± 0.04 | 14.88 ± 0.08 | |
| 3 | L | 40.43 ± 0.04 | 40.33 ± 0.07 |
| D | 13.17 ± 0.02 | 13.21 ± 0.06 | |
| 4 | L | 33.49 ± 0.03 | 32.96 ± 0.08 |
| D | 18.85 ± 0.02 | 18.40 ± 0.07 | |
| 5 | L | 79.80 ± 0.03 | 77.90 ± 0.09 |
| D | 69.02 ± 0.02 | 66.07 ± 0.07 | |
4. Conclusions
Two stochastic biosensors based on the immobilization of α-hemolysin and hemin in the paste of an exfoliated graphene doped with N and S were used for the enantioanalysis of aspartic acid in biological samples: whole blood, urine, saliva, and tissue. The proposed stochastic biosensors showed high sensitivity, low limits of determination, and high selectivity and enantioselectivity. The results obtained for the validation of the screening test showed that the proposed biosensors and test can be used for the enantioanalysis of aspartic acid in biological samples, and also for the fast detection of gastric cancer.Funding
This work was supported by a grant of the Ministry of Research, Innovation and Digitization, CNCS/CCCDI – UEFISCDI, project number PN-III-P4-ID-PCCF-2016-0006 within PNCDI III.Author contributions
Conceptualization, R. I. S. v. S., S. M. P., R. M. I. M.; methodology, R. I. S. v. S., D. C. G., R. M. I. M., and L. B. T.; validation, R. I. S. v. S., D. C. G., and R. M. I. M.; investigation, D. C. G., L. B. T. and R. M. I. M.; writing—original draft preparation, R. I. S. v. S., D. C. G., R. M. I. M., S. M. P. and L. B. T.; writing—review and editing, R. I. S. v. S., D. C. G., R. M. I. M., S. M. P. and L. B. T.; supervision, R. I. S. v. S. All authors have read and agreed to the published version of the manuscript. The authors are grateful to Dr Bogdan Cozar for the Raman investigation of the graphene sample and to Dr Irina Kacso for the FTIR investigation and useful discussions regarding the spectrum interpretation.Conflicts of interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.Acknowledgements
SEM investigation was partially supported through the infrastructure obtained in the Project: Research Center and Advanced Technologies for Alternative Energies—CETATEA-623/11.03.2014. The authors are grateful to Dr Bogdan Cozar for the Raman investigation of the graphene sample and to Dr Irina Kacso for the FTIR investigation and useful discussions regarding the spectrum interpretation.References
- J. Liu, S. Lin, Z. Li, L. Zhou, X. Yan, Y. Xue, L. Meng, J. Lu, B. Suo and W. Jiang, Free amino acid profiling of gastric juice as a method for discovering potential biomarkers of early gastric cancer, Int. J. Clin. Exp. Pathol., 2018, 11, 2323–2336 Search PubMed.
- J. J. A. J. Bastings, H. M. van Eijk, S. W. O. Damink and S. S. Rensen, D-amino acids in health and disease: a focus on cancer, Nutrients, 2019, 11, 2205 CrossRef CAS.
- S. Ritz-Timme and M. J. Collins, Racemization of aspartic acid in human proteins, Ageing Res. Rev., 2002, 1, 43–59 CrossRef CAS PubMed.
- A. D'Aniello, D-aspartic acid: an endogenous amino acid with an important neuroendocrine role, Brain Res. Rev., 2007, 53, 215–234 CrossRef PubMed.
- S. Xiao and L. Zhou, Gastric cancer: metabolic and metabolomics perspectives (review), Int. J. Oncol., 2017, 51, 5–17 CrossRef CAS PubMed.
- A. N. Fonteh, R. J. Harrington, A. Tsai, P. Liao and M. G. Harrington, et al., Free amino acid and dipeptide changes in the body fluids from Alzheimer's disease subjects, Amino Acids, 2007, 32, 213–224 CrossRef CAS PubMed.
- S. Nishiumi, T. Kobayashi and A. Ikeda, et al., A novel serum metabolomics-based diagnostic approach for colorectal cancer, PLoS One, 2012, 7, e40459 CrossRef CAS PubMed.
- B. S. Sekhon, Enantioseparation of chiral drugs—an overview, Int. J. Pharm. Technol. Res., 2010, 2, 1584–1594 CAS.
- P. K. Katiyar and R. K. Wala, Chiral separation of aspartic acid by thin layer chromatography, Asian J. Chem., 2010, 22, 4945–4946 CAS.
- P. J. van den Oetelaar, L. E. van Beijsterveldt, J. R. van Beckhoven and H. J. Hoenders, Detection of aspartic acid enantiomers by chiral capillary gas chromatography. Determination of in vivo racemisation and reduction of metal-induced background, J. Chromatogr. A, 1986, 386, 135–143 CrossRef.
- H. Yu, Y. S. Ding and S. F. Mou, Some factors affecting separation and detection of amino-acids by high performance anion-exchange chromatography with integrated pulsed amperometric detection, J. Chromatogr. A, 2003, 997, 145–153 CrossRef CAS PubMed.
- S. Wang, L. Fan and S. Cui, CE-LIF chiral separation of aspartic acid and glutamic acid enanatiomers using human serum albumin and sodium cholate as dual selectors, J. Sep. Sci., 2009, 32, 3184 CrossRef CAS PubMed.
- H. Guan, P. Zhou, X. Zhou and Z. He, Sensitive and selective detection of aspartic acid and glutamic acid based on polythiophene-gold nanoparticles composites, Talanta, 2008, 77, 319–324 CrossRef CAS PubMed.
- Y. H. Deng, H. Wang and H. S. Zhang, Determination of amino acid neurotransmitters inhuman cerebrospinal fluid and saliva by capillary electrophoresis with laser-induced fluorescence detection, J. Sep. Sci., 2008, 31, 3088–3097 CrossRef CAS PubMed.
- B. B. Prasad, A. Srivastava and M. P. Tiwari, Highly sensitive and selective hyphenated technique (molecularly imprinted polymer solid-phase microextraction–molecularly imprinted polymer sensor) for ultra trace analysis of aspartic acid enantiomers, J. Chromatogr. A, 2013, 1283, 9–19 CrossRef CAS PubMed.
- G. Herzog and D. W. Arrigan, Electrochemical strategies for the label-free detection of amino acids, peptides and proteins, Analyst, 2007, 132, 615–632 RSC.
- P. Jainish and P. Prittesh, Biosensors and biomarkers: promising tools for cancer diagnosis, Int. J. Biosens. Bioelectron., 2017, 3, 313–316 Search PubMed.
- R. I. Stefan-van Staden, I. Popa-Tudor, C. I. Tirgoviste and R. A. Stoica, Molecular Recognition and Determination of Interleukins 1β, 6, 12, and 17 in Whole Blood from Diabetic Patients, Anal. Lett., 2020, 53, 2021–2033 CrossRef CAS.
- R. I. Stefan-van Staden, A. Moscalu-Lungu and M. Badulescu, Disposable Stochastic Sensors Based on Nanolayer Deposition(s) of Silver, and AgC Composite on Plastic for the Assay of α-Amylase in Whole Blood and Saliva, Nanomaterials, 2020, 10, 1528 CrossRef CAS PubMed.
- R. I. Stefan-van Staden, D. C. Gheorghe, V. Jinga, C. S. Sima and M. Geanta, Fast Screening of Whole Blood and Tumor Tissue for Bladder Cancer Biomarkers Using Stochastic Needle Sensors, Sensors, 2020, 20, 2420 CrossRef PubMed.
- F. Pogacean, M. Coros, L. Magerusan, V. Mirel, A. Turza, G. Katona, R. I. Stefan-van Staden and S. Pruneanu, Exfoliation of graphite rods via pulses of current for graphene synthesis: Sensitive detection of 8-hydroxy-2-deoxyguanosine, Talanta, 2019, 196, 182–190 CrossRef CAS PubMed.
- R. A. Rochman, S. Wahyuningsih, A. H. Ramelan and Q. A. Hanif, Preparation of nitrogen and sulphur Co-doped reduced graphene oxide (rGO-NS) using N and S heteroatom of thiourea, IOP Conf. Ser.: Mater. Sci. Eng., 2019, 509, 012119, DOI:10.1088/1757-899X/509/1/012119.
- A. C. Ferrari and D. M. Basko, Raman spectroscopy as a versatile tool for studying the properties of graphene, Nat. Nanotechnol., 2013, 8, 235–246 CrossRef CAS.
- L. G. Caņado, K. Takai, T. Enoki, M. Endo, Y. A. Kim, H. Mizusaki, A. Jorio, L. N. Coelho, R. Magalhães-Paniago and M. A. Pimenta, General equation for the determination of the crystallite size La of nanographite by Raman spectroscopy, Appl. Phys. Lett., 2006, 88, 1–4 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1ra02066h |
| This journal is © The Royal Society of Chemistry 2021 |