Open Access Article
Open Access ArticleSynthesis of the pentasaccharide repeating unit from Ruminococcus gnavus and measurement of its inflammatory properties†
Teron Hayniea,
Shawn Gublera,
Christoph Dreesb,
Tanner Heatona,
Tanner Mitton a,
Quinn Gleavea,
Albert Bendelac
a,
Quinn Gleavea,
Albert Bendelac b,
Shenglou Denga and
Paul B. Savage
b,
Shenglou Denga and
Paul B. Savage *a
*a
aDepartment of Chemistry and Biochemistry, Brigham Young University, Provo, UT 84602, USA. E-mail: paul_savage@byu.edu
bCommittee on Immunology, Department of Pathology, University of Chicago, Chicago, IL 60637, USA
First published on 16th April 2021
Abstract
The roles played by the gut microbiome in human health are increasingly recognized, and the prevalence of specific microorganisms has been correlated with different diseases. For example, blooms of the Gram-positive bacterium Ruminococcus gnavus have been correlated with inflammatory bowel disease, and recently a polysaccharide produced by this organism was shown to stimulate release of inflammatory cytokines. This stimulation was proposed to signal through toll-like receptor 4 (TLR4). We have synthesized the pentasaccharide repeating unit of this polysaccharide and showed that it stimulates TNF-α and IL-6 release from bone marrow-derived dendritic cells (BMDCs) in a TLR4-dependent manner. A related glycan does not stimulate significant cytokine release, demonstrating TLR4 selectivity in glycan recognition.
Introduction
Interactions between gastrointestinal microbiota and immune responses control multiple aspects of human health.1 These complex interactions control inflammatory responses, and dysregulation of these mechanisms and/or introduction of microorganisms that produce compounds that stimulate inflammation can lead to disease.2 Incidence of inflammatory bowel diseases, such as Crohn's disease, continues to increase.3 One contributing cause to these diseases is the makeup of the microbiome. For example, multiple studies link the presence of the bacterium Ruminococcus gnavus in the gut to symptoms of Crohn's disease.4–8 However, these reports did not provide a pathway linking inflammatory responses to this organism. Recently, Clardy and coworkers9 reported that R. gnavus produces a glucorhamnan polysaccharide that triggers inflammatory responses through toll-like receptor 4 (TLR4), and they hypothesize that TLR4-mediated responses to this polysaccharide contribute to the inflammatory cascade associated with Crohn's disease.The role of TLR4 in innate immunity is well established; it has been best characterized as a receptor for lipopolysaccharide (LPS), also called endotoxin, produced by most Gram-negative bacteria.10 Upon recognition of LPS, TLR4-producing cells release inflammatory cytokines, including TNF-α and IL-6. A highly used means of treating Crohn's disease involves use of antibodies that bind TNF-α and thereby inhibit propagation of inflammatory signaling.11 Notably, R. gnavus is Gram-positive and does not produce LPS; consequently, TLR4 mediated-signaling must come through a different interaction.
Continuing evaluation of the range of microbial molecular patterns recognized by TLR4 (ref. 10) has led to identification of fungal polysaccharides that can stimulate cytokine release by TLR4-producing cells.12 The first of these identified were mannans produced by the fungus Saccharomyces cerevisiae.13 Relative to LPS, higher concentrations of these polysaccharides are required to cause cytokine release by stimulation through TLR4.
We have an ongoing interest in the immunogenicity of bacterially-produced polysaccharides.14 In general, these polysaccharides are made up of repeating units of two to six sugars, and our particular focus is the immunogenicity of individual repeating subunits. To confirm that the glucorhamnan produced by R. gnavus stimulates cytokine release through TLR4 and to determine that the individual repeating subunit, of five sugars, retains stimulatory properties, we synthesized the pentasaccharide and studied responses by murine cells producing TLR4 and with cells from TLR4 knockout mice. This is the first synthesis of this pentasaccharide, and we found that it stimulated TNF-α and IL-6 release from wild-type bone marrow-derived dendritic cells (BMDCs), and this response was abrogated in TLR4 knockout BMDCs. Furthermore, to verify that stimulation came from interactions with the pentasaccharide, a control tetrasaccharide was used, which did not stimulate significant cytokine release from the wild-type cells.
Results and discussion
The structure of the inflammatory polysaccharide proposed by Clardy and coworkers is shown in Fig. 1, along with the structure of the repeating subunit (1) we selected as a synthetic target and a tetrasaccharide (2) used as a control. The latter is the repeating unit from the capsular polysaccharide produced by Streptococcus pneumoniae serotype 14.14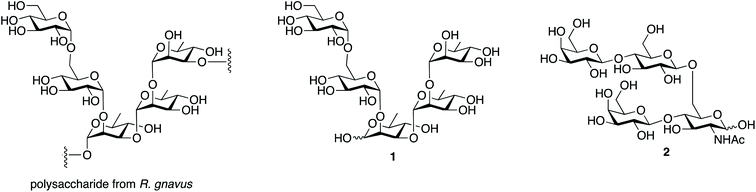 | ||
| Fig. 1 Structures of the repeating unit from the polysaccharide produced by R. gnavus, the pentasaccharide synthetic target and the repeating unit from S. pneumoniae serotype 14, used as a control. | ||
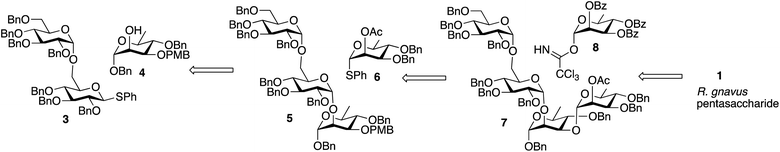
A convergent retrosynthetic dissection of the pentasaccharide target can give Glcα1-6Glc and Rhaα1-2Rha disaccharides bonded to a central rhamnose, and initially, we followed this approach. However, we found that stepwise addition of rhamnose groups gave higher yields than formation of the rhamnose disaccharide and glycoside formation with the central rhamnose. Consequently, the synthesis involved glycoside bond formation of disaccharide 3 with rhamnose 4, to give 5, followed by stepwise introduction of 6 and 8 to give the final pentasaccharide (Scheme 1).
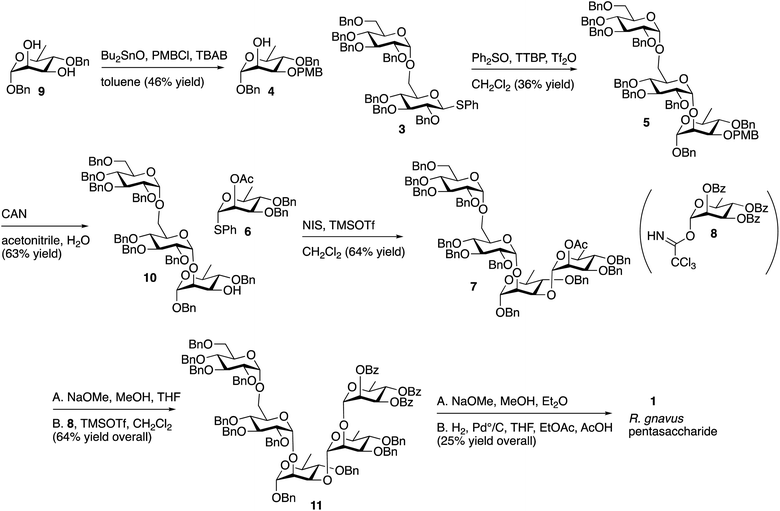
The route described in Scheme 1 utilizes known, protected sugar intermediates as much as possible. Acceptor 4 was prepared by selective protection of the equatorial C3 hydroxyl on 9 (ref. 15) (Scheme 2) using dibutyltin oxide and para-methoxybenzyl chloride (PBMCl). Donor 3, prepared using the procedure reported by Chu, et al.,16 was activated using the Crich17 method and coupled with 4 to give 5. Selective removal of PMB using ceric ammonium nitrate (CAN) gave 10 in moderate yield. Donor 6 was prepared as described by Mukherjee and Ghosh,18 and the compound was activated with N-iodosuccinimide/trimethylsilyl triflate (TMSOTf) and reacted with 10 to give 7 in moderate yield. The single ester in 7 was removed with sodium methoxide, and the Schmidt19 donor 8 (ref. 20) was incorporated using trimethylsilyl triflate as a promoter to give 11. The remaining benzoyl and benzyl group were removed sequentially using sodium methoxide and hydrogen with palladium to give the target pentasaccharide, as a mixture of anomers. The 1H NMR spectrum of this pentasaccharide was compared with that of the polysaccharide reported by Clardy and coworkers,9 and the two spectra are comparable (Fig. S1 in ESI†).
With pentasaccharide 1 in hand, we were prepared to evaluate stimulation through TLR4. However, in working with TLR4, care must be taken to avoid a false positive response from LPS contamination. TLR4 responds to LPS at low concentrations, and LPS contamination can occur in reagents, solvents and on labware and is a common problem among pharmaceuticals.21 Our final compound was purified chromatographically, and every precaution was made to avoid LPS contamination. Nevertheless, to ensure that TLR4-mediated signaling was not due to LPS contamination, we employed a tetrasaccharide glycan that had been prepared and purified previously in our laboratory. This glycan, 2 in Fig. 1, is the repeating unit of the polysaccharide produced by S. pneumoniae serotype 14, and we prepared this compound for structural studies of monoclonal antibodies generated to this tetrasaccharide.14 As an initial measure of the amount of LPS present in the samples of 1 and 2, the limulus amebocyte lysate (LAL) assay was used. This assay is highly sensitive and widely used for the quantification of LPS contamination. With 1 and 2, we detected 0.16 and 0.14 endotoxin units per mg, respectively. These amounts equate to approximately 15 pg of LPS per mg of carbohydrate or 1.5 × 10−8 weight%.
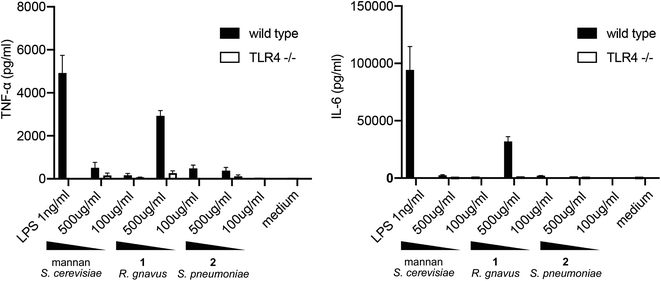
To evaluate the TLR4-mediated cytokine release stimulated by pentasaccharide 1, BMDCs were harvested from wild-type C57BL/J6 mice and from TLR4 knockout mice. These cells were exposed to LPS, mannan polysaccharide from S. cerevisiae, pentasaccharide 1 and control glycan 2 for 24 h. Concentrations of TNF-α and IL-6 were then measured via ELISA. Results are shown in Fig. 2.
As expected, relatively low concentrations of LPS stimulated release of TNF-α and IL-6 from BMDCs from wild-type mice. This release was abrogated with cells from the TLR4 knockout mice. As noted above, mannan from S. cerevisiae has been shown to stimulate cytokine release in a TLR4-dependent manner, and at 500 μg mL−1, a relatively small amount TNF-α was released from the cells. Cells lacking TLR4 released less cytokine. Very little IL-6 was released in response to the mannan. In contrast, a significant amount of both TNF-α and IL-6 were released upon exposure of BMDCs to the R. gnavus pentasaccharide, and the stimulation was largely TLR4 dependent. However, comparable TLR4-mediated responses to the glucorhamnan pentasaccharide and to LPS require very different concentrations of the molecules: 500 μg mL−1 of the pentasaccharide are required to stimulate a response similar to that triggered by 1 ng mL−1 of LPS. Notably, the control glycan from S. pneumoniae showed little to no stimulation of cytokine release. Considering the low and comparable amounts of endotoxin found in samples of 1 and 2, it is apparent that the pentasaccharide, and not LPS contamination, stimulates cytokine release from BMDCs in a TLR4-dependent manner.
Conclusions
Studies correlating the presence of R. gnavus with inflammatory bowel disease (e.g., Crohn's disease) provided incentive to determine if and how these organisms can trigger inflammation. The paper by Clardy and coworkers provided a potential mechanism by which glucorhamnan may contribute to inflammation through recognition by TLR4. Our work demonstrates that the individual repeating pentasaccharide is recognized by TLR4. This recognition results in release of inflammatory cytokines, and the synthesis supports the reported structure of the glucorhamnan repeating unit. The fact that pentasaccharide 1 stimulates a response through TLR4, while glycan 2 does not significantly stimulate cytokine release, suggests that TLR4 selectively responds to glycans with the pattern presented by 1.An understanding of the importance of glucorhamnan produced by R. gnavus must take into account the prevalence of this glycan among these bacteria. Pamer and coworkers22 sequenced the genomes of multiple R. gnavus isolates from healthy volunteers and found genomic diversity in the gene cluster encoding the enzymes for polysaccharide synthesis, which suggests that not all R. gnavus strains produce the same glucorhamnan polysaccharide. That is, only a subset of these organisms may produce the inflammatory glycan and contribute to IBD. Thus, it becomes important to not only identify the presence of R. gnavus, but also to identify the presence of organisms producing the inflammatory glycan. Development of high-affinity antibodies to this glycan or its subunits would allow this type of identification. This pentasaccharide may also be incorporated into a conjugate vaccine that would provide immune responses inhibiting growth of organisms producing this inflammatory glycan in the gastrointestinal tract.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
Funding from the National Institutes of Health (U01 AI125250 and R01 AI144094) is gratefully acknowledged including a fellowship from the German Research Foundation (DFG).References
- J. Y. Yoo, M. Groer, S. V. Ozorio Dutra, A. Sakar and D. I. McSkimming, Microorganisms, 2020, 8, 1587 CrossRef CAS PubMed.
- C. Caenepeel, N. S. S. Tabib, S. Vieira-Silva and S. Vermeire, Aliment. Pharmacol. Ther., 2020, 52, 1453–1468 Search PubMed.
- K. Cushing and P. D. R. Higgins, JAMA, J. Am. Med. Assoc., 2021, 325, 69–80 CrossRef CAS PubMed.
- B. P. Willing, J. Dicksved, J. Halfvarson, A. F. Andersson, M. Lucio, Z. Zheng, G. Jarnerot, C. Tysk, J. K. Jansson and L. Engstrand, Gastroenterology, 2010, 139, 1844 CrossRef PubMed.
- C. W. Png, S. K. Linden, K. S. Gilshenan, E. G. Zoetendal, C. S. McSweeney, L. I. Sly, M. A. McGuckin and T. H. J. Florin, Am. J. Gastroenterol., 2010, 105, 2420–2428 CrossRef CAS PubMed.
- M. Joossens, G. Huys, M. Cnockaert, V. De Preter, K. Verbeke, P. Rutgeerts, P. Vandamme and S. Vermeire, Gut, 2011, 60, 631–637 CrossRef PubMed.
- A. B. Hall, M. Yassour, J. Sauk, A. Garner, X. F. Jiang, T. Arther, G. K. Lagoudas, T. Vatanen, N. Forelos, R. Wilson, M. Bertha, M. Cohen, J. Garber, H. Khalili, D. Gevers, A. N. Ananthakrishnan, S. Kugathasan, E. S. Lander, P. Blainey, H. Vlamakis, R. J. Xavier and C. Huttenhower, Genome Med., 2017, 9, 103 CrossRef PubMed.
- K. Nishino, A. Nishida, R. Inoue, Y. Kawada, M. Ohno, S. Sakai, O. Inatomi, S. Bamba, M. Sugimoto, M. Kawahara, Y. Naito and A. Andoh, J. Gastroenterol., 2018, 53, 95–106 CrossRef PubMed.
- M. T. Henke, D. J. Kenny, C. D. Cassilly, H. Vlamakis, R. J. Xavier and J. Clardy, Proc. Natl. Acad. Sci. U. S. A., 2019, 116, 12672–12677 CrossRef CAS PubMed.
- M. M. Garcia, C. Goichoechea, M. Molina-Álvarez and D. Pascual, Eur. J. Pharmacol., 2020, 874, 172975 CrossRef CAS PubMed.
- N. Assasi, G. Blackhouse, F. Xie, J. K. Marshall, E. J. Ervine, K. Gaebel, D. Robertson, K. Campbell, R. Hopkins and R. Goeree, Expert Rev. Pharmacoecon. Outcomes Res., 2010, 10, 163–175 CrossRef PubMed.
- A. Roeder, C. J. Kirschning, R. A. Rupec, M. Challer, G. Weindl and H. C. Korting, Med. Mycol., 2004, 42, 485–498 CrossRef CAS PubMed.
- H. Tada, E. Nemoto, H. Shimauchi, T. Watanabe, T. Mikami, T. Matsumoto, N. Ohno, H. Tamura, K. Shibata, S. Akashi, K. Miyake, S. Sugawara and H. Takada, Microbiol. Immunol., 2002, 46, 503–512 CrossRef CAS PubMed.
- Z. Plonskaya, S. Deng, A. Sarkar, L. Kain, M. Comellas-Aragones, C. S. McKay, K. Kaczanowska, M. Holt, R. McBride, V. Palomo, K. M. Self, S. Taylor, A. Irimia, S. R. Mehta, J. M. Dan, M. Brigger, S. Crotty, S. P. Schoenberger, J. C. Polson, I. A. Wilson, P. B. Savage, M. G. Finn and L. Teyton, J. Clin. Invest., 2017, 127, 1491–1504 CrossRef PubMed.
- J. Mariño-Albernas, V. Verez-Bencomo, L. Gonzalez-Rodriguez and C. S. Perez-Martinez, Carbohydr. Res., 1988, 183, 175–182 CrossRef.
- A. A. Chu, S. H. Nguyen, J. A. Sisel, A. Minciunescu and C. S. Bennett, Org. Lett., 2013, 15, 2566–2569 CrossRef CAS PubMed.
- D. Crich and W. Li, Org. Lett., 2006, 8, 959–962 CrossRef CAS PubMed.
- M. M. Mukherjee and R. Ghosh, J. Org. Chem., 2017, 82, 5751–5760 CrossRef CAS PubMed.
- R. R. Schmidt and Z. Zhu, in Glycoscience, ed. B. Fraser-Reid, K. Tatsuta and J. Thiem, 2008, pp. 452–524 Search PubMed.
- C. Yu, H. Wang, L. Chiang and K. Pei, Synthesis, 2007, 9, 1412–1420 CrossRef.
- O. Perdijk, R. J. J. van Neerven, B. Meijer, H. F. J. Savelkoul and S. Brugman, Glycobiology, 2018, 28, 126–130 CrossRef CAS PubMed.
- M. T. Sorbara, E. R. Littmann, E. Fontana, T. U. Moody, C. E. Kahout, M. Gjonbalaj, V. Eaton, R. Seok, I. M. Leiner and E. G. Pamer, Cell Host Microbe, 2020, 28, 134–146 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1ra01918j |
| This journal is © The Royal Society of Chemistry 2021 |