Open Access Article
Open Access ArticleEnzymatic dephosphorylation-triggered self-assembly of DNA amphiphiles†
Jianmei Zou‡
 *ab,
Qian Gao‡a,
Jinfang Niea,
Yun Zhanga and
Cheng Jin
*ab,
Qian Gao‡a,
Jinfang Niea,
Yun Zhanga and
Cheng Jin *c
*c
aGuangxi Key Laboratory of Electrochemical and Magnetochemical Function Materials, College of Chemistry and Bioengineering, Guilin University of Technology, 12 Jiangan Road, Guilin 541004, China. E-mail: 2019136@glut.edu.cn
bMolecular Science and Biomedicine Laboratory, State Key Laboratory for Chemo/Bio-Sensing and Chemometrics, College of Chemistry and Chemical Engineering, College of Life Sciences, Aptamer Engineering Center of Hunan Province, Hunan University, Changsha, Hunan 410082, China
cInstitute of Molecular Medicine (IMM), Renji Hospital, Shanghai Jiao Tong University School of Medicine, Shanghai 200240, China. E-mail: jinch@hnu.edu.cn
First published on 21st May 2021
Abstract
In this work, phosphorylated lipid-conjugated oligonucleotide (DNA-lipid-P) has been synthesized to develop an enzyme-responsive self-assembly of DNA amphiphiles based on dephosphorylation-induced increase of hydrophobicity. Since elevated ALP level is a critical index in some diseases, ALP-triggered self-assembly of DNA amphiphiles shows promise in disease diagnosis and cancer treatment.
Lipid-conjugated oligonucleotides are DNA amphiphiles that can self-assemble into lipid-based DNA micelles. As such, lipid tails act as a hydrophobic core and DNA act as a hydrophilic corona.1–3 Because of their advantages of facile preparation, programmable design, small size (<100 nm) and biocompatibility, lipid-based DNA micelles show potential in the imaging of intracellular targets (mRNA and small molecules) and drug delivery.4–6
To further improve their potential for better drug delivery and disease diagnosis, stimuli-responsive control of the change of morphology or assembly/disassembly of DNA micelles has attracted enormous attention in practical applications. It was reported that photo-irradiation and nucleic acid hybridization have been used to trigger assembly/disassembly or the change of morphology of DNA micelles.7–10 For example, Jin et al. developed stability-tunable DNA micelles by using photo-controllable dissociation of an intermolecular G-quadruplex.7 The intermolecular parallel G-quadruplexes were introduced into lipid-based DNA micelles to lock the whole structure, resulting in enhanced structural stability against disruption by serum albumin. However, photo-controlled release of complementary DNA blocks the formation of G-quadruplexes and thus leads to the dissociation of micelles by the existence of serum albumin. In addition, Chien et al. reported stimuli-responsive programmable shape-shifting DNA micelles by controlling geometric structure and electrostatics.8 DNA hybridization and dissociation change the geometric structure and electrostatics of hydrophilic moiety, leads to the conversion of DNA assemblies between spherical and cylindrical structures.
In spite of these advances, enzyme-responsive regulation of self-assembly of DNA amphiphiles has scarcely reported. Since the hydrophobicity of lipid tail plays a critical role in the aggregation of DNA micelles, we speculated that the self-assembly of lipid-conjugated oligonucleotides can be regulated by controlling the hydrophobicity of lipid tails. Carry this idea forward, we noted that ALP is a hydrolase that removes phosphate groups from nucleic acids or proteins.11,12 Remarkably, ALP has been widely employed to convert hydrophilic phosphorylated small molecules to hydrophobic dephosphorylated products for molecular imaging, disease diagnosis and cancer therapy.13–16 Herein, therefore, enzymatic dephosphorylation-triggered self-assembly of DNA amphiphile is reported. As shown in Fig. 1, DNA-lipid-P is composed of four segments: DNA, linker, lipid and phosphate groups. Four negatively charged phosphate groups at lipid terminus decrease their hydrophobicity. Therefore, DNA-lipid-P exhibits weak self-assembly. However, ALP converts DNA-lipid-P to DNA-lipid by removing phosphate groups. The newly generated DNA-lipid shows greater hydrophobicity compared to DNA-lipid-P thus enables self-aggregation in aqueous solution.
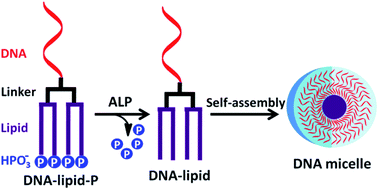 | ||
| Fig. 1 Schematic illustration of enzymatic dephosphorylation-triggered self-assembly of DNA amphiphile. | ||
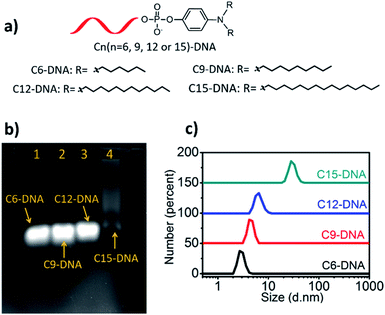
Lipid-conjugated oligonucleotides are amphiphiles which compose of two segments: hydrophobic lipid tails and hydrophilic oligonucleotides. Generally, lipid-conjugated oligonucleotides can self-assemble into aggregated DNA nanostructures, for example, DNA micelles, in aqueous solution by intermolecular hydrophobic interaction. To investigate whether the hydrophobicity affects the self-assembly of lipid-conjugated oligonucleotides, a series of lipid phosphoramidites with different length of alkyl chains (six, nine, twelve and fifteen) at the lipid tail were conjugated with oligonucleotide on a DNA synthesizer. The obtained lipid-conjugated oligonucleotides named as C6-DNA, C9-DNA, C12-DNA and C15-DNA, respectively (Fig. 2a). High-performance liquid chromatography (HPLC) is a universal tool to assess the hydrophobicity of DNA by comparing their retention times. Greater retention time indicates the stronger hydrophobicity. As shown in Table S2,† the retention time of DNA, C6-DNA, C9-DNA, C12-DNA and C15-DNA is 10.0, 19.1, 23.8, 27.3 and 30.8 minutes, respectively, suggesting that DNA with longer alkyl chains has stronger hydrophobicity. The result is consistent with the previous report.17 Next, the self-assembly of these lipid-conjugated oligonucleotides was investigated by agarose gel electrophoresis and dynamic light scattering (DLS) assays. As shown in Fig. 2b, only C15-DNA shows a tailed nucleic acids band which is belongs to the self-assembled nanostructure. In addition, results of DLS assays exhibit that the particle size of 10 μM C6-DNA, C9-DNA, C12-DNA and C15-DNA in buffer solution is 2.7 nm, 4.2 nm, 6.5 nm and 28.2 nm, respectively (Fig. 2c). Besides, the morphology of self-assembled C15-DNA micelles was visualized with atomic force microscopy (AFM) and the result shows the spherical nanostructure with diameter of 36.8 ± 6.1 nm (Fig. S7†). Both evidences support the self-assembly of C15-DNA in buffer solution. In another word, the self-assembly of lipid-conjugated oligonucleotides into DNA micelles is hydrophobicity-dependent. A greater hydrophobicity indicates a stronger tendency of aggregation.
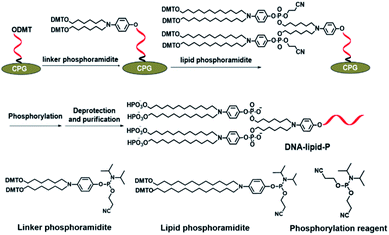
Next, we further synthesize DNA-lipid-P by solid-phase synthesis and phosphoramidite chemistry. In a previous literature, we developed a novel lipid phosphoramidite in which two DMT-protected hydroxyl groups was modified at the terminus of lipid tails thus enables further chemical phosphorylation during DNA synthesis.17 As shown in Fig. 3, linker and lipid phosphoramidites were successively conjugated at the 5′-terminus of DNA, followed by coupling with chemical phosphorylation reagent. After deprotection and purification, DNA-lipid-P was obtained and characterized by mass spectrum. As shown in Fig. S8,† the calculated molecular weight of DNA-lipid-P is 7789.8 Da, and the observed molecular weight is 7792.4 Da. The mass error is 2.6 Da (0.03%) which is within the mass error tolerance (0.03%), suggesting the successful synthesis of DNA-lipid-P. As such, DNA-lipid was also successfully synthesized with high purity (>98%) (Fig. S9†).
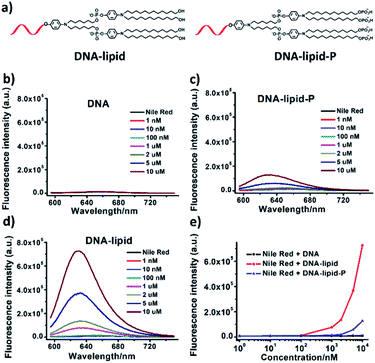
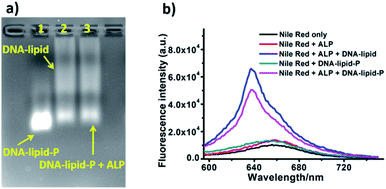
Having confirmed the successful synthesis of DNA-lipid-P, we further investigate the self-assembly of DNA-lipid and DNA-lipid-P in buffer solution. Nile red, a fluorescent dye that exhibits significant fluorescence in hydrophobic media, but negligible emission in aqueous solution, was used to determine the encapsulation of guest molecules to further assess the formation of the micellar structure.7,18 Nile red (1 μM) were incubated with various concentrations of DNA, DNA-lipid or DNA-lipid-P and the corresponding fluorescence spectroscopies were recorded. As shown in Fig. 4, both DNA (Fig. 4b) and DNA-lipid-P (Fig. 4c) show weaker fluorescence emission at 630 nm, even the concentration was upper to 10 μM. However, DNA-lipid (Fig. 4d) exhibits a bright fluorescence emission at 630 nm, suggesting the formation of hydrophobic core. After calculation, the critical micelle concentration (CMC) of DNA-lipid is 0.36 μM (Fig. 4e). The remarkable difference of CMC between DNA-lipid and DNA-lipid-P indicates that enzymatic conversion of DNA-lipid-P to DNA-lipid could trigger the spontaneous intermolecular aggregation.
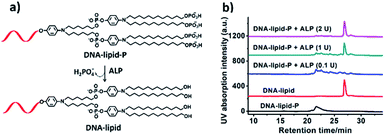
Next, ALP was used to convert DNA-lipid-P to DNA-lipid (Fig. 5a). As shown in Fig. 5b (black and red lines), the retention time of DNA-lipid and DNA-lipid-P is 26.5 and 20.9 minutes, respectively, indicates that the phosphorylation of lipid tail indeed decreases the hydrophobicity of lipid-conjugated oligonucleotides. Then, ALP was incubated with DNA-lipid-P (10 μM) at 37 °C for ten minutes and then 75 °C for five minutes to deactivate ALP, followed by subjected to fluorescence measurements. As shown in Fig. 5b (pink line), after the treatment of ALP (2 U), the DNA peak of DNA-lipid-P at 20.9 minutes disappeared; instead, a new DNA peak at 26.5 minutes was observed. Mass spectrum analysis indicates that the molecular weight of newly generated DNA peak is 7474.2 Da, which is consistent with the calculated molecular weight of DNA-lipid (7470.9 Da) (Fig. S10†). In a word, ALP enables enzymatic dephosphorylation of DNA-lipid-P; and the generated DNA-lipid has greater hydrophobicity than DNA-lipid-P thus facilitates the controllable self-assembly into DNA micelles.
Encouraged by the ALP-induced dephosphorylation of DNA-lipid-P to DNA-lipid, we further assess whether ALP enables activatable self-assembly of DNA-lipid-P. As shown in Fig. 6a, DNA-lipid-P (1 μM) shows weak self-assembly in buffer solution. However, DNA-lipid (1 μM) exhibits obvious aggregation band in gel electrophoresis assay. After incubation with ALP (1 U), DNA-lipid-P + ALP group also shows tailed band which suggests the formation of aggregated nanostructures. In addition, results of Nile red-encapsulated fluorescence experiments also support the conclusion of ALP-activate self-assembly of DNA-lipid-P (Fig. 6b). Therefore, ALP-induced enzymatic dephosphorylation triggers the self-assembly of DNA-lipid-P.
In summary, enzymatic dephosphorylation-triggered self-assembly of DNA amphiphile is developed by integrating enzymatic dephosphorylation-induced increase of hydrophobicity and intermolecular aggregation of lipid-conjugated oligonucleotides. The strategy may also suitable for many other amphiphiles. Since elevated ALP level is a critical index in some diseases and even cancers, we believe that ALP-triggered self-assembly of DNA-lipid-P shows potential in disease diagnosis and cancer therapy.
Conflicts of interest
The authors declare no competing financial interest.Acknowledgements
The authors gratefully acknowledge the financial support from Natural Science Foundation of Guangxi Province (2020GXNSFBA297114), Foundation of Guilin University of Technology (GUTQDJJ2019136), Open Foundation of State Key Laboratory for Chemo/Bio-Sensing and Chemometrics, National Science Foundation of China (No. 21874032 and 21765007), Guangxi Science Fund for Distinguished Young Scholars (No. 2018GXNSFFA281002).Notes and references
- H. Liu, Z. Zhu, H. Kang, Y. Wu, K. Sefan and W. Tan, Chem.–Eur. J., 2010, 16, 3791 CrossRef CAS PubMed.
- T. G. W. Edwardson, K. M. M. Carneiro, C. J. Serpell and H. F. Sleiman, Angew. Chem., Int. Ed., 2014, 53, 4567 CrossRef CAS PubMed.
- J. Zou, C. Jin, R. Wang, H. Kuai, L. Zhang, X. Zhang, J. Li, L. Qiu and W. Tan, Anal. Chem., 2018, 90, 6843 CrossRef CAS.
- T. Chen, C. S. Wu, E. Jimenez, Z. Zhu, J. G. Dajac, M. You, D. Han, X. Zhang and W. Tan, Angew. Chem., Int. Ed., 2013, 52, 2012 CrossRef CAS PubMed.
- C. Wu, T. Chen, D. Han, M. You, L. Peng, S. Cansiz, G. Zhu, C. Li, X. Xiong, E. Jimenez, C. J. Yang and W. Tan, ACS Nano, 2013, 7, 5724 CrossRef CAS PubMed.
- Y. Wu, K. Sefah, H. Liu, R. Wang and W. Tan, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 5 CrossRef CAS PubMed.
- C. Jin, X. Liu, H. Bai, R. Wang, J. Tan, X. Peng and W. Tan, ACS Nano, 2017, 11, 12087 CrossRef CAS PubMed.
- M.-P. Chien, A. M. Rush, M. P. Thompson and N. C. Gianneschi, Angew. Chem., Int. Ed., 2010, 122, 5202 CrossRef.
- D. Bousmail, P. Chidchob and H. F. Sleiman, J. Am. Chem. Soc., 2018, 140, 9518 CrossRef CAS.
- M. You, Z. Zhu, H. Liu, B. Gulbakan, D. Han, R. Wang, K. R. Williams and W. Tan, ACS Appl. Mater. Interfaces, 2010, 2, 3601 CrossRef CAS.
- A. Alonso, J. Sasin, N. Bottini, I. Friedberg, I. Friedberg, A. Osterman, A. Godzik, T. Hunter and J. Dixon, Cell, 2004, 117, 699 CrossRef CAS.
- Y. Shi, Cell, 2009, 139, 468 CrossRef CAS PubMed.
- Z. Zheng, P. Chen, M. Xie, C. Wu, Y. Luo, W. Wang, J. Jiang and G. Liang, J. Am. Chem. Soc., 2016, 138, 11128 CrossRef CAS PubMed.
- H. Wang, Z. Feng, S. J. Del, A. A. Rodal and B. Xu, J. Am. Chem. Soc., 2018, 140, 3505 CrossRef CAS PubMed.
- Z. Feng, H. Wang, S. Wang, Q. Zhang, X. Zhang, A. A. Rodal and B. Xu, J. Am. Chem. Soc., 2018, 140, 9566 CrossRef CAS PubMed.
- Z. Feng, H. Wang, R. Zhou, J. Li and B. Xu, J. Am. Chem. Soc., 2017, 139, 3950 CrossRef CAS.
- C. Jin, J. He, J. Zou, W. Xuan, T. Fu, R. Wang and W. Tan, Nat. Commun., 2019, 10, 2704 CrossRef.
- I. K. Kurniasih, H. Liang, P. C. Mohr, G. Khot, J. P. Rabe and A. Mohr, Langmuir, 2015, 31, 2639 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: Experimental section and additional results. See DOI: 10.1039/d1ra01668g |
| ‡ J. Z. and Q. G. are co-first authors. |
| This journal is © The Royal Society of Chemistry 2021 |