Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Latin American databases of natural products: biodiversity and drug discovery against SARS-CoV-2†
Marvin J. Núñez a,
Bárbara I. Díaz-Eufracio
a,
Bárbara I. Díaz-Eufracio b,
José L. Medina-Franco
b,
José L. Medina-Franco b and
Dionisio A. Olmedo
b and
Dionisio A. Olmedo *cd
*cd
aNatural Product Research Laboratory, School of Chemistry and Pharmacy, University of El Salvador, San Salvador, El Salvador. E-mail: marvin.nunez@ues.edu.sv
bDIFACQUIM Research Group, Department of Pharmacy, School of Chemistry, National Autonomous University of Mexico, Mexico City 04510, Mexico. E-mail: jose.medina.franco@gmail.com; dieb@comunidad.unam.mx
cCenter for Pharmacognostic Research on Panamanian Flora (CIFLORPAN), College of Pharmacy, University de Panama, Panama. E-mail: ciflorp4@up.ac.pa
dSistema Nacional de Investigación (SNI), SENACYT, Panamá
First published on 4th May 2021
Abstract
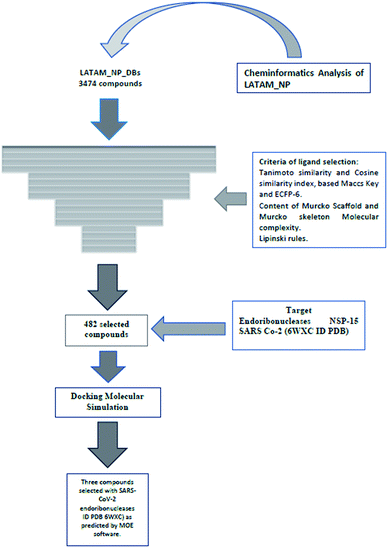
In this study, we evaluated 3444 Latin American natural products using cheminformatic tools. We also characterized 196 compounds for the first time from the flora of El Salvador that were compared with the databases of secondary metabolites from Brazil, Mexico, and Panama, and 42![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 969 compounds (natural, semi-synthetic, synthetic) from different regions of the world. The overall analysis was performed using drug-likeness properties, molecular fingerprints of different designs, two parameters similarity, molecular scaffolds, and molecular complexity metrics. It was found that, in general, Salvadoran natural products have a large diversity based on fingerprints. Simultaneously, those belonging to Mexico and Panama present the greatest diversity of scaffolds compared to the other databases. This study provided evidence of the high structural complexity that Latin America's natural products have as a benchmark. The COVID-19 pandemic has had a negative effect on a global level. Thus, in the search for substances that may influence the coronavirus life cycle, the secondary metabolites from El Salvador and Panama were evaluated by docking against the endoribonuclease NSP-15, an enzyme involved in the SARS CoV-2 viral replication. We propose in this study three natural products as potential inhibitors of NSP-15.
969 compounds (natural, semi-synthetic, synthetic) from different regions of the world. The overall analysis was performed using drug-likeness properties, molecular fingerprints of different designs, two parameters similarity, molecular scaffolds, and molecular complexity metrics. It was found that, in general, Salvadoran natural products have a large diversity based on fingerprints. Simultaneously, those belonging to Mexico and Panama present the greatest diversity of scaffolds compared to the other databases. This study provided evidence of the high structural complexity that Latin America's natural products have as a benchmark. The COVID-19 pandemic has had a negative effect on a global level. Thus, in the search for substances that may influence the coronavirus life cycle, the secondary metabolites from El Salvador and Panama were evaluated by docking against the endoribonuclease NSP-15, an enzyme involved in the SARS CoV-2 viral replication. We propose in this study three natural products as potential inhibitors of NSP-15.
Introduction
Natural products (NPs) and their derivatives continue to be an important source of chemical entities for the design and development of drugs, reporting a total of 355 molecules approved for clinical use between the years 1981 to 2019 and placing them at an intermediate point between biological products (262).1 Databases of publicly available NPs have been used in process discoveries, and to develop drug design and contain metabolites of plants, fungi, marine organisms, and bacteria.2 These databases contain a variety of chemical types that have various pharmacological activities.3 Some natural products from these databases have been evaluated by chemo- and bioinformatic methods, concluding that they have large structural and scaffold diversities.4Latin American database of natural products (LATAM_DBS_NPs) has been evaluated using chemoinformatics and bioinformatics tools. Such analysis allowed us to create and manipulate molecules by representing and visualizing the chemical structures of small molecules. Furthermore, we calculate physicochemical properties based on chemical descriptors, chemical space, fingerprints, similarity studies, the content of cyclic systems, fragments of aliphatic and alicyclic skeletons applied to the bases of natural products.5–14 The increasing use of cheminformatics in NPs research has led to the sub-discipline “natural product informatics.”15 In this study we analyzed and compared four databases of Latin American NPs: LAIPNUDLSAV, UPMA_2V, BIOFACQUIM_2V, NUBBE_2V and eleven databases of natural products available for free at http://zinc12.docking.org/browse/catalogs/. These include the following datasets: AfroDb NPs; Herbal ingredients in vivo metabolism database (HIM); Herbal Ingredients' Targets (HIT); ICC Indofine NPs DBs (Indofine Chemical Company); Naturally occurring Plant based Anti-cancerous Compound-Activity-Target Database (NPACT Database); AnalytiCon Discovery NP (ACDISC_NP); Specs Natural Products; The Nuclei of Bioassays, Ecophysiology and Biosynthesis of Natural Products Database (NuBBE DB 2013 and 2017); InterBioScreen Ltd (IBScreen NP); Chemical Entities of Biological Interest (ChEBI); and Princeton BioMolecular Research.
This is the first systematic cheminformatic and bioinformatic study of LATAM_DBS_NPs against NPS-15 endoribonuclease of SARS CoV-2. The acronym refers to severe acute respiratory syndrome (SARS) and the CoV-2 refers to type 2 coronavirus.
NPs have been found to be highly potent in blocking enzyme function and membrane receptors of human coronavirus.16 In addition, the evolution of COVID-19 is featured with uncontrolled inflammation, threeway anti-inflammatory herbal compounds will be a potential tool to repress such fatal symptoms.17 Nsp-15, a hexamer endoribonuclease that cleaves the remains of uridines. This protein is homologous to the Nsp15 endoribonucleases of SARS-CoV and MERS-CoV but differs from them in that it can contribute to the increased virulence of SARS-CoV-2.18
Materials and methods
Databases of natural products
LATAM_DBS_NPs and REF_NPs were analyzed for their physicochemical properties, chemical space, scaffold content, and molecular complexity. These include two datasets prepared in house and another two NPs DBs published in journals: LAIPNUDELSAL, UMPA_NP_2V, BIOFACQUIM_2V, and NUBBE_2V DB, that were compared with eleven NPs of the reference data set (REF_NPs) of public access: AFRON_NP, AnalytiCon DB ACDISC_NP, CHEBI, HITM_NP, HIT_NP, IBSCREEN_NP, INDOFINE_NP, NPACT_NP, PRINCETON_NP, SPECS_NP and NUBBEDB V1 and V2. Table 1 indicates the compounds analyzed and Table 2 shows the description and sources of the databases used in this work.| Dataset of NPs | Number of initial compounds | Number of compounds after data curation |
|---|---|---|
| LAIPNUDELSAL_NP | 214 | 196 |
| UPMA_2V_NP | 485 | 462 |
| BIOFACQUIM_NP | 531 | 507 |
| NUBBE_2V_NP | 2366 | 2279 |
| IBSCREEN_NP subset | 8444 | 8413 |
| CHEBI subset | 6054 | 5622 |
| AFRO_NP | 550 | 494 |
| ACDISC_NP | 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 245 245 |
10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 859 859 |
| HIM_NP | 663 | 637 |
| HIT_NP | 801 | 583 |
| INDOFINE_NP | 143 | 99 |
| NPACT_NP | 1422 | 1093 |
| SPECS_NP subset | 1488 | 1349 |
| PRINCETON subset | 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 083 083 |
13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 672 672 |
| NUBBE_1V | 587 | 148 |
49![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 076 076 |
46![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 413 413 |
| Dataset of NPs | Description | Sources |
|---|---|---|
| LAIPNUDELSAL_NP | This database was built by making a bibliographic compilation of the compounds isolated and characterized by spectroscopic methods of the flora of El Salvador during the period 1981 to 2019 | In house (Laboratorio de Investigación en Productos Naturales de la Universidad de El Salvador, El Salvador) |
| UPMA_2V_NP | A collection actualized of NPs isolated from Panamanian flora from 1972 to 2019 | In house (CIFLORPAN, Facultad de Farmacia, Universidad de Panamá, Panamá) |
| BIOFACQUIM_NP_2V | Database with NPs isolated and characterized in Mexico from 2020 | DOI: 10.3390/biom90100317 |
| NUBBE_2V_NP | NuBBE_DB_2V has compounds obtained from plants, semi-synthetic products microorganisms, products of biotransformation, and marine environment modified in 2017 | DOI: 10.1038/s41598-017-07451-x,19 available in: https://nubbe.iq.unesp.br/portal/nubbe-search.html |
| IBSCREEN_NP subset | Natural products and derivatives NPs were generated for institutes of the Soviet Union from 2015 | Available in: http://zinc12.docking.org/browse/catalogs/natural-products, h commercially available: https://www.ibscreen.com/natural-compounds20 |
| CHEBI subset | Chemical entities of biological interest (ChEBI). This dictionary includes natural products, synthetic compounds, drugs, and chemicals environment | DOI: 10.1093/nar/gkp886,21 commercially available in: https://www.ebi.ac.uk/chebi/ |
| AFRO_NP | AfroDb represents a collection of NPs from the African continent from 2013 | DOI: 10.1371/journal.pone.0078085,22 available in: http://zinc12.docking.org/browse/catalogs/natural-products |
| ACDISC_NP | Natural products bioactive from 2013, AnalytiCon Discovery NP | Available in: http://zinc12.docking.org/browse/catalogs/natural-products,23 http://zinc.docking.org/catalogs/acdiscnp/ |
| HIM_NP | HIM Db is set up for the in vivo metabolite's information of the active ingredients for Chinese herbs from 2013 | DOI: 10.1186/1758-2946-5-28,24 available in: http://zinc12.docking.org/browse/catalogs/natural-products |
| HIT_NP | HIT is collecting target information of herbal active compounds from 2013 | DOI: 10.1093/nar/gkq1165,25 available in: http://zinc12.docking.org/browse/catalogs/natural-products |
| INDOFINE_NP | Databases of NPs from 2013 | Available in: http://zinc12.docking.org/browse/catalogs/natural-products,26 http://zinc12.docking.org/catalogs/indofinenp |
| NPACT_NP | Database of plant-derived natural compounds that exhibit anti-cancerous activity from 2013 | DOI: org/10.1093/nar/gks1047,27 http://zinc12.docking.org/catalogs/npactnp |
| SPECS_NP | A collection of NPs from 2015 | Available in: http://zinc12.docking.org/catalogs/specsnp,28 commercially available in http://www.specs.net |
| PRINCETON subset | Compounds obtained for high throughput screening | Available in: http://zinc12.docking.org/browse/catalogs/natural-products,29 commercially available in: http://www.princetonbio.com |
| NUBBE_1V | The Nuclei of Bioassays, Ecophysiology, and Biosynthesis of Natural Products Database (NuBBEDB_1V) was created as the first natural product library from Brazilian biodiversity in 2013 | DOI: 10.1021/np3006875,30 available in: http://zinc12.docking.org/catalogs/nubbenp |
The molecular descriptors were determined by MOE v.2018.0102 (ref. 31) and DataWarrior v.5.2.1:32 hydrogen bond acceptors (HBAs), hydrogen bond donors (HBDs), number of rotatable bonds (NRBs), the octanol/water partition coefficient (w/o; S![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log
log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P), molecular weight (MW), and topological polar surface area (TPSA) (data in ESI†). These descriptors have been widely used to describe properties of interest in research and development departments in the pharmaceutical industries.5–14
P), molecular weight (MW), and topological polar surface area (TPSA) (data in ESI†). These descriptors have been widely used to describe properties of interest in research and development departments in the pharmaceutical industries.5–14
Chemical space
A visual representation of the chemical space was done using the principal component analysis (PCA)5–14 of the six physicochemical properties with DataWarrior v.5.2.1.32Molecular fingerprints
The molecular fingerprints have been used as indicators of structural diversity in different data sets.33–35 Two fingerprint keys were calculated: Extended Connectivity Fingerprints of Radius six (ECFP-6) and Molecular Access System (MACCS) Key (166-bits) with the Python script.36 The correlation of similarity pairs was calculated with the correlation-like-similarity index: Tanimoto coefficient and cosine-like similarity index were analyzed with a cumulative distribution function (CDF).Molecular scaffolds diversity
The Bemis–Murcko scaffold references were calculated with molecular equivalent indices (MEQI)37–39 and DataWarrior.31 MEQI was used to obtain the cyclic system ring of the compounds in the different databases of NPs for their scaffold content and diversity.40–43The scaffold distribution of the NPs dataset was explored, evaluated, and plotted in RStudio44,45 for the cyclic system retrieval curves (CSR) for indicated the scaffold diversity of different databases.7–14 These curves were plotted with the fraction of scaffolds in the x-axis and the fraction of compounds in the y-axis. The CSR curves provide information on the scaffold diversity of all the data sets.
Molecular complexity
The molecular complexity of NP datasets was explored and calculated using the descriptors: a fraction of sp3 hybridized carbons (Fsp3), fraction of chiral carbon (FCC), fraction of atoms aromatic (Fa_Aro), fraction of aromatic bond (Far_b), molecular flexibility, shape index and globularity molecular.11,14,46,47Molecular docking simulation
The binding site was checked with the option (Site Finder), which is listed by the size of the possible binding sites of the ligand to the protein, in the first four rows appear the amino acid residues that maintain the interaction with the co-crystallized ligand. This step is important since other sites where there is no compound can be selected to simulate docking molecular. Finally, we isolated the atoms and the region of proximity to the selected site in the protein, reviewed the structure of the ligand in CPK format, and generated the surface area of the binding site.49
The database was created in the excel format and later transformed into a CVS file. This was imported into the MOE window, where we checked if the structures were correctly drawn in their 2D model. Then, we saved it in a format with moe extension. This constituted the working database for chemoinformatic analysis mbd format.
For curing of the databases, we removed the compound duplicates, washed for removing protonated forms of acidic and basic, disconnected the metals present, reviewed the presence of partial loads, and finished minimizing energy state structures and generated conformations 3D, for subsequent analyzes.49
In the tab that indicates the method and scoring function, we chose put in the placement method: Triangle Matcher con the score London dG and number pose in thirty, while that in refinement, we selected a rigid receptor and the scoring GBVI/WSA with five poses. The forcefield used were MMFF94x.49,50
Results and discussion
Chemical space
The space chemicals in 46![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 413 unique compounds were visualized using PCA of descriptors physiochemical properties. The visualization of space chemicals shows that LATAM_DBS_NPs, and REF_NPs occupy similar chemical spaces. Fig. 1 and 2 show a visual representation of the chemical space of 3440 compounds of LATAM_DBS_NPs. The difference in the variance of the six physicochemical properties analyzed by PCA, is present in ESI.†
413 unique compounds were visualized using PCA of descriptors physiochemical properties. The visualization of space chemicals shows that LATAM_DBS_NPs, and REF_NPs occupy similar chemical spaces. Fig. 1 and 2 show a visual representation of the chemical space of 3440 compounds of LATAM_DBS_NPs. The difference in the variance of the six physicochemical properties analyzed by PCA, is present in ESI.†
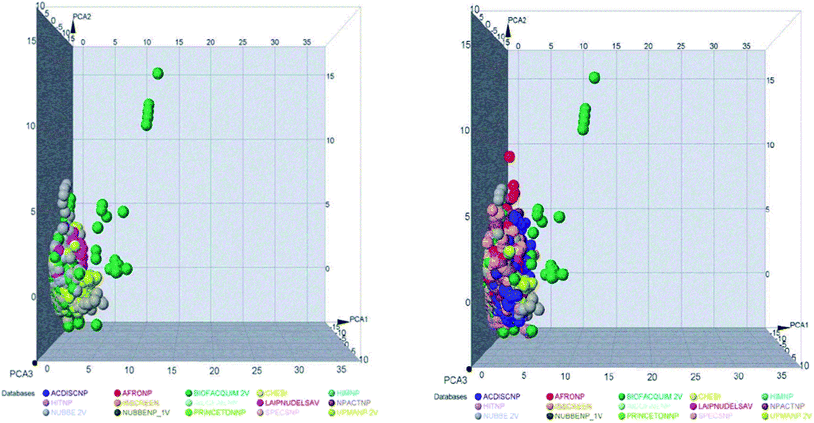 | ||
| Fig. 1 Representation of the chemical space of four LATAM_DBS_NPs (left) and LATAM_DBS_NPs with the eleven REF_NPs (right). | ||
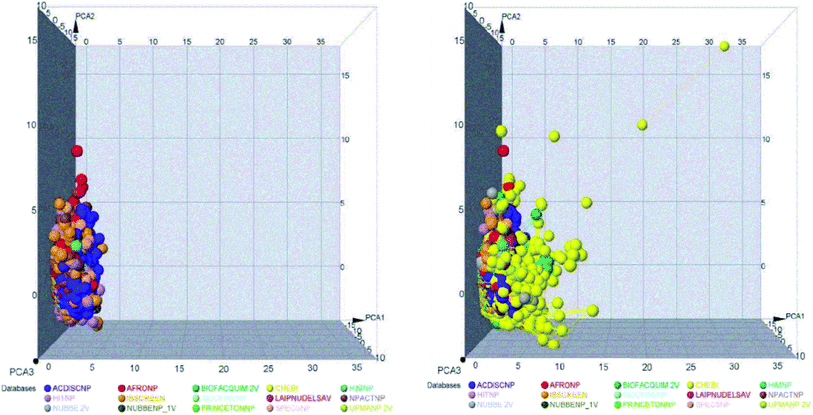 | ||
| Fig. 2 Representation of the chemical space of all NPs, REF_NPs (left) and overlap of all databases evaluated (right). | ||
These data indicate that the partition coefficient (S![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log
log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P) and TPSA have the greatest effect on the variance statistic with a value of 66.69% and 22.39%, respectively, while others descriptor of physicochemical properties has a much lower influence on the envelope, showing a value of 6.27%.
P) and TPSA have the greatest effect on the variance statistic with a value of 66.69% and 22.39%, respectively, while others descriptor of physicochemical properties has a much lower influence on the envelope, showing a value of 6.27%.
Fingerprint-based diversity
The molecular diversity of LATAM_DBS_NP and REF_NPs were calculated using the MACCS and ECFP-6 fingerprint (FP) and the Tanimoto and Cosine similarity index.The graphic of diversity analysis on the x-axis is the similarity value pairs based on ECFP-6/Tanimoto, ECFP-6/Cosine, MACCS/Tanimoto/, MACCS/Cosine index, while on the axis of the ordinate we plotted the CDF values for each database evaluated. The CDF realized with Tanimoto/ECFP-6 indicated that LAIPNUDELSAV and PRINCETON_NP datasets are the most diverse, while that with Cosine/ECFP-6 the INDOFINE_NP, LAIPNUDELSAV, PRINCETON_NP and UPMA_2V databases demonstrated better diversity based on fingerprint (ESI†).
The analysis of MACCS fingerprint and Tanimoto similarity indicated that LAIPNUDELSAV, PRINCETON_NP and INDOFINE_NP are more diverse and HIT NP and CHEBI are the least diverse; meanwhile, the metric MACSS/Cosine similarity index, HIMNP indicated greater diversity as the database reference while LATAM_DBS_NPs and REF_NP showed median values greater than 0.50, except for NUBBE_2V, SPECSNP and UPMANP2V which were less diverse, according to MACCS keys/Cosine similarity index. Tables 3 and 4 show summary statistics of the pairwise similarity computed with MACCS/Tanimoto and MACCS/Cosine.
| NPS_DBS | Mean | SD | Min | 1st Q | Median | 3rd Q | Max |
|---|---|---|---|---|---|---|---|
| a Min: minimum; Max: maximum; Q: quartile; SD, standard deviation. | |||||||
| ACDISC NP | 0.4883 | 0.1426 | 0.0000 | 0.3860 | 0.4821 | 0.5849 | 1.0 |
| AFRO NP | 0.4634 | 0.1709 | 0.0000 | 0.3462 | 0.4694 | 0.5818 | 1.0 |
| BIOFACQUIM | 0.4534 | 0.1447 | 0.0172 | 0.3529 | 0.4444 | 0.5429 | 1.0 |
| CHEBI | 0.2935 | 0.1473 | 0.0 | 0.1875 | 0.2821 | 0.3857 | 1.0 |
| HIM NP | 0.4547 | 0.1647 | 0.0294 | 0.3333 | 0.4340 | 0.5614 | 1.0 |
| HIT NP | 0.3836 | 0.1845 | 0.00 | 0.2537 | 0.375 | 0.5000 | 1.0 |
| IBSCREEN | 0.4614 | 0.1309 | 0.0000 | 0.3704 | 0.4627 | 0.5507 | 1.0 |
| INDOFINENP | 0.5022 | 0.1973 | 0.0870 | 0.3684 | 0.4681 | 0.5778 | 1.0 |
| LAIPNUDELSAV | 0.5978 | 0.1539 | 0.1277 | 0.4902 | 0.5870 | 0.7045 | 1.0 |
| NPACTNP | 0.4842 | 0.1633 | 0.0000 | 0.3778 | 0.4884 | 0.5962 | 1.0 |
| NUBBE 1V | 0.4362 | 0.1653 | 0.0000 | 0.3214 | 0.4286 | 0.5385 | 1.0 |
| NUBBE 2V | 0.4243 | 0.1728 | 0.0000 | 0.3061 | 0.4259 | 0.5410 | 1.0 |
| PRINCETONNP | 0.5176 | 0.1329 | 0.0429 | 0.4237 | 0.5000 | 0.6000 | 1.0 |
| SPECSNP | 0.4790 | 0.1598 | 0.0400 | 0.3594 | 0.4638 | 0.5926 | 1.0 |
| UPMANP 2V | 0.4885 | 0.1608 | 0.0000 | 0.3768 | 0.4744 | 0.5926 | 1.0 |
| NPS_DBS | Mean | SD | Min | 1st Q | Median | 3rd Q | Max |
|---|---|---|---|---|---|---|---|
| a Min: inimum; Max: maximum; Q: quartile; SD, standard. | |||||||
| ACDISC NP | 0.5674 | 0.2310 | 0.0211 | 0.3747 | 0.5628 | 0.7968 | 1.0000 |
| AFRO NP | 0.5590 | 0.2287 | 0.0294 | 0.3711 | 0.5483 | 0.7757 | 1.0000 |
| BIOFACQUIM | 0.5231 | 0.2224 | 0.0416 | 0.3425 | 0.5038 | 0.7280 | 1.0000 |
| CHEBI | 0.4783 | 0.2070 | 0.0047 | 0.3214 | 0.4616 | 0.6259 | 1.0000 |
| HIM NP | 0.6139 | 0.2166 | 0.0197 | 0.4299 | 0.6406 | 0.8071 | 1.0000 |
| HIT NP | 0.5573 | 0.2238 | 0.0178 | 0.3730 | 0.544 | 0.7177 | 1.000 |
| IBSCREEN | 0.5666 | 0.2576 | 0.1987 | 0.3136 | 0.5502 | 0.8004 | 0.9083 |
| INDOFINE NP | 0.5059 | 0.2558 | 0.1004 | 0.2961 | 0.5511 | 0.6273 | 1.0000 |
| LAIPNUDELSAV | 0.5743 | 0.2645 | 0.1916 | 0.3705 | 0.5423 | 0.8362 | 0.9785 |
| NPACT NP | 0.5298 | 0.1606 | 0.2839 | 0.4056 | 0.5639 | 0.6240 | 0.8189 |
| NUBBE 1V | 0.5261 | 0.1577 | 0.2557 | 0.3869 | 0.5219 | 0.6464 | 0.8839 |
| NUBBE 2V | 0.4475 | 0.2088 | 0.0972 | 0.3094 | 0.4252 | 0.5601 | 0.8756 |
| PRINCETONNP | 0.5411 | 0.1530 | 0.2328 | 0.4680 | 0.5262 | 0.6440 | 0.8626 |
| SPECSNP | 0.4191 | 0.2066 | 0.1567 | 0.2651 | 0.3416 | 0.5419 | 0.9026 |
| UPMANP 2V | 0.5212 | 0.1842 | 0.1731 | 0.3789 | 0.4936 | 0.6729 | 0.9360 |
Scaffold diversity
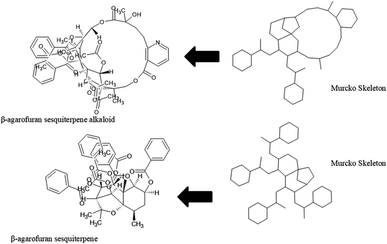
The diversity of scaffolds of Latin American NPs was based on the Murcko scaffold and Murcko skeleton was obtained with RStudio.45,46 The Murcko scaffold contains all plain ring systems of the given molecules, plus all direct connections between them. Substituents, which do not contain ring systems are removed from rings and ring connecting chains, while that in the Murcko skeleton was a generalized Murcko scaffold, which has all heteroatoms replaced with carbon atoms (Fig. 3).31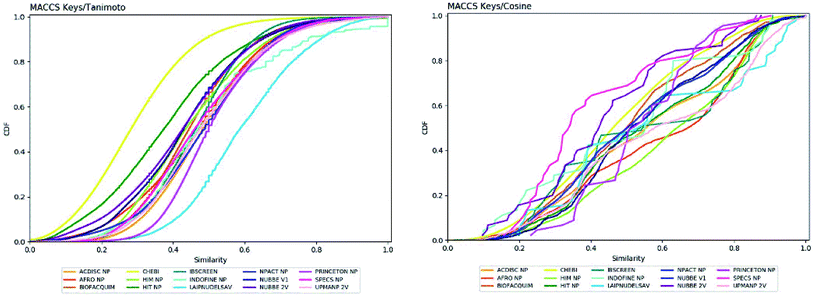 | ||
| Fig. 3 Cumulative distribution function of the pairwise similarity distribution of the different data sets computed with MACCS-Tanimoto and Cosine. | ||
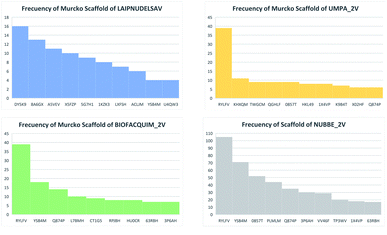
Table 5 summarizes the results of the scaffold diversity of the fifteen databases. In this table, the number and fraction of scaffolds are reported, the number and fraction of scaffolds containing only one compound (singletons), and metrics were obtained from CSR curves (AUC and F50). According to these metrics, databases: NPACT_NP, AFRODB, BIOFACQUIM_2V, and UPMA_2V are those having the largest fractions of scaffolds (FN/M) with values equal to or greater than 0.46.
| Databases | M | N | FN/M | Nsing | FNsing/M | FNsing/N | AUC | F50 |
|---|---|---|---|---|---|---|---|---|
| a N: number of chemotypes; M: number of molecules; Nsing: number of singletons; AUC: area under the curve; F50: fraction of chemotypes that contains 50% of the data set. | ||||||||
| LAIPNUDELSAV | 196 | 76 | 0.3878 | 40 | 0.2041 | 0.5263 | 0.7301 | 0.1579 |
| UPMA_2V | 462 | 215 | 0.4654 | 129 | 0.2792 | 0.6000 | 0.7096 | 0.1907 |
| BIOFACQUIM_2V | 507 | 239 | 0.4714 | 144 | 0.2840 | 0.6025 | 0.7739 | 0.1042 |
| NUBBE_2V | 2279 | 720 | 0.3159 | 349 | 0.1531 | 0.4847 | 0.7633 | 0.1177 |
| IBSCREEN | 8413 | 2881 | 0.3424 | 1449 | 0.1722 | 0.5030 | 0.7890 | 0.0415 |
| CHEBI | 5622 | 2171 | 0.3862 | 1637 | 0.2912 | 0.7540 | 0.7096 | 0.1621 |
| AFRONP | 494 | 253 | 0.5121 | 180 | 0.3644 | 0.7115 | 0.7988 | 0.0950 |
| ACDISCNP | 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 859 859 |
1663 | 0.1531 | 405 | 0.0373 | 0.2435 | 0.7208 | 0.2059 |
| HIM_NP | 637 | 136 | 0.2135 | 30 | 0.0471 | 0.2206 | 0.7287 | 0.1959 |
| HIT_NP | 583 | 194 | 0.3328 | 77 | 0.1321 | 0.3969 | 0.7115 | 0.2000 |
| INDOFINE_NP | 99 | 30 | 0.3030 | 13 | 0.1313 | 0.4333 | 0.7052 | 0.1702 |
| NPACT_NP | 1093 | 564 | 0.5160 | 390 | 0.3568 | 0.6915 | 0.7499 | 0.1593 |
| SPECS_NP | 1349 | 364 | 0.2698 | 133 | 0.0986 | 0.3654 | 0.9046 | 0.0255 |
| PRINCETON_NP | 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 672 672 |
1020 | 0.0746 | 396 | 0.0290 | 0.3882 | 0.7736 | 0.0962 |
| NUBBE_1V | 148 | 52 | 0.3514 | 31 | 0.2095 | 0.5962 | 0.7100 | 0.1841 |
Fig. 4(a) shows the metric of Murcko scaffold (ring system) in the fifteen databases that correlated with CSR curves, F50 (a fraction of chemotypes that contains 50% of the data set), and areas under the curve (AUC) are reported in Table 5. These curves indicate that the Latin American dataset UPMA_2V and NUBBE_1V are those with the greatest structural diversities with F50 0.19 and 0.18, respectively. On the contrary, when comparing the values of AUC 0.90, 0.79. 0.78 and 0.77, we observe that the databases SPECS_NP, AFRODB, IBSCREEN, BIOFACQUIM_2V and PRINCETON_NP are the least diverse.
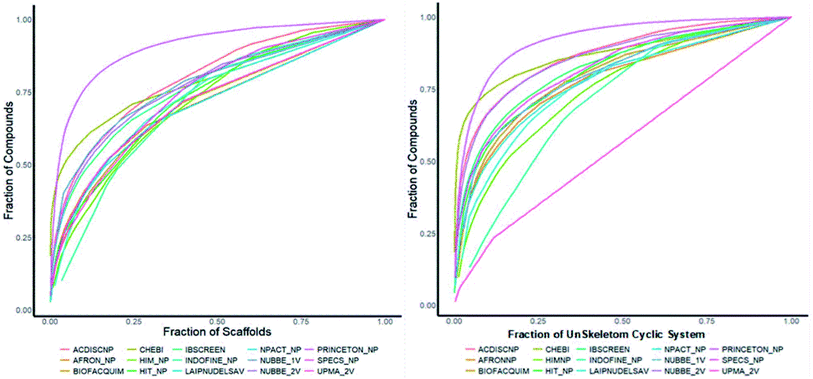 | ||
| Fig. 4 Murcko scaffold (a) and Murcko skeleton (b) retrieval curves for the data sets studied in this work. | ||
However, Fig. 4(b) shows the CRS curves of skeleton cyclic system and AUC and F50 metrics. The CSR curves indicate that SPECS_NP, AFRONNP, IBSCREEN, AFRONNP, BIOFACQUIM_2V and NUBBE_2V are the least diverse when comparing their values of AUC 0.92, 0.87, 0.86, 0.85 and 0.82, respectively, while when comparing the values of F50, it was observed that the database UPMA_2V (0.42) and HIT_NP are the most diverse in its content from the Murcko skeleton, and ACDISCNP, LAIPNUDELSAV and INDOFINE_NP have diversity Murcko skeleton intermediate values from 0.10 to 0.15.
Meanwhile, Table 6 summarizes the diversity of the Murcko skeleton (unskeleton cyclic system) of the fifteen databases. This table presents the number and fraction of unskeleton cyclic systems (Mucko skeleton), the number and fraction of containing only one compound (singletons), and metrics obtained from CSR curves (AUC: area under the curve; F50: fraction of chemotypes that contains 50% of the data set.)
| Databases | M | N | FN/M | Nsing | FNsing/M | FNsing/N | AUC | F50 |
|---|---|---|---|---|---|---|---|---|
| a N: number of chemotypes; M: number of molecules; Nsing: number of singletons; AUC: area under the curve; F50: fraction of chemotypes that contains 50% of the data set. | ||||||||
| LAIPNUDELSAV | 196 | 46 | 0.2347 | 17 | 0.0867 | 0.3696 | 0.7717 | 0.1304 |
| UPMA_2V | 462 | 401 | 0.8680 | 355 | 0.7684 | 0.8853 | 0.5601 | 0.4239 |
| BIOFACQUIM_2V | 507 | 134 | 0.2643 | 66 | 0.1302 | 0.4925 | 0.8574 | 0.0395 |
| NUBBE_2V | 2279 | 354 | 0.1553 | 146 | 0.0641 | 0.4124 | 0.8240 | 0.0624 |
| IBSCREEN | 8413 | 1668 | 0.1983 | 645 | 0.0767 | 0.3867 | 0.8723 | 0.0075 |
| CHEBI | 5622 | 1206 | 0.2145 | 830 | 0.1476 | 0.6882 | 0.7777 | 0.0964 |
| AFRONNP | 494 | 166 | 0.3360 | 99 | 0.2004 | 0.5964 | 0.8665 | 0.0334 |
| ACDISCNP | 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 859 859 |
898 | 0.0827 | 186 | 0.0171 | 0.2071 | 0.7512 | 0.1566 |
| HIMNP | 637 | 83 | 0.1303 | 8 | 0.0126 | 0.0964 | 0.8081 | 0.0796 |
| HIT_NP | 583 | 113 | 0.1938 | 42 | 0.0720 | 0.3717 | 0.7079 | 0.2273 |
| INDOFINE_NP | 99 | 22 | 0.2222 | 7 | 0.0707 | 0.3182 | 0.7767 | 0.1034 |
| NPACT_NP | 1093 | 348 | 0.3184 | 184 | 0.1683 | 0.5287 | 0.8217 | 0.0698 |
| SPECS_NP | 1349 | 215 | 0.1594 | 70 | 0.0519 | 0.3256 | 0.9202 | 0.0247 |
| PRINCETON_NP | 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 672 672 |
486 | 0.0355 | 145 | 0.0106 | 0.2984 | 0.7945 | 0.0833 |
| NUBBE_1V | 148 | 36 | 0.2432 | 14 | 0.0946 | 0.3889 | 0.7956 | 0.0896 |
Scaffold content in Latin American databases of natural product
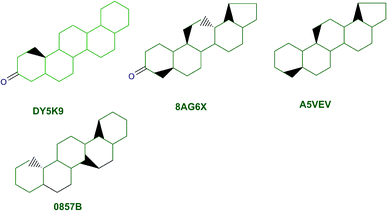
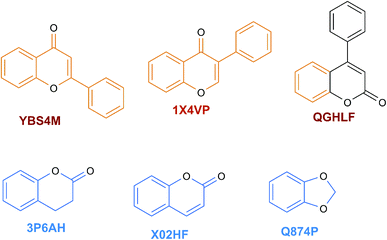
The content of the scaffold in LATAM_NPs has a great structural variety. The chemotypes: DY5K9, 8A6GX, A5VEV and 0857T are exclusively found in LAIPNUDELSAV_DBs. They possess the pentacyclic triterpene skeleton as shown in Fig. 5. In contrast, Fig. 6 shows some of the similar scaffolds present in LATAM_DBs: YSB4M and 1X4VP that have a benzopyran-4-one. However, these skeletons are contained in other chemotypes such as RPJBH, 63RBH, and KHXQM, which have a low frequency in BIOFACQUIM and UPMA_2V, respectively. Other chemotypes present is benzopyran-2-one identified by the codes are Q874P, 3P6AH and X02HF that are shown in Fig. 6. These nuclei are present in CT1G5, QGHLF and PLMLM, identified in the databases BIOFACQUIM, NUBBE_2V and UPMA_2V. This group constitutes the most abundant compounds in LATAM_NPs, which are shown in Fig. 7. Murcko skeletons included in the four Latin American natural product databases are similar to the nuclei present in Fig. 5 and 6. However, the β-agarofuran skeletons: X5FZP, 5G7H1, 1KZK3, LXFSH and ACLIM represent the second group of compounds in LAINPUDELSAV DBs. Fig. 8 shows two beta-agarofurans present in high frequency in the databases of the Natural Products Laboratory of the University of El Salvador, we observe their high structural complexity.Molecular complexity
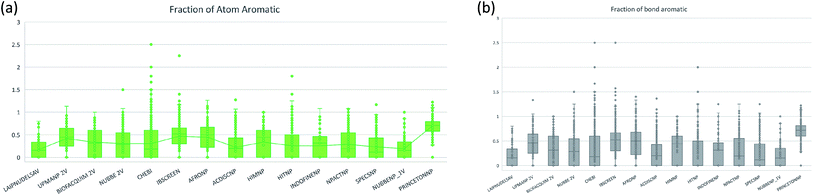
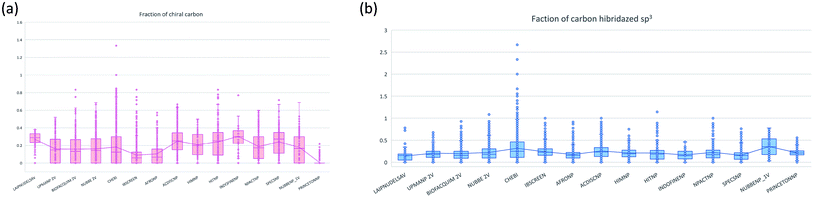
The molecular complexity of LATAM_DBS_NPs and eleven reference databases were calculated by means of five metrics of molecular properties: a fraction of sp3 hybridized carbons (Fsp3), fraction of chiral carbon (FCC), fraction of atoms aromatic (Fa_Ar), fraction of aromatic bond (Far_b), globularity molecular (glob_m), flexibility in molecular and shape index.Fig. 9 and 10 show box plots for the distributions of Fa_aro, F_sp3, Fb_aro, and FCC. The globularity molecular, molecular flexibility and shape index (ESI†). Tables 7–10 summarize the statistics of Fa_aro, Fb_aro, F_sp3, and FCC.
| Fa_aro | Min | 1Qst | Median | 3Qst | Max | Mean | SD |
|---|---|---|---|---|---|---|---|
| a Min: minimum; Max: maximum; Q: quartile; SD: standard deviation. | |||||||
| LAIPNUDELSAV | 0.00 | 0.00 | 0.15 | 0.33 | 0.80 | 0.17 | 0.19 |
| UPMANP 2V | 0.00 | 0.25 | 0.44 | 0.64 | 1.13 | 0.41 | 0.27 |
| BIOFACQUIM_2V | 0.00 | 0.00 | 0.31 | 0.60 | 1.00 | 0.33 | 0.29 |
| NUBBE_2V | 0.00 | 0.00 | 0.28 | 0.54 | 1.50 | 0.30 | 0.28 |
| CHEBI | 0.00 | 0.00 | 0.18 | 0.60 | 2.50 | 0.30 | 0.34 |
| IBSCREEN | 0.00 | 0.30 | 0.51 | 0.65 | 2.25 | 0.46 | 0.25 |
| AFRONP | 0.00 | 0.22 | 0.50 | 0.66 | 1.26 | 0.44 | 0.28 |
| ACDISCNP | 0.00 | 0.00 | 0.20 | 0.42 | 1.27 | 0.22 | 0.24 |
| HIMNP | 0.00 | 0.00 | 0.44 | 0.60 | 1.00 | 0.33 | 0.30 |
| HITNP | 0.00 | 0.00 | 0.00 | 0.50 | 1.80 | 0.25 | 0.31 |
| INDOFINENP | 0.00 | 0.00 | 0.31 | 0.45 | 1.08 | 0.25 | 0.28 |
| NPACTNP | 0.00 | 0.00 | 0.19 | 0.54 | 1.08 | 0.28 | 0.29 |
| SPECSNP | 0.00 | 0.00 | 0.11 | 0.42 | 1.16 | 0.23 | 0.26 |
| NUBBE_1V | 0.00 | 0.00 | 0.15 | 0.33 | 1.00 | 0.19 | 0.23 |
| PRINCENTONNP | 0.00 | 0.57 | 0.70 | 0.79 | 1.22 | 0.69 | 0.15 |
| Fb_aro | Min | 1Qst | Median | 3Qst | Max | Mean | SD |
|---|---|---|---|---|---|---|---|
| a Min: minimum; Max: maximum; Q: auartile; SD: standard deviation. | |||||||
| LAIPNUDELSAV | 0.00 | 0.00 | 0.15 | 0.33 | 0.80 | 0.17 | 0.19 |
| UPMANP 2V | 0.00 | 0.25 | 0.46 | 0.64 | 1.33 | 0.42 | 0.27 |
| BIOFACQUIM_2V | 0.00 | 0.00 | 0.31 | 0.60 | 1.00 | 0.34 | 0.30 |
| NUBBE_2V | 0.00 | 0.00 | 0.28 | 0.54 | 1.50 | 0.30 | 0.28 |
| CHEBI | 0.00 | 0.00 | 0.18 | 0.60 | 2.50 | 0.31 | 0.35 |
| IBSCREEN | 0.00 | 0.30 | 0.52 | 0.66 | 2.50 | 0.48 | 0.26 |
| AFRONP | 0.00 | 0.22 | 0.50 | 0.67 | 1.40 | 0.45 | 0.29 |
| ACDISCNP | 0.00 | 0.00 | 0.20 | 0.42 | 1.36 | 0.22 | 0.24 |
| HIMNP | 0.00 | 0.00 | 0.44 | 0.60 | 1.00 | 0.33 | 0.31 |
| HITNP | 0.00 | 0.00 | 0.00 | 0.50 | 2.00 | 0.26 | 0.32 |
| INDOFINENP | 0.00 | 0.00 | 0.31 | 0.45 | 1.25 | 0.26 | 0.29 |
| NPACTNP | 0.00 | 0.00 | 0.19 | 0.55 | 1.25 | 0.29 | 0.30 |
| SPECSNP | 0.00 | 0.00 | 0.11 | 0.44 | 1.25 | 0.23 | 0.27 |
| NUBBE_1V | 0.00 | 0.00 | 0.15 | 0.35 | 1.00 | 0.19 | 0.23 |
| PRINCENTONNP | 0.00 | 0.60 | 0.72 | 0.81 | 1.23 | 0.70 | 0.15 |
| F_sp3 | Min | 1Qst | Median | 3Qst | Max | Mean | SD |
|---|---|---|---|---|---|---|---|
| a Min: minimum; Max: maximum; Q: quartile; SD: standard deviation. | |||||||
| LAIPNUDELSAV | 0.00 | 0.05 | 0.15 | 0.19 | 0.78 | 0.14 | 0.11 |
| UPMANP 2V | 0.00 | 0.11 | 0.18 | 0.25 | 0.68 | 0.20 | 0.15 |
| BIOFACQUIM_2V | 0.00 | 0.09 | 0.16 | 0.25 | 0.93 | 0.20 | 0.18 |
| NUBBE_2V | 0.00 | 0.10 | 0.18 | 0.30 | 1.09 | 0.22 | 0.18 |
| CHEBI | 0.00 | 0.11 | 0.25 | 0.46 | 2.67 | 0.32 | 0.28 |
| IBSCREEN | 0.00 | 0.16 | 0.23 | 0.30 | 1.00 | 0.24 | 0.12 |
| AFRONP | 0.00 | 0.10 | 0.16 | 0.22 | 0.94 | 0.18 | 0.14 |
| ACDISCNP | 0.00 | 0.13 | 0.23 | 0.33 | 1.04 | 0.26 | 0.18 |
| HIMNP | 0.00 | 0.13 | 0.19 | 0.27 | 0.75 | 0.21 | 0.13 |
| HITNP | 0.00 | 0.07 | 0.19 | 0.27 | 1.14 | 0.21 | 0.19 |
| INDOFINENP | 0.00 | 0.08 | 0.15 | 0.25 | 0.46 | 0.17 | 0.12 |
| NPACTNP | 0.00 | 0.10 | 0.18 | 0.27 | 1.00 | 0.22 | 0.19 |
| SPECSNP | 0.00 | 0.07 | 0.15 | 0.22 | 0.76 | 0.16 | 0.12 |
| NUBBE_1V | 0.03 | 0.17 | 0.35 | 0.52 | 0.78 | 0.36 | 0.20 |
| PRINCENTONNP | 0.00 | 0.17 | 0.21 | 0.26 | 0.56 | 0.21 | 0.07 |
| FCC | Min | 1Qst | Median | 3Qst | Max | Mean | SD |
|---|---|---|---|---|---|---|---|
| a Min: minimum; Max: maximum; Q: quartile; SD: standard deviation. | |||||||
| LAIPNUDELSAV | 0.00 | 0.23 | 0.29 | 0.33 | 0.38 | 0.27 | 0.08 |
| UPMANP 2V | 0.00 | 0.00 | 0.15 | 0.27 | 0.52 | 0.16 | 0.15 |
| BIOFACQUIM_2V | 0.00 | 0.00 | 0.13 | 0.27 | 0.83 | 0.16 | 0.16 |
| NUBBE_2V | 0.00 | 0.00 | 0.14 | 0.28 | 0.69 | 0.16 | 0.14 |
| CHEBI | 0.00 | 0.00 | 0.13 | 0.30 | 1.33 | 0.18 | 0.20 |
| IBSCREEN | 0.00 | 0.00 | 0.06 | 0.13 | 0.83 | 0.09 | 0.10 |
| AFRONP | 0.00 | 0.00 | 0.07 | 0.16 | 0.57 | 0.09 | 0.10 |
| ACDISCNP | 0.00 | 0.15 | 0.25 | 0.34 | 0.68 | 0.10 | 0.12 |
| HIMNP | 0.00 | 0.10 | 0.20 | 0.32 | 0.50 | 0.21 | 0.14 |
| HITNP | 0.00 | 0.09 | 0.25 | 0.35 | 0.83 | 0.24 | 0.18 |
| INDOFINENP | 0.00 | 0.22 | 0.30 | 0.37 | 0.77 | 0.30 | 0.19 |
| NPACTNP | 0.00 | 0.05 | 0.17 | 0.30 | 0.60 | 0.19 | 0.15 |
| SPECSNP | 0.00 | 0.11 | 0.27 | 0.35 | 0.71 | 0.24 | 0.14 |
| NUBBE_1V | 0.00 | 0.00 | 0.17 | 0.30 | 0.69 | 0.18 | 0.16 |
| PRINCENTONNP | 0.00 | 0.00 | 0.00 | 0.00 | 0.23 | 0.01 | 0.02 |
The analysis of the fraction of atoms and bond aromatics having this database shows median values between 0.44 and 0.71 for PRINCETON_NP, IBSCREEN_NP, HIM_NP, AFRONP and UPMA_NP_2V, indicating high aromaticity and rigidity in these databases. However, the analysis of the carbon fraction with sp3 hybridization indicates that the databases IBSCREEN_NP, CHEBI, ACDISC_NP, PRINCETON_NP and NUBBE_1V are the most complex, showing values from 0.22 to 0.35, while LAIPNUDELSAV, AFRO_NP, BIOFACQUIM_2V INDOFINE_NP, UPMA_NP 2V, and NPACT_NP are the least complex with values between 0.15 and 0.17. Otherwise, the chiral carbon fraction indicates that HIM_NP, HIT_NP, ACDIS_NP, SPECS_NP, LAIPNUDELSAV and INDOFINE_NP have a greater number of chiral carbons with values of 0.20 to 0.30.
The analysis of the molecular flexibility and the shape index of LATAM_DBS_NPs and REF_NP suggests that the compounds have high structural rigidity and spherical shape (non-linear) since they present values less than 0.50 for most of the analyzed databases (ESI†).
The globularity molecular is not a good metric of complexity because it does not differentiate the data sets analyzed in this work (ESI†).
Molecular docking
NSP-15 encodes a uridylate-specific endoribonuclease enzyme (EndoU), indispensable for the viral cycle of coronavirus (COVID-19). This facilitates the breakdown of RNA at the ends of the uridylates. The loss of NSP-15 directly impacts RNA replication processes and their pathogenesis. Therefore, NSP-15 constitutes a potential therapeutic target for the development of inhibitors of endoribonuclease-dependent viral replication.52
NSP-15 (endoribonuclease) was selected for this work since there is little research on this target of the coronavirus (SARS-CoV-2).
The structure NSP-15 (endoribonuclease) complex with tipiracid was obtained from the Protein Data Bank. This viral protein of the severe acute respiratory syndrome coronavirus 2 (SARS CoV-2) was encoded as 6WXC PDB and possesses uridylate-specific endoribonuclease with A and B chains.53
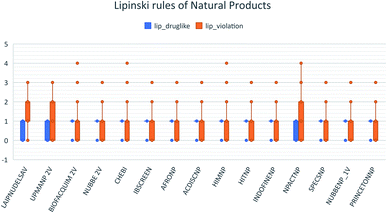
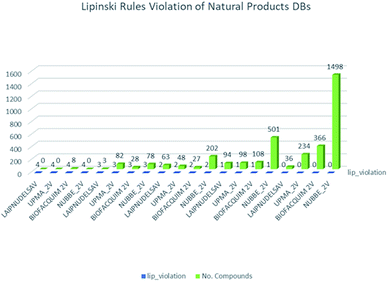
These data indicate that LAIPNUDELSAV and UMPA_2V are the two most diverse, have more fraction contents of Murcko scaffold and Murcko skeleton, and overly complex ring systems. These 482 compounds were selected using inclusion criteria, one violation of the Lipinski rules. Fig. 12 shows the comparison of drug-like properties and the violation of Lipinski's rule for the fifteen databases of natural products evaluated, meanwhile, Fig. 13 presents an illustration of distribution by the number of compounds in all databases of natural products that meet and break the Lipinski rule (Fig. 14).
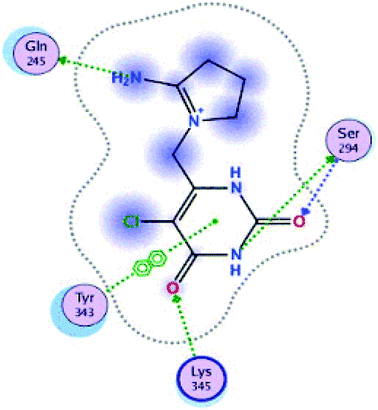
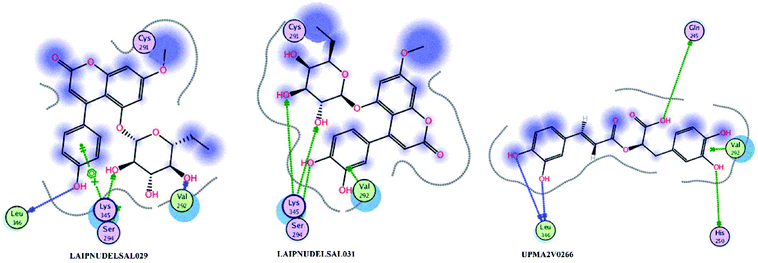
Fig. 15 and 16 visualize the binding mode between the ligands, LAIPNUDELSAV_029, LAIPNUDELSAL_031 and UPMA_2V_0266 in the active site of endoribonucleases NSP-15 in model 2D and 3D.
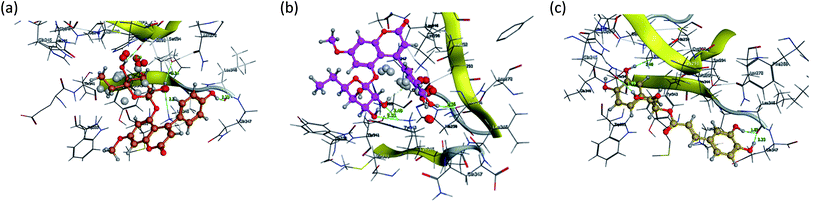 | ||
| Fig. 16 Binding interaction of LAIPNUDELSAV_029, LAIPNUDELSAV_031 and UPMA_2V_0266 within the active site of endoribonucleases NSP-15 in 3D. | ||
LAIPNUDELSAV_029 and LAIPNUDELSAV_031 corresponding a 4-phenylcoumarins with O-glycosidic bonds in C-7 and the hydroxyl in C-1′ of sugar residues,54 while UPMA-2V_0266 a derivative of caffeic acid.55
These three compounds share with the cocrystallized ligand (CMU) the hydrogen bridge interaction with Ser294A and Lys345A residues, while we observed additional interactions with Val292, Leu346 and His 250 residues with these natural products.
Phenylcoumarins and rosmarinic acid interact in a similar way at the binding site of the NSP-15 protein but occupy a greater volume at this site, therefore, we propose that these substances could act as inhibitors of this viral protein, but these statements must be validated in subsequent studies of this work.
Table 11 shows the functional groups of the three ligands that interact with amino acid residues in the active site of NSP-15. In addition, it includes the docking score, RMSD, and binding energy values obtained from the molecular docking simulation with the MOE v2019.01 software.49
| Ligand group | Receptor | Residue of amino acids | Types of interaction | Energy of bond interaction, kcal mol−1 | Docking score | RMSD refine | Binding energy, kcal mol−1 |
|---|---|---|---|---|---|---|---|
| LAIPNUDELSAV_029 | −5.1983 | 2.1983 | −10.1 | ||||
| H–O–C-4′ | O | LEU346 A | H-Donor | −1.7 | |||
| H–O–C-3′′ | O | SER294 A | H-Donor | −2.3 | |||
| H–O–C-5′′ | O | VAL292 A | H-Donor | −3.0 | |||
| H–O–C-3′′ | N | LYS345 A | H-Acceptor | −2.4 | |||
| 6-Ring | N | LYS345 A | Pi-Cation | −0.8 | |||
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||||||
| LAIPNUDELSAV_031 | −5.0742 | 1.6492 | −4.9 | ||||
| H–O–C-4′′ | N | LYS345 A | H-Acceptor | −1.7 | |||
| H–O–C-3′′ | O | LYS345 A | H-Acceptor | −1.3 | |||
| H–O–C-5′′ | O | SER294 A | H-Donor | −1.7 | |||
| 6-Ring | N | VAL292 A | Pi-Cation | −0.2 | |||
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||||||
| UPMA-2V_0266 | −5.7371 | 2.6559 | −8.7 | ||||
| H–O–C-14 | N | HIS250 A | H-Donor | −2.1 | |||
H–O–C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O (C-18) O (C-18) |
O | GLN245 A | H-Donor | −1.4 | |||
| H–O–C-4 | O | LEU346 A | H-Donor | −1.3 | |||
| H–O–C-3 | O | LEU346 A | H-Donor | −3.7 | |||
| 6-Ring | N | VAL292 A | Pi-Cation | −0.2 | |||
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||||||
| TIPIRACID (CMU) | −5.4824 | 1.7440 | −27.2 | ||||
These skeletons have biological activity against HIV, and tuberculosis, and has antioxidant activity.
The docking study has allowed the identification of two 4-phenylcoumarins and catechol derivatives as potential inhibitors of riboendonuclease NPS-15 of SARS-CoV-2. A variety of NPs (alkaloid, catechol derivative, benzofuran, benzopyran, polyphenols)56 and 4-methyl-coumarin derivatives57 have been reported as inhibitors of structural and no structural proteins of COVID-19.
Conclusions
In this cheminformatics study, we analysed 46![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 413 compounds from four unique NPs from Latin American natural products and eleven published NPs databases used as references. These NPs occupy similar chemical spaces, therefore share properties of interest in the bioprospecting project of NPs for the discovery and development of lead molecules of therapeutic potential.
413 compounds from four unique NPs from Latin American natural products and eleven published NPs databases used as references. These NPs occupy similar chemical spaces, therefore share properties of interest in the bioprospecting project of NPs for the discovery and development of lead molecules of therapeutic potential.
The databases of the University of El Salvador and the University of Panama present the greatest structural diversities based on the fingerprint. However, when comparing the chemotype relationship, NPs of Africa, Mexico, and Panama have the highest scaffold diversity. The University of El Salvador database, herbal ingredients active targets, and ICC indofine present the greatest diversity in the Murcko skeleton content bases at F50. The content of the scaffold in three LATAM_NPs predominated compound of types benzopyran-one with modification in positions 2 and 4 of this cyclic system with anti-radical and antioxidant effects, meanwhile pentacyclic triterpenes and β-agarofuran prevailed in LAIPNUDELSAV. This last group of compounds has a reported an anti-inflammatory effect by inhibiting NOS.
From the analysis of molecular complexity, it is concluded that the compounds of the University of Panama are the ones with the highest aromaticity and structural rigidity. The opposite case occurs with NUBBE_1V_2013, which has a large fraction of sp3 carbons, while the databases of the University of El Salvador, Mexico, and Panama have fewer types of carbons; additionally, the LAIPNUDELSAV contains the highest number of chiral carbons. By contrast, the comparison of the molecular flexibility and the shape index of the Latin American NPs and the reference NPs show that all evaluated compounds have high structural rigidity.
Molecular docking simulation has allowed identification of 5-O-β-D-glucopyranosyl-4′-hydroxy-7-methoxy-4-phenylcoumarin, 5-O-β-D-galactopyranosyl-3′,4′-dihydroxy-7-methoxy-4-phenylcoumarin and rosmarinic acid as potential inhibitors of riboendonuclease NPS-15 of SARS-CoV-2. This is the first in the silico study evaluating the potential of Latin American natural products against COVID-19. Furthermore, this work can contribute to helping in the future design of new candidate anti-SARS-CoV-2.
Author contributions
M. J. N. participated in the conceptualization, design, and creation of databases of the University of El Salvador. He was also actively involved in the analysis of results, writing, reviewing, and editing of the manuscript. B. I. D. E. T. was involved in data curation, computational analysis, designing computer program, supporting algorithms and validation of the results. She also collaborated in the writing, review, and edition of the manuscript. J. L. M. F. has worked in the conceptualization of research, review of methodological design, analysis, visualization, and statistical analysis of the results. Also, he participated in the writing, review, and edition of the manuscript. D. A. O. A. participated in the conceptualization, creation, conducting, methodological design of the research and obtaining funds. He also actively collaborated on data curation, statistical evaluation, computational analysis, writing, reviewed editing manuscript.Conflicts of interest
Authors declare that there are no financial or commercial conflicts of interest.Acknowledgements
DAO thanks the Vice-rectory of Research and Postgraduate Studies of the University of Panama for the funds awarded through the VIP Project 01-14-2019-05. This work was made possible thanks to the support of the Sistema Nacional de Investigación (SNI) of Secretaría Nacional de Ciencia, Tecnología e Innovación of Panama (2018–2021). This research is part of the International Collaborative Project: The Rise of Natural Product Libraries in Latin America and Computer Aided Drug Discovery.References
- D. J. Newman and G. M. Cragg, J. Nat. Prod., 2020, 83(3), 770–803 CrossRef CAS PubMed.
- M. Sorokina and C. Steinbeck, J. Cheminf., 2020, 12, 20, DOI:10.1186/s13321-020-00424-9.
- J. L. Medina-Franco and F. I. Saldívar-González, Biomolecules, 2020, 10, 1566 CrossRef CAS PubMed.
- F. D. Prieto-Martínez and U. Norinder, Cheminformatics explorations of natural products, in Progress in the Chemistry of Organic Natural Products, ed. A. Kinghorn, H. Falk, S. Gibbons, J. Kobayashi and Y. Asakawa, Springer, Cham, Switzerland, 2019, vol. 110 Search PubMed.
- J. L. Medina-Franco, Future Sci. OA, 2020, 6, FSO468 CrossRef PubMed.
- K. Martínez-Mayorga, A. Madariaga-Mazon, J. L. Medina-Franco and G. Maggiora, Expert Opin. Drug Discovery, 2020, 15(3), 293–306 CrossRef PubMed.
- N. Sánchez-Cruz, B. A. Pilón-Jiménez and J. L. Medina-Franco, F1000Research, 2020, 8, 2071, DOI:10.12688/f1000research.21540.2.
- A. L. Chávez-Hernández, N. Sánchez-Cruz and J. L. Medina-Franco, Mol. Inf., 2020, 39, 2000050 CrossRef PubMed.
- A. L. Chávez-Hernández, N. Sánchez-Cruz and J. L. Medina-Franco, Biomolecules, 2020, 10(11), 1518 CrossRef PubMed.
- F. I. Saldívar-González and J. L. Medina-Franco, in Small Molecule Drug Discovery, ed. A. L. Trabocchi, Elsevier, Amsterdam, Netherlands, 2020, pp. 83–102 Search PubMed.
- D. A. Olmedo and J. L. Medina-Franco, in Cheminformatics and its Applications, ed. A. Stefaniu, A. Rasul and G. Hussain, London, United Kingdom, 2020, section 1, ch. 6, pp. 83–106 Search PubMed.
- B. A. Pilón-Jiménez, F. I. Saldívar-González, B.I Diaz-Eufracio and J. L. Medina-Franco, Biomolecules, 2019, 9(1), 31 CrossRef PubMed.
- F. I. Saldívar-González, M. Valli, A. D. Andricopulo, V. Da Silva Bolzani and J. L. Medina-Franco, J. Chem. Inf. Model., 2019, 59(1), 74–85 CrossRef PubMed.
- D. A. Olmedo, M. González-Medina, M. P. Gupta and J. L. Medina-Franco, Mol. Diversity, 2017, 21(4), 779–789 CrossRef CAS PubMed.
- E. López-López, J. Bajorath and J. L. Medina-Franco, J. Chem. Inf. Model., 2021, 61(1), 26–35 CrossRef PubMed.
- N. A. M. Al-Rashedi, I. M. G. Munahi and L. A. H. AH ALObaidi, J. Biomol. Struct. Dyn., 2020, 1–14 CrossRef PubMed.
- J. Huang, G. Tao, J. Liu, J. Cai, Z. Huang and J.-X. Chen, Front. Pharmacol., 2020, 11, 1635 Search PubMed.
- A. Da Silva-Antonio, L. S. Silveira-Moreira-Wiedemann and V. F. Veiga-Junior, RSC Adv., 2020, 10, 23379–23393 RSC.
- A. C. Pilon, M. Valli, A. C. Dametto, M. E. F. Pinto, R. Freire, I. Castro-Gamboa, A. D. Andricopulo and V. S. Bolzani, Sci. Rep., 2017, 7(1), 7215 CrossRef PubMed.
- InterBioScreen, https://www.ibscreen.com/natural-compounds. accessed April 30, 2017.
- P. de Matos, R. Alcántara, A. Dekker, M. Ennis, J. Hastings, K. Haug, I. Spiteri, S. Turner and C. Steinbeck, Nucleic Acids Res., 2010, 38(Database issue), D249–D254, DOI:10.1093/nar/gkp886.
- F. Ntie-Kang, D. Zofou, S. B. Babiaka, R. Meudom, M. Scharfe, L. L. Lifongo, J. M. Mbah, L. M. Mbaze, W. Sippl and S. M. N. Efange, PLoS One, 2013, 8(10), e78085 CrossRef CAS PubMed.
- AnalytiCon Discovery NP, http://zinc.docking.org/catalogs/acdiscnp/, accessed April 30, 2017.
- H. Kang, K. Tang, Q. Liu, Y. Sun, Q. Huang, R. Zhu, J. Gao, D. Zhang, C. Huang and Z. Cao, J. Cheminf., 2013, 5(1), 28 CAS.
- H. Ye, L. Ye, H. Kang, D. Zhang, L. Tao, K. Tang, X. Liu, R. Zhu, Q. Liu, Y. Z. Chen, Y. Li and Z. Cao, Nucleic Acids Res., 2011, 39(Database issue), D1055–D1059, DOI:10.1093/nar/gkq1165.
- Indofine Natural Products ICC, http://zinc12.docking.org/catalogs/indofinenp, accessed April 30, 2017.
- M. Mangal, P. Sagar, H. Singh, G. P. S. Raghava and S. M. Agarwal, Nucleic Acids Res., 2013, 41, D1124–D1129, DOI:10.1093/nar/gks1047.
- Specs Natural Products, http://zinc12.docking.org/catalogs/specsnp, accessed May 30, 2018.
- Princeton BioMolecular Research, http://zinc12.docking.org/browse/catalogs/natural-products, accessed May 30, 2018.
- M. Valli, R. N. Dos Santos, L. D. Figueira, C. H. Nakajima, I. Castro-Gamboa, A. D. Andricopulo and V. S. Bolzani, J. Nat. Prod., 2013, 76(3), 439–444 CrossRef CAS PubMed.
- Molecular Operating Environment (MOE), 2018.01; Chemical Computing Group ULC, 1010 Sherbrooke St. West, Suite #910, Montreal, QC, Canada, H3A 2R7, 2019 Search PubMed.
- T. Sander, J. Freyss, M. VonKorff and C. Rufener, J. Chem. Inf. Model., 2015, 55, 460–473 CrossRef CAS PubMed.
- M. G. Santibánez-Morán, E. López-López, F. D. Prieto-Martínez, N. Sánchez-Cruz and J. L. Medina-Franco, RSC Adv., 2020, 10, 25089, 10.1039/d0ra04922k.
- N. A. Durán-Iturbide, B. I. Díaz-Eufracio and J. L. Medina-Franco, ACS Omega, 2020, 5, 16076–16084 CrossRef PubMed.
- B. I. Díaz-Eufracio, O. Palomino-Hernández, R. A. Houghten and J. L. Medina-Franco, Mol. Diversity, 2018, 22(2), 259–267 CrossRef PubMed.
- Python 3.6.12 documentation, https://docs.python.org/6.3, accessed February 16, 2021.
- M. Johnson, 2006, An introduction to the MeqiSuite Indices. Pannanugget Consulting L.L.C, http://www.pannanugget.com/MeqiSuiteIntro.pdf Search PubMed.
- Y.-J. Xu and M. Johnson, J. Chem. Inf. Comput. Sci., 2002, 42, 912–926 CrossRef CAS PubMed.
- Y.-J. Xu and M. Johnson, J. Chem. Inf. Comput. Sci., 2001, 41, 181–185 CrossRef CAS PubMed.
- E. López-López, J. J. Naveja and J. L. Medina-Franco, Expert Opin. Drug Discovery, 2019, 14, 335–341 CrossRef PubMed.
- J. L. Medina-Franco, K. Martínez-Mayorga, A. Bender and T. Scior, QSAR Comb. Sci., 2009, 28, 1551–1560 CrossRef CAS.
- M. González-Medina, J. Owen, T. El-Elimat, C. Pearce, N. Oberlies, M. Figueroa and J. L. Medina-Franco, Front. Pharmacol., 2017, 8, 180 Search PubMed.
- E. Lenci, G. Menchi, F. I. Saldívar-Gonzalez, J. J. Medina-Franco and A. Trabocchi, Org. Biomol. Chem., 2019, 17, 1037–1052 RSC.
- R RStudio Team, RStudio: Integrated Development for R. RStudio, PBC, Boston, MA, http://www.rstudio.com/.Studio, Version 1.3.1093, 2009–2020 Search PubMed.
- D. S. Murrell, I. Cortes-Ciriano, G. J. van Westen, I. P. Stott, A. Bender, T. E. Malliavin and R. C. Glen, J. Cheminf., 2015, 7, 45 Search PubMed.
- O. Méndez-Lucio and J. L. Medina-Franco, Drug Discovery Today, 2017, 22, 120–126 CrossRef PubMed.
- R. Todeschini and V. Consonni, in Methods and Principles in Medicinal Chemistry, WILEY-VCH Verlag GmbH, 2000, online ISBN:9783527613106, DOI:10.1002/9783527613106.
- H. M. Berman, J. Westbrook, Z. Feng, G. Gilliland, T. N. Bhat, H. Weissig, I. N. Shindyalov and P. E. Bourne, Nucleic Acids Res., 2000, 28(1), 235–242 CrossRef CAS PubMed.
- Molecular Operating Environment (MOE), 2019.0102; Chemical Computing Group ULC, 1010 Sherbrooke St. West, Suite #910, Montreal, QC, Canada, H3A 2R7, 2019 Search PubMed.
- P. Labute, J. Comput. Chem., 2008, 29(10), 1693–1698, DOI:10.1002/jcc.
- C. Wu, Y. Liu, Y. Yang, P. Zhang, W. Zhong, Y. Wang, Q. Wang, Y. Xu, M. Li, X. Li, M. Zheng, L. Chen and H. Li, Acta Pharm. Sin. B, 2020, 10(5), 766–788 CrossRef CAS PubMed.
- A. Chandra, V. Gurjarb, I. Qamara and N. Singha, J. Biomol. Struct. Dyn., 2020, 1–16 CrossRef CAS PubMed.
- K. Youngchang, J. Wower, N. Maltseva, C. Chang, R. Jedrzejczak, M. Wilamowski, S. Kang, V. Nicolaescu, G. Randall, K. Michalska and A. Joachimiak, bioRxiv, 2020 DOI:10.1101/2020.06.26.173872.
- S. Froelich, K. M. A. Siems, M. A. Hernández, R. A. Ibarra, W. G. Berendsohn and K. Jenett-Siems, Pharmazie, 2006, 61, 641–644 CAS.
- A. Lasure, B. Van Poel, L. Pieters, M. Claeys, M. Gupta, D. VandenBerghe and A. Vlietinck, Planta Med., 1994, 60, 276–277 CrossRef CAS PubMed.
- A. A.-R. Gamal El-Din, F. A. M. Mamdouh, S. I. Tarek, E. S. Mai, S. Ebtihal and R. M. Abd El-Baky, RSC Adv., 2020, 10, 26895 RSC.
- M. Özdemira, B. Köksoy, D. Ceyhan, K. Sayın, E. Erçağ, M. Bulut and B. Yalçın, Design and in silico study of the novel coumarin derivatives against SARS-CoV-2 main enzymes, J. Biomol. Struct. Dyn., 2020, 1–16 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1ra01507a |
| This journal is © The Royal Society of Chemistry 2021 |