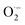Open Access Article
Open Access ArticleUltrasmall copper nanoclusters with multi-enzyme activities†
Yangbin Peng‡
a,
Ying Ren‡b,
Hao Zhua,
Yu Anc,
Baisong Chang *a and
Taolei Sun
*a and
Taolei Sun *ac
*ac
aState Key Laboratory of Advanced Technology for Materials Synthesis and Processing, Wuhan University of Technology, Wuhan 430070, PR China. E-mail: chang@whut.edu.cn; suntl@whut.edu.cn
bDepartment of Radiology, Shengjing Hospital of China Medical University, Shenyang, 110004, P. R. China
cSchool of Chemistry, Chemical Engineering and Life Science, Wuhan University of Technology, Wuhan 430070, PR China
First published on 19th April 2021
Abstract
Reactive oxygen species (ROS) as a key messenger of signal transduction mediate physiological activities, however, oxidative stress produced by excessive ROS can cause the destruction of cell homeostasis, which will result in a series of diseases. Therefore, effective control of ROS level is critical to the homeostasis of the cell. Here, we reported that glutathione (GSH)-stabilized copper nanoclusters (CuNCs) with about 9 Cu atoms can functionally mimic three major antioxidant enzymes, namely catalase (CAT), glutathione peroxidase (GPx) and superoxide dismutase (SOD). The rate of H2O2 decomposition was calculated to be ∼0.23 mg L−1 s−1 when the concentration of CuNCs was 100 μg mL−1. The SOD-like activity by catalyzing the disproportionation of superoxide  to H2O2 and O2 reached 25.6 U mg−1 when the effective inhibition rate was ∼55.4%. Intracellular ROS scavenging studies further identified that CuNCs can obviously protect cells from oxidative stress and the cell viability recovered to above 90%. Hence, we expect that ultrasmall CuNCs will provide good therapeutic potential in the future treatment of ROS-related diseases.
to H2O2 and O2 reached 25.6 U mg−1 when the effective inhibition rate was ∼55.4%. Intracellular ROS scavenging studies further identified that CuNCs can obviously protect cells from oxidative stress and the cell viability recovered to above 90%. Hence, we expect that ultrasmall CuNCs will provide good therapeutic potential in the future treatment of ROS-related diseases.
1 Introduction
Aerobic organisms produce reactive oxygen species (ROS) such as superoxide , hydrogen peroxide (H2O2) and hydroxyl radicals (˙OH) in order to maintain normal life activities.1–3 ROS mediate the transduction mechanisms of various signals, which play an important role in regulating myriad physiological functions in cells.4 Excessive production of ROS leads to oxidative stress, causing the destruction of biomolecules.5 These events cause a variety of pathological and physiological processes involved in neurodegeneration, diabetes, chronic kidney disease, cardiovascular disease and cancer.6,7 Therefore, the maintenance of ROS level against oxidative stress is particularly important for sustaining cellular homeostasis.8
, hydrogen peroxide (H2O2) and hydroxyl radicals (˙OH) in order to maintain normal life activities.1–3 ROS mediate the transduction mechanisms of various signals, which play an important role in regulating myriad physiological functions in cells.4 Excessive production of ROS leads to oxidative stress, causing the destruction of biomolecules.5 These events cause a variety of pathological and physiological processes involved in neurodegeneration, diabetes, chronic kidney disease, cardiovascular disease and cancer.6,7 Therefore, the maintenance of ROS level against oxidative stress is particularly important for sustaining cellular homeostasis.8
Redox equilibrium in cells is achieved by complex endogenous antioxidant defense systems, which include endogenous antioxidant enzymes, e.g., superoxide dismutase (SOD), catalase (CAT) and glutathione peroxidase (GPx).9,10 The regulation of oxidative stress caused by the imbalance of ROS has been recognized as a vital therapeutic target for cancers and neurological diseases. However, the development of natural antioxidant enzymes is still hampered owing to the intrinsic shortcomings such as difficulty in storage, easy inactivation, poor stability and relatively limited sources. To overcome these shortcomings of natural enzymes, researchers are working to develop ideal alternatives to biological enzymes.11,12
Scientists have developed various enzymatically active materials including metal complexes, redox polymers and ceria.13–15 Since the discovery of Fe3O4 nanoparticles with peroxidase activity in 2007,16 the material, widely known as nanozymes, has attracted considerable attention.17 Vernekar et al.18 demonstrated that V2O5 nanowires displayed remarkable GPx-like antioxidant activity under physiologically conditions. Mechanism study illustrated that the internalization of V2O5 nanowires can equilibrate the ROS and control oxidative stress by catalytically reducing H2O2 in the presence of glutathione (GSH). Zhang and colleagues identified that platinum nanoparticles distributed homogeneously on metal–organic frameworks possessed CAT-like catalytic activity.19 This material relied on the O2 that was catalyzed by H2O2 to facilitate production of 1O2 in hypoxic tumor site, playing a critical role for tumor treatments.
In fact, antioxidant enzymes have different regulatory mechanisms for scavenging ROS. For example, SOD has a very high specificity for dismutase  into O2 and H2O2 in vivo. Although the presence of H2O2 is still harmful, CAT and peroxidase will immediately break it into harmless water. CAT is responsible for the catalysis of excess H2O2 during oxidative stress, while GPx shows the function of fine-tune the concentration of H2O2 for cell signalling.20 Inspired by these findings, scientists are no longer satisfy with the development of materials with single enzyme activity and designing material with multi-enzyme activities deserves more attention.
into O2 and H2O2 in vivo. Although the presence of H2O2 is still harmful, CAT and peroxidase will immediately break it into harmless water. CAT is responsible for the catalysis of excess H2O2 during oxidative stress, while GPx shows the function of fine-tune the concentration of H2O2 for cell signalling.20 Inspired by these findings, scientists are no longer satisfy with the development of materials with single enzyme activity and designing material with multi-enzyme activities deserves more attention.
Studies demonstrated that the CeO2 and MnO2 nanoparticles showed inherent multi-enzyme activities.21,22 Fan et al.23 synthesized highly stable platinum nanoparticles and identified that this material showed CAT and peroxidase activities for various substrates. Furthermore, Huang et al.24 constructed a novel multi-nanozyme cooperative platform consisting of V2O5 nanowire and MnO2 nanoparticles. V2O5 nanowire acted as a GPx mimic, while MnO2 nanoparticle was used to mimic SOD and CAT. In a cellular model of Parkinson disease, Mn3O4 nanozymes protected cells from oxidative damage by its redox regulation, thus preventing neurological diseases caused by ROS imbalance.16
Copper, an essential micronutrient in human body, plays a biological role in many enzymes.25 Cu-based nanoparticles, especially cuprous and cupric oxides (Cu2O and CuO), have been studied extensively. Recently, Liu et al.26 prepared ultrasmall Cu-based nanoparticles for scavenging ROS and alleviating inflammation-related diseases. Hao et al.10 synthesized chiral molecules-mediated porous CuxO nanoclusters with CAT, SOD and GPx activities for ameliorating Parkinson's disease. In this context, we attempted to design and construct ultrasmall copper nanoclusters (CuNCs) by a one-step method (Scheme 1). Of particular importance was that these CuNCs could functionally mimic three major cellular antioxidant enzymes (namely CAT, GPx and SOD). To the best of our knowledge, developing ultrasmall CuNCs with excellent multi-enzyme catalytic activities has not been reported before.
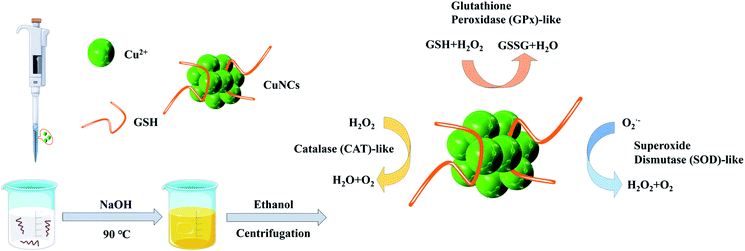 | ||
| Scheme 1 Schematic illustration of the preparation of ultrasmall CuNCs with multi-enzyme activities. | ||
2 Experiment
2.1 Materials
All the chemicals were commercially available and used without further purification. Glutathione in the reduced form (GSH) was obtained from ACROS ORGANICS. Copper nitrate trihydrate, sodium citrate, 3,3′,5,5′-tetramethylbenzidine (TMB), sodium hydroxide and ethanol were purchased from Sinopharm Chemical Reagent Co. Ltd. Terephthalic acid (TPA) was from URCHEM (Shanghai, China). SOD assay kit (WST-1 method) and total antioxidant capacity detection kit (T-AOC) were purchased from Nanjing Jiancheng Bioengineering Institute. Human embryonic kidney (HEK 293), Dulbecco's modified Eagle's medium (DMEM), Fetal Bovine Serum (FBS) and Penicillin-Streptomycin were bought from GIBCO. SOD from bovine erythrocytes and 2′,7′-dichlorodifluorofluorescin diacetate (DCFH-DA) were obtained from Sigma-Aldrich. Cell Counting Kit-8 (CCK-8) was purchased in Beyotime Biotechnology Co. Ltd. 5,5′-Dithiobis-(2-nitrobenzoic acid) (DTNB) was obtained from Aladdin Reagent Co. Ltd. Milli-Q ultrapure water was used in all needed experiments.2.2 Synthesis of ultrasmall CuNCs
In a typical procedure,27 GSH (1.22 g) and Cu(NO3)2 (188 mg) were dissolved in 50 mL of deionized water. The solution was stirred at 25 °C until a white hydrogel was formed. Then, the hydrogel solution was heated to 90 °C for 10 min under magnetic stirring. Thereafter, the white hydrogel solution turned into a light yellow by dropping 1 mL of NaOH (1 M) solution. After cooling to 25 °C, the products were collected by precipitating with ethanol and centrifugation at 8000 rpm. By discarding the supernatant liquid, the solid at the bottom was dissolved in water and then precipitated with ethanol. The entire purification process was repeated three times. The resultant CuNCs were freeze-dried under vacuum and stored in 4 °C refrigerators for further use.2.3 Cytotoxicity experiment
After inserting HEK 293 cells into 96-well plate for 24 h, CuNCs (0, 10, 20, 50, 100 ng mL−1) were added and incubated for 30 min. Then, the cells were treated with H2O2 (100 μM) and cultured at 37 °C for 24 h. CCK-8 assay was used to detect cell viability. The group without H2O2 was set as a negative control.
2.4 Characterizations
Fourier transform infrared spectroscopy (FT-IR) spectra were recorded in the wavelength range of 4000–400 cm−1 with Vertex 80v (Bruker, Germany). The UV-Vis optical spectra were recorded with UV-2000 (Shimadzu, Japan). High-resolution TEM images were obtained from JEM-2100F STEM operating at 200 kV (JEOL, Japan). Photoluminescence (PL) experiments were performed with a FLS1000 fluorescence spectrometer (Edinburgh Instruments, England). X-ray photoelectron spectroscopy (XPS) study was performed on an ESCALAB 250Xi spectrometer (ThermoFisher Scientific, USA). Mass spectrometry studies were performed on an ESI-Q-TOF MS spectrometer (Bruker, Germany). The isotopic simulation pattern was calculated by the IsoPro 3.1 MS/MS software. Dynamic light scattering was were performed on a NanoZS ZEN3600 (Malvern England). Dissolved oxygen was recorded by JPBJ-609 Dissolved Oxygen Meter (Shanghai INESA Scientific Instrument Co. Ltd.). Microplate reader was conducted on a Cytation 5 (Biotek, USA). Electron paramagnetic resonance (EPR) spectra were measured by a Bruker A300 (X-band) spectrometer (Bruker, Germany). Atomic force microscope (AFM) was recorded on Fastscan (Bruker, Germany). Inductively coupled plasma spectrometry (ICP) was performed on a Prodigy 7 (Liman-Leebers, USA).2.5 Catalase-like activity of CuNCs
Of note, the dissolved oxygen meter was activated for 1 h before measurement, then the full and zero oxygen calibrations were performed by using ultrapure water and 5% sodium sulfite solution, respectively. After calibration, the test interval was set as 5 s. At 25 °C, 1.7 mL of PBS buffer and 2 mL of 10 mM H2O2 were added to an amber laboratory bottle. After that, electrode probe was immersed below the liquid level and the test was started. The testing of the variation of O2 was accomplished within 5 min after addition of 0.3 mL of CuNCs. This operation was repeated three times and took the average values.To verify the mechanism of CuNCs-induced decomposition of H2O2, TPA was adopted as a fluorescence probe, which reacts with ˙OH from H2O2, forming highly fluorescent 2-hydroxyterephthalic acid with an emission peak at 425 nm upon exposure to the excitation wavelength of 315 nm. The solutions contained CuNCs (0–100 μg mL−1), TPA (5 mM) and H2O2 (10 mM). The mixtures were incubated for 4 h.
2.6 GPx-like activity of CuNCs
GPx-like catalytic activity was determined by a modified Hafeman method.28 Sulfhydryl compounds can react with DTNB to break the disulfide bonds of DTNB to produce 2-nitro-5-thiobenzoic acid (NTB−), which can be ionized in neutral and alkaline environments to generate NTB2−. This response can be quantified by the optical absorbance at 412 nm. In a typical assay, GSH (1 mM), H2O2 (1 mM) and CuNCs (20 μg mL−1) were added and reacted for 6 min at 37 °C. After the reaction, 1 mL of sodium citrate buffer of DTNB (1 mM) was added and recorded by UV-Vis spectrophotometer. The control experiment was performed by replacing CuNCs with PBS buffer under otherwise identical experiment factors.2.7 SOD-like activity of CuNCs
The addition of SOD and nanozymes with SOD activity will cause a decrease of absorption at 450 nm. The SOD inhibition rate of nanozymes was obtained by this method based on the UV absorption values at 450 nm before and after catalysis.SOD-like activity was also verified by EPR. In a typical assay, PBS (10 mM), xanthine (5 mM), and xanthine oxidase (0.5 U mL−1) were incubated at 37 °C. CuNCs (0, 10, 30, 50, 100 μg mL−1) and DMPO were added 10 min later and then recorded.
2.8 Peroxidase-like activity of CuNCs
The peroxidase activity was characterized by chromogenic substrate TMB and H2O2. CuNCs catalyzed H2O2 to oxidize TMB to form a blue mixture with maximum absorbance at 652 nm. Typically, TMB (5 mM), H2O2 (5 mM) and CuNCs (0–200 μg mL−1) were mixed evenly and detected on the microplate reader.2.9 ˙OH scavenging activity of CuNCs
The combination of ˙OH and DMPO usually results in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2
2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2
2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 peak area by the EPR spectroscopy. Typically, H2O2 (10 mM) and FeSO4 (20 μM) were added into the PBS (10 mM, pH 7.2) and incubated for 1.5 min. Then, 50 μL of DMPO and CuNCs (0, 10, 30, 50, 100 μg mL−1) were mixed and recorded immediately.
1 peak area by the EPR spectroscopy. Typically, H2O2 (10 mM) and FeSO4 (20 μM) were added into the PBS (10 mM, pH 7.2) and incubated for 1.5 min. Then, 50 μL of DMPO and CuNCs (0, 10, 30, 50, 100 μg mL−1) were mixed and recorded immediately.
2.10 ABTS radical scavenging activity of CuNCs
ABTS radical scavenging capacity was measured by the T-AOC kit. ABTS can be oxidized into ABTS˙+ with maximum absorption at 405 nm. The presence of antioxidants will inhibit the production of ABTS˙+. The total antioxidant capacity of CuNCs can be calculated by the absorbance of ABTS˙+ at 405 nm. The ABTS radical scavenging activity was calculated as follows:where Ablank represents the absorbance without CuNCs, and Asample represents the absorbance after adding different concentrations of CuNCs.
3 Results and discussion
3.1 Characterizations of CuNCs
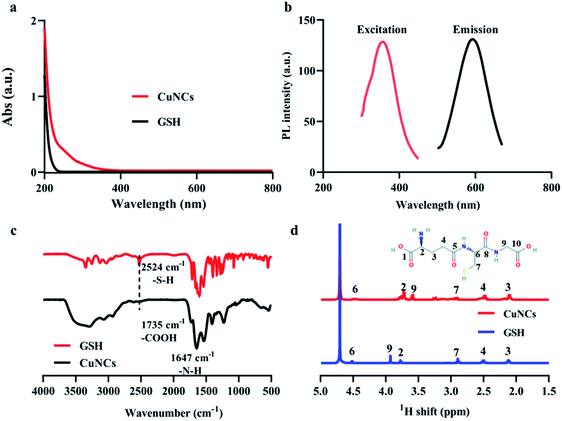
UV-Vis and photoluminescence spectra of as-prepared CuNCs were shown in Fig. 1a and b. There was no absorption peak at about 610 nm corresponding to the surface plasmon resonance of common copper nanoparticles (Fig. S1†), suggesting the small size of obtained CuNCs. Besides, the presence of absorption signals below 300 nm indicated the formation of CuNCs. The emission and excitation spectra were shown in Fig. 1b. In aqueous solution, CuNCs exhibited a symmetrical emission at ∼595 nm. The corresponding excitation peak was ∼365 nm. In order to obtain the optimal synthetic recipes, a series of molar ratios of GSH/Cu2+ were selected to synthesize CuNCs and the corresponding fluorescence emission spectra were recorded. In Fig. S2a,† we can find that when the ratio of GSH/Cu2+ was lower than 4, the fluorescence intensity was proportional to the increase of GSH/Cu2+ ratios. Subsequently, when the ratios of GSH/Cu2+ continued to increase, the fluorescence emission intensity was decreased. We speculated that once the amount of GSH was in excess, most of free GSH molecules remained in solution, producing a cloudy mixture and causing the fluorescence quenching of CuNCs. Moreover, the as-prepared CuNCs showed obvious aggregation-induced emission enhancement.29 With the increase of ethanol concentrations, CuNCs in the solution continued to aggregate, accompanying with the significant increase of fluorescence intensity of CuNCs (Fig. S2b, S3 and S4†).Fourier transform infrared spectroscopy (FT-IR) and proton nuclear magnetic resonance (1H-NMR) measurements were used to analyse the surface chemical bonding of CuNCs. Owing to the presence of thiol groups in the molecular structure, GSH exhibits metal-chelating properties that ensure the formation of high-affinity metal–ligand clusters.30 Furthermore, the thiol group plays a critical role in reducing size and improving size distribution, while amino and carboxyl groups in GSH can protect the CuNCs from aggregation and increase their colloidal stability.31 Compared with the ν(S–H) absorption band of GSH at 2524 cm−1, this peak disappeared for the as-obtained CuNCs (Fig. 1c), indicating that GSH combined with Cu atoms in the form of Cu–S bonds. Fig. 1d showed the 1H-NMR spectra of GSH and CuNCs in the range of 1.5–5.0 ppm, respectively. The peak at 4.69 ppm can be attributed to the residual D2O solvent.32 Based on the chemical structure of GSH (inset of Fig. 1d), the signals at 2.12 and 2.51 ppm were assigned to the protons on 3 and 4. Besides, the peak from proton on 7 can split into two peaks at 3.25 and 2.91 ppm, which may be caused by the adjacent chiral 6. Because the protons on 6 and 7 were very close to the copper core, the corresponding proton signals significantly downfield shifted.33
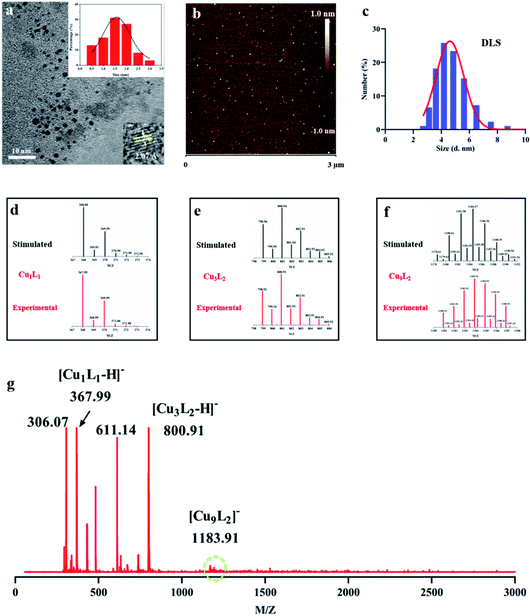
For a more intuitive observation of CuNCs, high-resolution TEM (HR-TEM) image (Fig. 2a) showed that the average size was 1.5 ± 0.5 nm. Under the intense e-beam from TEM instruments, the fusion of metal core may occur due to the e-beam-induced heating and the breaking of metal–S bonds, resulting in an increased polydispersity of CuNCs' diameters.34 AFM was also employed to show a panoramic view of CuNCs (Fig. 2b and S5†). The size distribution of CuNCs coincided well with the analysis of HR-TEM. Owing to the rapid measurement and good repeatability, zeta potential was further used to study the dispersibility of CuNCs in aqueous solution (Fig. 2c and S6†). The dynamic diameter of CuNCs was determined to be larger than that obtained from HR-TEM (Fig. 2a) because of the presence of hydrated shells.
Mainly owing to the ultrasmall size of CuNCs, mass spectroscopy (MS) was then utilized to analyse their chemical compositions and the exact number of atoms. The mass spectrum of ultrasmall CuNCs was measured by the middle molecular weight mode in the range from 0 to 3000 (Fig. 2g). The largest peak (m/z = 1183.91) can be attributed to the Cu9L2 (L = C10H16O6N3S). In view of the diameter of a single Cu atom centering at about 0.255 nm,35 the size of CuNCs with compositions of Cu9L2 was very consistent with the size analysis with the HR-TEM image. It was worth mentioning that large energy ionization process may produce the formation of ion fragments.36 Hence, the peaks with lower mass values including Cu3L2 (m/z = 800.91) and Cu1L1 (m/z = 367.99) were assigned to the fragment ion signals. Moreover, this assignment was further supported by the precise match of simulated and experimental isotopic distributions indicated that the attribution of peak was correct (Fig. 2d–f and S7†). Besides, Cu loading capacity of CuNCs was up to 10.9% determined by ICP measurement.
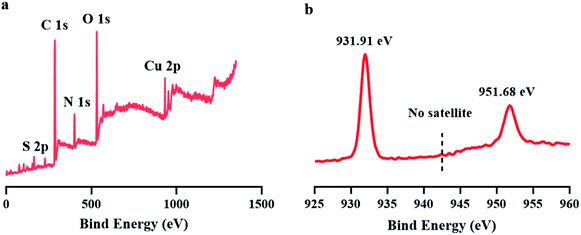
To verify the oxidation states of copper in CuNCs, X-ray photoelectron spectroscopy (XPS) was carried out. The XPS survey spectrum shown in Fig. 3a indicated the presence of Cu, C, N and S elements in the product. The absence of satellite peak around 942.0 eV implied the lack of Cu(II) (Fig. 3b). Meanwhile, two peaks at 931.98 and 951.88 eV can be easily observed, corresponding to the binding energies of the 2p3/2 and 2p1/2 of the electrons of Cu(0), respectively. Another point worth noting was that it was difficult to distinguish Cu(0) and Cu(I) because the binding energy of Cu(0) is very close to Cu(I),37 Accordingly, we cannot rule out the possible existence of Cu(I) in CuNCs.
3.2 CAT-like activity of CuNCs
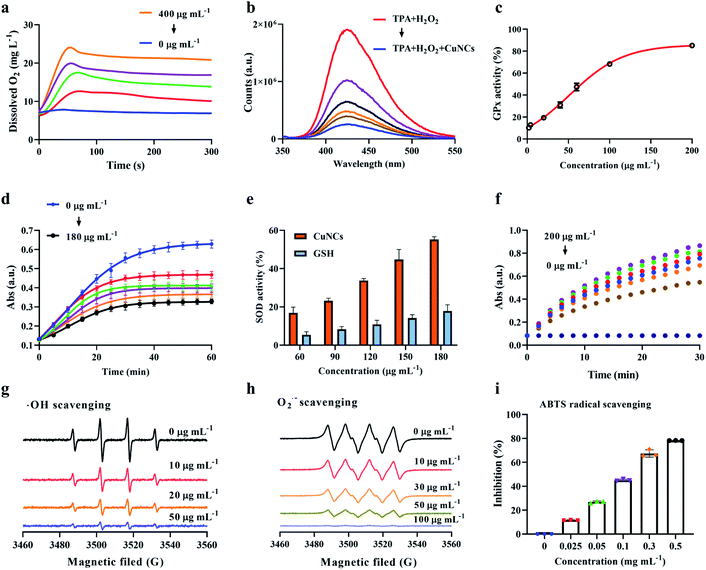
The ability of CuNCs to functionally mimic cellular antioxidant enzymes was investigated as follows. Based on the mechanism that the CAT catalyses the decomposition of H2O2 into O2 and H2O, CAT-like activity of CuNCs was determined by monitoring the contents of dissolved O2 in solution. As shown in Fig. 4a, when the concentration of CuNCs increased from 0 to 400 μg mL−1 at a fixed concentration of H2O2, there was a significant increase in the dissolved O2, indicating the CAT-like catalytic function of CuNCs. For example, the rate of H2O2 decomposition was calculated to be 0.23 mg L−1 s−1 when the concentration of CuNCs was 100 μg mL−1 (Fig. S8a†). More importantly, we found that catalytic efficiency almost maintained constant when the reaction temperature varied in the range of 25–40 °C, showing excellent catalytic stability of CuNCs (Fig. S8b and S9a†). In order to rule out the contribution of GSH on CAT-like activity of CuNCs, we further replaced CuNCs with GSH (0–300 μg mL−1) and performed the catalytic tests under the same conditions (Fig. S9b†). GSH could hardly catalyse H2O2 to produce O2 compared with the CuNCs (Fig. S10†), which suggested that the CAT-like activity of CuNCs mainly originated from CuNCs rather than the surrounding ligand molecules of GSH.We used TPA as a fluorescent probe, which can generate strong fluorescent substances in the presence of ˙OH, to verify the mechanism of CuNCs catalysing H2O2. If the content of ˙OH in the system decreases, the corresponding fluorescence intensity will be weakened. As shown in Fig. 4b, the increased concentrations of CuNCs caused a gradient drop in the fluorescence intensity. Such results revealed that CuNCs did not promote the generation of ˙OH during the CuNCs-induced decomposition of H2O2, directly identifying the chemical pathways of CAT-like activity.
3.3 GPx-like activity of CuNCs
Generally, GPx takes advantage of cellular GSH as a reducing agent for catalysing the reduction of H2O2 to water along with the production of GSSG. The free thiol group decreases with the increase of GSSG, in which the reduction of thiol group can be linearly related to NTB2− with typical optical absorbance at 412 nm. As shown in Fig. 4c, the GPx activities of CuNCs were investigated by independently varying the concentrations of CuNCs from 0 to 200 μg mL−1. As the majority of GSH was oxidized into GSSG by H2O2 in reaction system, the content of the remaining GSH that can react with DTNB will be decreased. In addition, we investigated the variation trend of DTNB absorbance with time in the presence of CuNCs (0–60 μg mL−1), and found that with the increase of concentrations of CuNCs, UV absorption at 412 nm decreased significantly, indicating the good GPx-like activity of CuNCs (Fig. S11†). Furthermore, in the presence of CuNCs, the kinetic constants of CuNCs nanozymes were determined by changing the concentrations of H2O2 and GSH, showing typical Michaelis Menten kinetics (Fig. S12†). For example, the Km value of CuNCs with H2O2 as substrate was ∼0.10 mM, and the Vmax value was ∼6.78 μM min−1. The Km and Vmax values of CuNCs with GSH as the substrate were ∼0.91 mM and ∼4.76 μM min−1, respectively.3.4 SOD-like activity of CuNCs
CuNCs exhibited SOD-like activity by catalysing the disproportionation of superoxide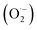 to H2O2 and O2. As displayed in Fig. 4d, the inhibition rate of SOD increased rapidly with the increased concentration of CuNCs (60–180 μg mL−1), indicating the SOD-like activity of CuNCs under physiologically relevant conditions. Notably, two groups with inhibition rates of 44.8% and 55.4% were selected and the final SOD activity was calculated to be 25.6 U mg−1. Considering the possible contribution of GSH in the SOD-like activity of CuNCs, we replaced the CuNCs with GSH to further determine the corresponding SOD activity. As shown in the Fig. 4e, we observed that irrespective of concentrations, the SOD inhibition rate of CuNCs was about 3 times higher than that of GSH. For example, at 180 μg mL−1, the SOD inhibition rate of CuNCs was about 55.4%, while this value obviously dropped to about 17.7% for GSH. Furthermore, the effect of catalytic time on SOD activity was also investigated. The UV absorption values of the system solution gradually decreased responding to the increased concentration of CuNCs (Fig. 4d). In the first 20 min, the UV absorption values of the solutions for all selected concentrations of CuNCs showed a positive correlation growth trend. As the catalytic reaction time continued to elongate, the increase in UV absorption will slow down and remained constant after 35 min. The possible reason was that due to the certain content of superoxide anion free radicals produced by the reaction system, accompanying with the gradual completion of catalytic reaction, the UV absorption value gradually decreased and eventually approached a fixed value.
to H2O2 and O2. As displayed in Fig. 4d, the inhibition rate of SOD increased rapidly with the increased concentration of CuNCs (60–180 μg mL−1), indicating the SOD-like activity of CuNCs under physiologically relevant conditions. Notably, two groups with inhibition rates of 44.8% and 55.4% were selected and the final SOD activity was calculated to be 25.6 U mg−1. Considering the possible contribution of GSH in the SOD-like activity of CuNCs, we replaced the CuNCs with GSH to further determine the corresponding SOD activity. As shown in the Fig. 4e, we observed that irrespective of concentrations, the SOD inhibition rate of CuNCs was about 3 times higher than that of GSH. For example, at 180 μg mL−1, the SOD inhibition rate of CuNCs was about 55.4%, while this value obviously dropped to about 17.7% for GSH. Furthermore, the effect of catalytic time on SOD activity was also investigated. The UV absorption values of the system solution gradually decreased responding to the increased concentration of CuNCs (Fig. 4d). In the first 20 min, the UV absorption values of the solutions for all selected concentrations of CuNCs showed a positive correlation growth trend. As the catalytic reaction time continued to elongate, the increase in UV absorption will slow down and remained constant after 35 min. The possible reason was that due to the certain content of superoxide anion free radicals produced by the reaction system, accompanying with the gradual completion of catalytic reaction, the UV absorption value gradually decreased and eventually approached a fixed value.
3.5 Peroxidase-like activity of CuNCs
To prove the peroxidase activity of CuNCs, the catalysis of TMB was tested in the presence of H2O2. As shown in Fig. 4f, different concentrations of CuNCs can catalyse H2O2 to oxidate TMB, while no oxidation reaction was detected without CuNCs or H2O2, verifying that both components were necessary. We employed steady-state kinetics to further explore the peroxidase-like activity of CuNCs and observed that the oxidation reaction catalysed by CuNCs followed the typical Michaelis–Menten kinetics for TMB and H2O2 (Fig. S13†). The Km value of CuNCs with H2O2 as the substrate was significantly lower than that of Horseradish peroxidase (HRP, Km = 3.70),38 revealing that the catalysis of TMB by CuNCs only required a lower concentration of H2O2. Turnover number (TON), also termed as Kcat, was defined as the maximum number of chemical conversions of the substrate molecules per second.39 It can be calculated from the following equation:where Vmax is the maximum reaction rate; [E] is the concentration of enzymes or CuNCs.40,41
The Vmax were obtained from Lineweaver–Burk plot and calculated to be 8.8 × 10−8 M s−1 and 5.15 × 10−8 M s−1 for H2O2 and TMB substrates. From Table S1,† we can find that Vmax of CuNCs was even higher than HRP.38 Overall, CuNCs performed good catalytic activities compared with other previously reported nanozymes (Table S2 and S3†).
3.6 Free radical scavenging of CuNCs
We further selected three representative free radicals (e.g., ˙OH, and ABTS) to verify the antioxidant performance of CuNCs. It is difficult to detect free radicals since the life span was extremely short, so DMPO was used to capture free radicals to form stable adducts (DMPO/˙OH), which results in a 1
and ABTS) to verify the antioxidant performance of CuNCs. It is difficult to detect free radicals since the life span was extremely short, so DMPO was used to capture free radicals to form stable adducts (DMPO/˙OH), which results in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2
2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2
2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 peak area on the EPR. From Fig. 4g, we can find that the signal was the strongest in absence of CuNCs, indicating that H2O2 produces a large number of ˙OH through Fenton effect under the action of Fe2+. When CuNCs (10 μg mL−1) were added into the system, the intensity of DMPO/˙OH was significantly reduced, indicating that CuNCs could effectively scavenging ˙OH. When the final concentration of CuNCs reaches 50 μg mL−1, the signal strength is only 10% of the original, indicating that CuNCs have been able to remove most ˙OH. Similarly, DMPO was also used to capture
1 peak area on the EPR. From Fig. 4g, we can find that the signal was the strongest in absence of CuNCs, indicating that H2O2 produces a large number of ˙OH through Fenton effect under the action of Fe2+. When CuNCs (10 μg mL−1) were added into the system, the intensity of DMPO/˙OH was significantly reduced, indicating that CuNCs could effectively scavenging ˙OH. When the final concentration of CuNCs reaches 50 μg mL−1, the signal strength is only 10% of the original, indicating that CuNCs have been able to remove most ˙OH. Similarly, DMPO was also used to capture  to form stable adducts
to form stable adducts 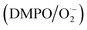 , which results in a 1
, which results in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 peak area on the EPR. As shown in Fig. 4h, the scavenging efficiency of
1 peak area on the EPR. As shown in Fig. 4h, the scavenging efficiency of 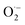 was proportional to the concentrations of CuNCs. CuNCs with the concentration of 100 μg mL−1 could drastically eliminate the
was proportional to the concentrations of CuNCs. CuNCs with the concentration of 100 μg mL−1 could drastically eliminate the  . In addition, we also compared the SOD natural enzyme scavenging
. In addition, we also compared the SOD natural enzyme scavenging  under the same conditions (Fig. S14†), demonstrating that 50 μg mL−1 of CuNCs was close to the SOD at 5 U mL−1. Finally, we verified the scavenging ability of CuNCs on ABTS free radicals and established that CuNCs (0.5 mg mL−1) can reach 78% of scavenging effect (Fig. 4i).
under the same conditions (Fig. S14†), demonstrating that 50 μg mL−1 of CuNCs was close to the SOD at 5 U mL−1. Finally, we verified the scavenging ability of CuNCs on ABTS free radicals and established that CuNCs (0.5 mg mL−1) can reach 78% of scavenging effect (Fig. 4i).
3.7 Intracellular ROS scavenging of CuNCs
We selected HEK 293 cell line to identify the protective ability of CuNCs against ROS injury in vitro. First, HEK 293 cells with different concentrations of CuNCs were incubated for 24 and 48 h, respectively (Fig. 5b and S15†). The cell viability studies showed that CuNCs had no cytotoxicity to HEK 293 cells. Moreover, we used H2O2 (100 μM) to simulate the damage caused by ROS to cells. After incubating the cells with H2O2, the ROS signals (green fluorescence) increased significantly. Interestingly, the decline of fluorescence indicated that the ROS could be gradually eliminated after adding CuNCs. Almost all the ROS signals disappeared and returned to the level of negative control when the CuNCs concentration reached 100 ng mL−1 (Fig. 5a and S16†). After that, we used the CCK-8 assay to quantify the cell viability. Nearly 50% of cells were damaged after incubating with H2O2, and the damaged cells can be successfully repaired after introducing CuNCs. The cell viability recovered to above 91% (Fig. 5c), confirming that CuNCs can protect cells from oxidative stress.4 Conclusion
In summary, this study described a one-pot method to prepare ultrasmall GSH-capped CuNCs with obvious aggregation-induced emission. We, for the first time, demonstrated that ultrasmall CuNCs had remarkable multienzyme catalytic activities, which can mimic three major structurally and functionally different antioxidant enzymes. Intracellular ROS scavenging of CuNCs indicated that CuNCs could protect neuronal cells from oxidative damage. Taken together, CuNCs with ROS scavenging capability show great potential in the future treatment of ROS-related diseases.Author contributions
B. C. and T. S. conceived and designed the study, supervised the project and wrote the manuscript. Y. P. performed all the experiments. Y. R. and H. Z. helped the design of the study and analyzed the data. Y. A. contributed to structural analysis by TEM. All authors discussed the results and commented on the manuscript.Conflicts of interest
The authors declare no competing financial interests.Acknowledgements
This work was supported by the National Natural Science Foundation of China (51803161, 518773168 and 51533007) and the Wuhan University of Technology fund for first-class university and first-class discipline construction projects (472-20162008).Notes and references
- B. D'Autréaux and M. B. Toledano, Nat. Rev. Mol. Cell Biol., 2007, 8, 813–824 CrossRef PubMed.
- C. A. Ferreira, D. Ni, Z. T. Rosenkrans and W. Cai, Nano Res., 2018, 11, 4955–4984 CrossRef CAS PubMed.
- H. Sies and D. P. Jones, Nat. Rev. Mol. Cell Biol., 2020, 21, 363–383 CrossRef CAS PubMed.
- M. Schieber and N. S. Chandel, Curr. Biol., 2014, 24, R453–R462 CrossRef CAS PubMed.
- L. Ferrucci and E. Fabbri, Nat. Rev. Cardiol., 2018, 15, 505–522 CrossRef CAS PubMed.
- A. Rimessi, M. Previati, F. Nigro, M. R. Wieckowski and P. Pinton, Int. J. Biochem. Cell Biol., 2016, 81, 281–293 CrossRef CAS PubMed.
- H. Guo, J. B. Callaway and J. P. Ting, Nat. Med., 2015, 21, 677–687 CrossRef CAS PubMed.
- N. Singh, M. A. Savanur, S. Srivastava, P. D'Silva and G. Mugesh, Angew. Chem., Int. Ed., 2017, 56, 14267–14271 CrossRef CAS PubMed.
- B. Poljsak and D. Šuput, Oxid. Med. Cell. Longevity, 2013, 956792 Search PubMed.
- C. Hao, A. Qu, L. Xu, M. Sun, H. Zhang, C. Xu and H. Kuang, J. Am. Chem. Soc., 2019, 141, 1091–1099 CrossRef CAS PubMed.
- X. Wang, W. Guo, Y. Hu, J. Wu and H. Wei, Nanozymes: next wave of artificial enzymes, Springer, 2016 Search PubMed.
- M. D. Okusa, M. H. Rosner, J. A. Kellum, C. Ronco and X. W. Acute Dialysis Quality Initiative, J. Am. Soc. Nephrol., 2016, 27, 44–48 CrossRef CAS PubMed.
- M. Soh, D. W. Kang, H. G. Jeong, D. Kim, D. Y. Kim, W. Yang, C. Song, S. Baik, I. Y. Choi, S. K. Ki, H. J. Kwon, T. Kim, C. K. Kim, S. H. Lee and T. Hyeon, Angew. Chem., Int. Ed., 2017, 56, 11399–11403 CrossRef CAS PubMed.
- A. Watanabe, M. Kajita, J. Kim, A. Kanayama, K. Takahashi, T. Mashino and Y. Miyamoto, Nanotechnology, 2009, 20, 455105 CrossRef PubMed.
- J. Zhao, W. Gao, X. Cai, J. Xu, D. Zou, Z. Li, B. Hu and Y. Zheng, Theranostics, 2019, 9, 2843–2855 CrossRef CAS PubMed.
- L. Gao, J. Zhuang, L. Nie, J. Zhang, Y. Zhang, N. Gu, T. Wang, J. Feng, D. Yang and S. Perrett, Nat. Nanotechnol., 2007, 2, 577–583 CrossRef CAS PubMed.
- N. Kamaly, J. C. He, D. A. Ausiello and O. C. Farokhzad, Nat. Rev. Nephrol., 2016, 12, 738–753 CrossRef CAS PubMed.
- A. A. Vernekar, D. Sinha, S. Srivastava, P. U. Paramasivam, P. D'Silva and G. Mugesh, Nat. Commun., 2014, 5, 1–13 Search PubMed.
- Y. Zhang, F. Wang, C. Liu, Z. Wang, L. Kang, Y. Huang, K. Dong, J. Ren and X. Qu, ACS Nano, 2018, 12, 651–661 CrossRef CAS PubMed.
- B. Jiang, D. Duan, L. Gao, M. Zhou, K. Fan, Y. Tang, J. Xi, Y. Bi, Z. Tong, G. F. Gao, N. Xie, A. Tang, G. Nie, M. Liang and X. Yan, Nat. Protoc., 2018, 13, 1506–1520 CrossRef CAS PubMed.
- C. Xu and X. Qu, NPG Asia Mater., 2014, 6, e90 CrossRef CAS.
- Y. Li, X. He, J. J. Yin, Y. Ma, P. Zhang, J. Li, Y. Ding, J. Zhang, Y. Zhao and Z. Chai, Angew. Chem., Int. Ed., 2015, 54, 1852–1855 CrossRef.
- J. Fan, J. J. Yin, B. Ning, X. Wu, Y. Hu, M. Ferrari, G. J. Anderson, J. Wei, Y. Zhao and G. Nie, Biomaterials, 2011, 32, 1611–1618 CrossRef CAS PubMed.
- Y. Huang, Z. Liu, C. Liu, E. Ju, Y. Zhang, J. Ren and X. Qu, Angew. Chem., Int. Ed., 2016, 55, 6646–6650 CrossRef CAS PubMed.
- R. A. Festa and D. J. Thiele, Curr. Biol., 2011, 21, R877–R883 CrossRef CAS PubMed.
- T. Liu, B. Xiao, F. Xiang, J. Tan, Z. Chen, X. Zhang, C. Wu, Z. Mao, G. Luo, X. Chen and J. Deng, Nat. Commun., 2020, 11, 2788 CrossRef CAS PubMed.
- C. Wang, L. Ling, Y. Yao and Q. Song, Nano Res., 2015, 8, 1975–1986 CrossRef CAS.
- Y. Weng, Q. Song, Y. Zhou, L. Zhang, J. Wang, J. Chen, Y. Leng, S. Li and N. Huang, Biomaterials, 2011, 32, 1253–1263 CrossRef CAS PubMed.
- J. Mei, Y. Hong, J. W. Lam, A. Qin, Y. Tang and B. Z. Tang, Adv. Mater., 2014, 26, 5429–5479 CrossRef CAS PubMed.
- X. Jia, X. Yang, J. Li, D. Li and E. Wang, Chem. Commun., 2014, 50, 237–239 RSC.
- X. Gao, Y. Lu, M. Liu, S. He and W. Chen, J. Mater. Chem. C, 2015, 3, 4050–4056 RSC.
- S. Kumar, M. D. Bolan and T. P. Bigioni, J. Am. Chem. Soc., 2010, 132, 13141–13143 CrossRef CAS PubMed.
- H. Qian, M. Zhu, U. N. Andersen and R. Jin, J. Phys. Chem. A, 2009, 113, 4281–4284 CrossRef CAS PubMed.
- Y. Huang, W. Liu, H. Feng, Y. Ye, C. Tang, H. Ao, M. Zhao, G. Chen, J. Chen and Z. Qian, Anal. Chem., 2016, 88, 7429–7434 CrossRef CAS PubMed.
- A. J. Heinrich, C. P. Lutz, A. Gupta and D. M. Eigler, Science, 2002, 298, 1381–1387 CrossRef CAS PubMed.
- W. Wei, Y. Lu, W. Chen and S. Chen, J. Am. Chem. Soc., 2011, 133, 2060–2063 CrossRef CAS PubMed.
- X. Jia, J. Li and E. Wang, Small, 2013, 9, 3873–3879 CrossRef CAS PubMed.
- L. Gao, J. Zhuang, L. Nie, J. Zhang, Y. Zhang, N. Gu, T. Wang, J. Feng, D. Yang, S. Perrett and X. Yan, Nat. Nanotechnol., 2007, 2, 577 CrossRef CAS PubMed.
- C. Lin, E. Kätelhön, L. Sepunaru and R. G. Compton, Chem. Sci., 2017, 8, 6423–6432 RSC.
- H. Wang, C. Liu, Z. Liu, J. Ren and X. Qu, Small, 2018, 14, 1703710 CrossRef PubMed.
- M. Liang, Y. Wang, K. Ma, S. Yu, Y. Chen, Z. Deng, Y. Liu and F. Wang, Small, 2020, 16, 2002348 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1ra01410b |
| ‡ These authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2021 |