Open Access Article
Open Access ArticleCombined mass spectrometry-guided genome mining and virtual screening for acaricidal activity in secondary metabolites of Bacillus velezensis W1†
Xingyu Li‡
*ab,
Shahzad Munir‡ c,
Yan Xub,
Yuehu Wangd and
Yueqiu He*c
c,
Yan Xub,
Yuehu Wangd and
Yueqiu He*c
aCollege of Science, Yunnan Agricultural University, Kunming 650201, China. E-mail: lixingyu@ynau.edu.cn
bDepartment of Chemistry, Cleveland State University, Cleveland, OH 44115, USA
cState Key Laboratory for Conservation and Utilization of Bio-resources in Yunnan, Yunnan Agricultural University, Kunming 650201, Yunnan, China. E-mail: ynfh2007@163.com
dKey Laboratory of Economic Plants and Biotechnology, Chinese Academy of Sciences, Kunming 650201, China
First published on 21st July 2021
Abstract
A comprehensive analytic strategy was performed to study the acaricidal activity ingredients of Bacillus velezensis W1, a strain for biological control of Tetranychus urticae. Through genome mining, 14 biosynthetic gene clusters were identified, which encode secondary metabolites, and these were further confirmed by MALDI-TOF-MS or LC-ESI-MS/MS, including bacillomycin D C13–C17, macrolactin A, 7-O-malonyl-macrolactin A, surfactin C14, and surfactin C15. Moreover, 27 volatile compounds were identified by GC-MS, mainly including cyclodipeptides, alkanes, organic acids, and esters. Finally, 43 compounds identified from W1 were used in the virtual screening of acaricidal activity. The results showed that 16 compounds, including cyclodipeptides, bacillomycins, macrolactins, and surfactins, have acaricidal potential. This work provides a base for studying the mechanism of acaricidal action of B. velezensis W1 and a comprehensive strategy for the study of active ingredients from biocontrol strains.
Introduction
Bacillus velezensis is a Gram-positive and endospore-forming bacterium that is enriched in various environments, including soil, plants, marine habitats, and intestinal microflora.1 A variety of strains have been used as an ecologically safe biological pesticide and plant growth promoter, such as B. velezensis FZB42, which has been developed as a natural commercial agent Rhizivoital® to control a variety of soil-borne diseases, and B. velezensis strain has been commercialized in the form of fungicide Botrybel.2 The secondary metabolites produced by B. velezensis play a vital role in biocontrol activities, such as cyclic lipopeptides (i.e., surfactin, bacillomycin, fengycin, iturin, and bacillibactin), polyketides (i.e., macrolactin, bacillaene, and difficidin, and various low molecular weight metabolites), some of which can directly or indirectly control plant pathogens, promote plant growth, and induced systemic acquired resistance in plants.3Studies on the secondary metabolism of Bacillus are usually started with screening crude extract for biological activity and then isolate, purify, identify, and characterize the active ingredients. This process has been proved to be effective and can promote the acquisition of active natural products. However, today, this is considered infeasible, mainly due to the high rate of rediscovery and frequent lack of trace components. With the development of modern chromatography technology and high-throughput genome sequencing technology, people have more options for the research methods of secondary metabolites of Bacillus, e.g., the antibiotics and secondary metabolite analysis shell (antiSMASH) can quickly identify wide genome, annotate and analyze secondary metabolite biosynthetic gene clusters, and helps to estimate the types of compounds encoded by the gene clusters.4 Combining mass spectrometry and genome mining is an advantageous strategy for targeting new molecule backbones or harvesting metabolic profiles to identify analogs from known compounds.5 The prediction of activity spectra for substances (PASS)6 and Binding DB7 helps to estimate the compound's possible biological activity based on its structural formulae.
Bacillus velezensis W1, former name B. amyloliquefaciens W1, later referred to as W1, can biologically control the phytophagous mite Tetranychus urticae. Our previous studies found that W1 gene occupies about 116 kb, equivalent to 2.63% of its total genetic capacity, to be responsible for biosynthesis, transport, and catabolism of secondary metabolites,8 and eight cyclodipeptides with acaricidal activities have been identified through the bioassay-guided fractionation of W1 cell-free supernatant.9 There is scarce information about the acaricidal activity of the secondary metabolites of the biocontrol strains. Therefore, to completely understand the secondary metabolites of W1 and its acaricidal activity, genome mining, MS-based untargeted metabolite analysis, and virtual screening for acaricidal activity were used in the current work.
Experimental and instrumentation
Bacillus velezensis W1
The storage conditions, culture, and fermentation procedures of biocontrol strain W1 are the same as those of the previously published articles from our group,8,9 and our group has got the whole-genome sequences of W1 that was deposited in GenBank with an accession number CP028375.8,9Genome mining of secondary metabolite gene clusters
The whole-genome sequence of W1 was undergone antibiotic and secondary metabolite analysis shell (antiSMASH, version 5.1.2), used to check the biosynthetic gene clusters (BGCs),4 and further aligned using National Center for Biotechnology Information (NCBI) database though BLASTtool (https://www.ncbi.nlm.nih.gov/) against different databases. The antiSMASH web service for bacterial version is available from https://antismash.secondarymetabolites.org/ (accessed on April 6, 2020), which can compare the identified BGCs in genomes submitted by the user with a large number of other microbial BGCs or compiled the Minimum Information for Biosynthetic Gene Clusters (MIBiG) database (https://mibig.secondarymetabolites.org/, accessed on April 6, 2020), and then pointed out whether similar pathways exist in other organisms.Mass spectrometry-based untargeted metabolite analysis
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 70 (v/v) acetonitrile/TFA 0.1% in water). The matrix and analyte were allowed to air dry and co-crystallized at room temperature, and then the target plate was inserted into the MALDI-TOF MS instrument. The spectrum was obtained through positive ion detection and reflector mode by accumulating 1000 laser shots in the m/z range 500–2000, and setting the acceleration and reflector voltage set at 20 and 23.5 kV, respectively.
70 (v/v) acetonitrile/TFA 0.1% in water). The matrix and analyte were allowed to air dry and co-crystallized at room temperature, and then the target plate was inserted into the MALDI-TOF MS instrument. The spectrum was obtained through positive ion detection and reflector mode by accumulating 1000 laser shots in the m/z range 500–2000, and setting the acceleration and reflector voltage set at 20 and 23.5 kV, respectively.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm at ambient temperature. The supernatant was absorbed through D101-type macroporous adsorption resin column chromatography with 95% EtOH as the eluent, and then EtOH was removed with a vacuum rotary evaporator at 55 °C to obtain a crude extract (5 g). Three 0.3 mg crude extracts were extracted with petroleum ether, ethyl acetate, and acetone, respectively, were then analyzed by GC-MS, which was performed by GC-MS spectrometry (Hp6890GC/5973MS, Agilent Technologies, USA) with an HP-5MS fused-silica capillary column (30 m long, 0.25 mm id, 0.25 μm film thickness; Agilent Technologies, USA). For GC-MS detection, the electron ionization system was conducted with ionization energy of 70 eV in ionization mode. Helium gas (99.999%) was used as the carrier gas at a constant flow rate of 1.0 mL min−1, and an injection volume of 1.0 μL (a split ratio of 10
000 rpm at ambient temperature. The supernatant was absorbed through D101-type macroporous adsorption resin column chromatography with 95% EtOH as the eluent, and then EtOH was removed with a vacuum rotary evaporator at 55 °C to obtain a crude extract (5 g). Three 0.3 mg crude extracts were extracted with petroleum ether, ethyl acetate, and acetone, respectively, were then analyzed by GC-MS, which was performed by GC-MS spectrometry (Hp6890GC/5973MS, Agilent Technologies, USA) with an HP-5MS fused-silica capillary column (30 m long, 0.25 mm id, 0.25 μm film thickness; Agilent Technologies, USA). For GC-MS detection, the electron ionization system was conducted with ionization energy of 70 eV in ionization mode. Helium gas (99.999%) was used as the carrier gas at a constant flow rate of 1.0 mL min−1, and an injection volume of 1.0 μL (a split ratio of 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) was operated. Temperature of the injector was kept at 250 °C, the temperature of the ion source was maintained at 230 °C, and the column pressure was 100 kPa. The oven temperature was 40 °C for 2 min, then gradually went up to 80 °C at a rate of 3 °C min−1, then increased to 280 °C at a rate of 5 °C min−1, and maintained at 280 °C for 30 min. Mass spectrum was collected at 70 eV, the scan interval was 0.5 s, and the size of a fragment was 35–500 Da. The solvent delay was 0–2 min, and the total GC-MS run time was 36 min. By comparing the average peak area of each element with the total area, the relative percentage of each component was calculated. A quadrupole mass spectrometer was used to obtain mass spectrometry data that were further matched with the Wiley 7n.1 compound library.
1) was operated. Temperature of the injector was kept at 250 °C, the temperature of the ion source was maintained at 230 °C, and the column pressure was 100 kPa. The oven temperature was 40 °C for 2 min, then gradually went up to 80 °C at a rate of 3 °C min−1, then increased to 280 °C at a rate of 5 °C min−1, and maintained at 280 °C for 30 min. Mass spectrum was collected at 70 eV, the scan interval was 0.5 s, and the size of a fragment was 35–500 Da. The solvent delay was 0–2 min, and the total GC-MS run time was 36 min. By comparing the average peak area of each element with the total area, the relative percentage of each component was calculated. A quadrupole mass spectrometer was used to obtain mass spectrometry data that were further matched with the Wiley 7n.1 compound library.Virtual screening for acaricidal active secondary metabolites
By comparing the structural similarities between known acaricides and compounds identified from biocontrol strain W1, based on compounds with similar structures having similar biological activities,10 BindingDB Web service (http://www.bindingdb.org/, accessed on April 6, 2020) was used to screen virtual acaricidal activity of W1 secondary metabolites. In detail, 32 known acaricidal compounds (Table S1†) were extracted from the DrugBank database (https://www.drugbank.ca/, accessed on April 6, 2020) by searching for the keywords “acaricidal” or “acaricide”. The SDfile of acaricidal pesticide as an active group was uploaded to BindingDB web service, and upload the SDfile of compounds identified from W1 as a set of query group. Finally, virtual screening results were obtained by selecting maximum similarity as a suitable screening model for using chemical fingerprints to quickly find similar compounds.7Results and discussion
Genome mining of Bacillus velezensis W1 BGCs and corresponding secondary metabolites compound classes
Through genome mining, a total of 15 regions composing of the secondary metabolite BGCs and boundary genes were found in the W1 genome, 9 of which matched the 14 BGCs of known secondary metabolites. It is shown in Table 1, these regions accumulate BGCs that encode the known secondary metabolite enzymes, of which 3 BGCs (including srf, dhb, and bac) belong to non-ribosomal peptide synthetases (NRPSs) for surfactin, bacillibactin, and bacilysin, 4 BGCs (including btr, mln, fen, and pps) are polyketide synthase genes (PKS) for butirosin, macrolactin, fengycin, and plipastatin, 5 BGCs (including bae, bmy, myc, itu, and dif) are PKS–NRPS hydride synthetases for bacillaene, bacillomycin D, mycosubtilin, iturin, and difficidin, 2 BGCs (including can and lan) belong to head-to-tail cyclized peptide (RiPP) synthase genes for amylocyclicin and mersacidin. Notably, in addition to compounds surfactin, butirosin, and iturin, the BGCs similarities of 11 other compounds matched 100% of known strains showed in Table 1. The potential strain Bacillus velezensis W1 was used previously with significant acaricidal activity against phytophagous mite Tetranychus urticae.9 Whole genome sequence of W1 strain was analyze through antiSMASH to investigate the BGCs. A total of 15 regions comprising secondary metabolites BGCs were revealed. B. amyloliquefaciens FZB42 also encoded a hybrid type I PKS–NRPS gene cluster called bae.11 Notably, 5 BGCs including bae was also found in W1 strain that encode for PKS–NRPS hydride synthetases for bacillaene. Integrated approach of genome mining was used for B. velezensis LM2303 strain 13 BGCs encoding secondary metabolites having potential biocontrol functions.12 In another study, 19 candidate genes were recorded using this technology in B. siamensis SCSIO 05746.13| Region | Synthetase style | BGCs | Metabolite | MIBiGa | Similarity | Strains |
|---|---|---|---|---|---|---|
a Biosynthetic gene cluster-ID in the MIBiG database.b B. velezensis FZB42.c B. circulans SANK 72073.d B. subtilis XF-1, B. subtilis ATCC6633, B. subtilis RB14, B. subtilis 168.e B. sp. HIL-Y85/54![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 728. — indicates unknown information. 728. — indicates unknown information. |
||||||
| Region 1 | Lassopeptide | — | — | — | — | — |
| Region 2 | NRPS | srf | Surfactin | BGC0000433 | 82% | FZB42b |
| Region 3 | Phosphonate | — | — | — | — | — |
| Region 4 | PKS | btr | Butirosin | BGC0000693 | 7% | SANKc |
| Region 5 | Terpene | — | — | — | — | — |
| Region 6 | PKS | mln | Macrolactin | BGC0000181 | 100% | FZB42b |
| Region 7 | PKS–NRPS | bae | Bacillaene | BGC0001089 | 100% | FZB42b |
| Region 8 | PKS | fen | Fengycin | BGC0001095 | 100% | FZB42b |
| PKS | pps | Plipastatin | BGC0000407 | 100% | XF-1d | |
| PKS–NRPS | bmy | Bacillomycin D | BGC0001090 | 100% | FZB42b | |
| myc | Mycosubtilin | BGC0001103 | 100% | ATCC6633d | ||
| itu | Iturin | BGC0001098 | 88% | RB14d | ||
| Region 9 | Terpene | — | — | — | — | — |
| Region 10 | PKS | — | — | — | — | — |
| Region 11 | PKS–NRPS | dif | Difficidin | BGC0000176 | 100% | FZB42b |
| Region 12 | NRPS | dhb | Bacillibactin | BGC0000309 | 100% | 168d |
| RiPP | can | Amylocyclicin | BGC0000616 | 100% | FZB42b | |
| Region 13 | NRPS | — | — | — | — | — |
| Region 14 | NRPS | bac | Bacilysin | BGC0001184 | 100% | FZB42b |
| Region 15 | RiPP | lan | Mersacidin | BGC0000527 | 100% | HIL-Y85e |
MALDI-TOF MS analysis
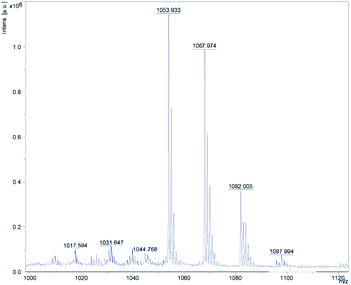
By analyzing the mass spectrometry data detected by MALDI-TOF-MS and comparing it with the mass data of known Bacillus metabolites, four cyclic lipopeptide compounds of ituin family were identified from the whole cells of W1, which are homologues of bacillomycin D, consisting of a heptapeptide molecule linked to a β-amino fatty acid chain of 13–17 carbons atoms.14,15 The eight well-resolved clusters of peaks were detected, as shown in Fig. 1, and the precursor ions were assigned as proton, sodium, or potassium ion adducts of the four molecules. For example, the precursor ions at m/z 1031.65 [M + H]+, and 1053.93 [M + Na]+ were assigned as the protonated molecule and sodiated molecule of bacillomycin D C14 respectively, as shown in Table 2.| Mass peaks (m/z) | Assignment | Ref. |
|---|---|---|
| 1017.594 | C13 bacillomycin D [M + H]+ | (Thasana, et al., 2010) |
| 1039.665 | C13 bacillomycin D [M + Na]+ | |
| 1031.647 | C14 bacillomycin D [M + H]+ | (Jin, et al., 2020) |
| 1053.933 | C14 bacillomycin D [M + Na]+ | |
| 1045.768 | C15 bacillomycin D [M + H]+ | (Ben Ayed, et al., 2017) |
| 1067.974 | C15 bacillomycin D [M + Na]+ | |
| 1082.005 | C16 bacillomycin D [M + Na]+ | |
| 1097.994 | C16 bacillomycin D [M + K]+ |
GC-MS analysis
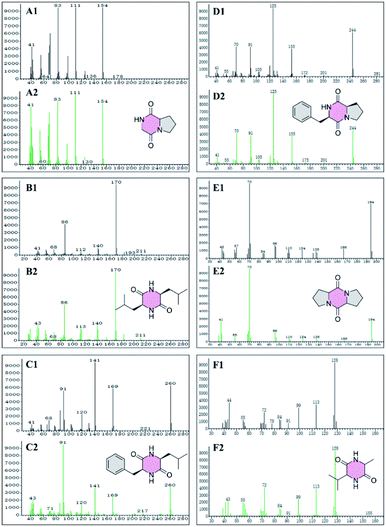
GC-MS was used to detect the petroleum ether, ethyl acetate, and acetone extraction from the crude extract of W1, generating 41, 64, and 39 peaks, respectively (Fig. S1–S3†). Among them, 27 compounds were identified (Table 3) with a quality match of 63% minimum, including 6 cyclic dipeptides, 10 alkanes, 5 esters, 3 acids, 2 ketones, and one alcohol. Fig. 2 showed the matching quality of 6 cyclic dipeptides and their standard GC-MS spectra. For the spectra of other compounds, please refer to the attachment (Fig. S1–S3†). Our previous studies have identified 8 cyclic dipeptides using NMR and MS from the supernatant of W1 by column chromatography and HPLC.9 In this study, 6 cyclic dipeptides were identified by GC-MS as cyclo (Gly–Pro), cyclo (Leu–Leu), cyclo (Phe–Leu), cyclo (Phe–Pro), cyclo (Pro–Pro), and cyclo (Ala–Val), of which cyclo (Gly–Pro), and cyclo (Pro–Pro) are consistent with previous reports, while the rest are first discovered in strain W1.| No. | Compound name | Exact mass | Formula | Qa (%) | Extractionb |
|---|---|---|---|---|---|
| a Q for match quality.b PE for petroleum ether extract, EA for ethyl acetate extract, AT for acetone extract. GC/MS was used for this analysis. | |||||
| 1 | Benzeneacetic acid | 136.05 | C8H8O2 | 91 | PE, AT |
| 2 | Benzenebutanoic acid | 164.08 | C10H12O2 | 86 | PE |
| 3 | Benzenepropanoic acid | 150.07 | C9H10O2 | 96 | PE, EA, AT |
| 4 | 3-(O-Azidophenyl)propanol | 177.09 | C9H11N3O | 78 | EA |
| 5 | Docosane | 310.36 | C22H46 | 93 | PE, AT |
| 6 | Eicosane, 7-hexyl- | 366.42 | C26H54 | 90 | AT |
| 7 | Heneicosane | 296.34 | C21H44 | 97 | PE, AT |
| 8 | Heptacosane | 380.44 | C27H56 | 80 | AT |
| 9 | Hexadecane, 2,6,10,14-tetramethyl- | 282.33 | C20H42 | 86 | AT |
| 10 | Eicosane | 282.33 | C20H42 | 96 | AT |
| 11 | Nonadecane | 268.31 | C19H40 | 97 | PE, AT |
| 12 | Octadecane | 254.30 | C18H38 | 96 | AT |
| 13 | Pentacosane | 352.41 | C25H52 | 97 | AT |
| 14 | Tricosane | 324.38 | C23H48 | 93 | AT |
| 15 | Cyclo (Gly–Pro) dipeptide | 154.07 | C7H10N2O2 | 95 | PE, AT |
| 16 | Cyclo (Leu–Leu) dipeptide | 226.17 | C12H22N2O2 | 63 | PE |
| 17 | Cyclo (Phe–Leu) dipeptide | 260.15 | C15H20N2O2 | 68 | PE |
| 18 | Cyclo (Phe–Pro) dipeptide | 244.12 | C14H16N2O2 | 98 | PE |
| 19 | Cyclo (Pro–Pro) dipeptide | 194.11 | C10H14N2O2 | 94 | PE, EA |
| 20 | Cyclo (Ala–Val) dipeptide | 170.11 | C8H14N2O2 | 64 | PE |
| 21 | Benzenepropanoic acid, methyl ester | 164.08 | C10H12O2 | 95 | PE |
| 22 | Ethyl citrate | 276.12 | C12H20O7 | 83 | PE, AT |
| 23 | Hexadecanonic acid, ethyl ester | 284.26 | C18H36O2 | 93 | AT |
| 24 | Hexadecanonic acid, methyl ester | 270.26 | C17H34O2 | 99 | PE, EA, AT |
| 25 | Octadecanoic acid, methyl ester | 298.29 | C19H38O2 | 98 | EA, AT |
| 26 | 2-Piperidinone | 99.07 | C5H9NO | 91 | PE, EA, AT |
| 27 | 2-Pyrrolidinone | 85.05 | C4H7NO | 90 | EA |
LC-ESI-MS and ESI-MS/MS analysis
The secondary metabolites produced by W1 strain were characterized through LC-ESI-MS in a positive full scan mode. Eight pairs of peaks were observed (Table 4), of which six pairs of peaks grouped into two clusters with a fold difference of 14 Da, indicating the presence of two groups of homolog molecules.| No. | tR (min) | Observed peaks (m/z) | Compound name | Exact mass | ||
|---|---|---|---|---|---|---|
| [M + H]+ | [M + Na]+ | [M + K]+ | ||||
| 1 | 31.462 | 1031.54 | 1053.53 | — | Bacillomycin D C14 | 1030.53 |
| 2 | 33.013 | — | 425.23 | 441.21 | Macrolactin A | 402.24 |
| 3 | 33.298 | 1045.55 | 1067.55 | — | Bacillomycin D C15 | 1044.54 |
| 4 | 35.167 | — | 511.23 | 527.21 | 7-O-Malonyl-macrolactin A | 488.25 |
| 5 | 35.357 | 1059.56 | 1081.57 | — | Bacillomycin D C16 | 1058.55 |
| 6 | 37.288 | 1073.60 | 1095.57 | — | Bacillomycin D C17 | 1072.59 |
| 7 | 57.175 | 1022.71 | 1044.66 | — | Surfactin C14 | 1021.70 |
| 8 | 59.170 | 1036.69 | 1058.67 | — | Surfactin C15 | 1035.68 |
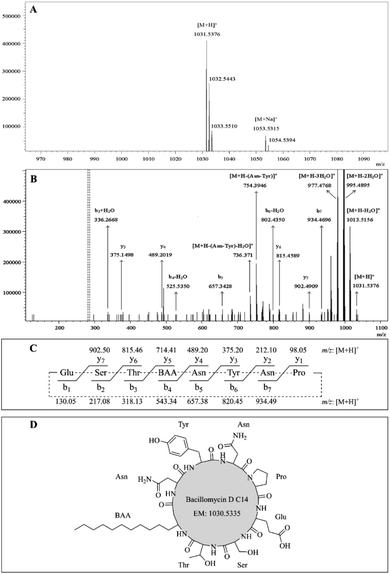
The first group includes four pairs of peaks corresponding to the protonate adducts and sodium adducts, at m/z 1031.54 [M + H]+ and 1053.53 [M + Na]+, 1045.55 [M + H]+ and 1067.55 [M + Na]+, 1059.56 [M + H]+ and 1081.57 [M + Na]+, and 1073.60 [M + H]+ and 1095.57 [M + Na]+. Therefore, the molecular masses of these four molecules were calculated as 1030.53, 1044.54, 1058.55, and 1072.59, respectively. First group consist of four peaks which are consistent with the molecular mass of bacillomycin D C14, bacillomycin D C15, bacillomycin D C16, and bacillomycin D C17. Its chemical structures consist of a heptapeptide molecule connected to a β-amino acid fatty chain of 14–17 carbon atoms.16
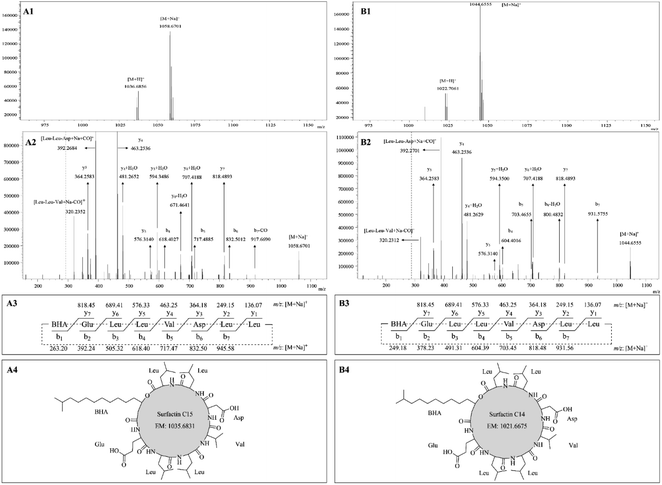
The protonated molecules [M + H]+ at m/z 1031.54, 1045.55, 1059.56, and 1073.60 were selected as precursor ions for further ESI-MS/MS analyses. Fig. 3A showed the ESI-MS spectrum with peaks at m/z 1031.54 [M + H]+, 1053.53 [M + Na]+, and its isotope ion peaks. Fig. 3B showed the ESI-MS/MS spectrum of the most abundant precursor ion at m/z 1031.54, showing fragment ions, where the ions at m/z 1031.54, 1013.52, 995.49, and 977.48, respectively, corresponded to the formations of [M + H]+ [M + H − H2O]+, [M + H − 2H2O]+, and [M + H − 3H2O]+. The remaining fragment ions were identified as y-ion series and b-ion series, which originated from the initial opening of the peptide bond between glutamate and proline in the ring (Fig. 3C). The y-ion series protonated ions at m/z 902.49 (y7), 815.46 (y6), 489.20 (y4), 375.15 (y3), the b-ion series protonated ions at m/z 934.47 (b7), 802.44 (b6 − H2O), 657.34 (b5), 525.54 (b4 − H2O), 336.27 (b3 + H2O) were observed in ESI-MS/MS spectrum (Fig. 3B). The other fragment protonated ions at m/z 754.39 and 736.37 were corresponded to [M + H − Asn–Tyr]+ and [M + H − Asn–Tyr − H2O]+, respectively. According to these regular CID fragments, the sequence can be deduced as Glu–Ser–Thr–BAA–Asn–Tyr–Asn–Pro, where BAA shorts for β-amino fatty acid. Therefore, the structure could be identified as bacillomycin D C14 (Fig. 3D). Similarly, the other homolog molecules of the first set were identified as bacillomycin D C15, bacillomycin D C16, and bacillomycin D C17 (Table 4). The other spectra were shown in Fig. S4–S6.† The second group has two pairs of peaks at m/z 1022.71 [M + H]+ and 1044.66 [M + Na]+, 1036.69 [M + H]+ and 1058.67 [M + Na]+ (Table 4). Therefore, the mass of the two molecules were, respectively, 1021.70 and 1035.68. Second group with two peaks were consistent with molecular mass of surfactin C14 and surfactin C15.17
The sodium adducts [M + Na]+ at m/z 1044.66 and 1058.67 were taken as precursor ions for ESI-MS/MS study. Fig. 4A1 showed ESI-MS spectrum with peaks at m/z 1036.68 [M + H]+, 1058.67 [M + Na]+. The ESI-MS/MS spectrum of the precursor ion at m/z 1058.67 was shown in Fig. 4A2 with the appearance of fragment ions. The ions at m/z 1058.67, 392.27, and 320.24 corresponded to ion formations [M + Na]+, [Leu–Leu–Asp + Na + CO]+, and [Leu–Leu–Val + Na − CO]+, respectively. The remaining fragment ions were assigned as y-ion and b-ion series deriving from the initial opening of the lactone ring (Fig. 4A3). As shown in Fig. 4A2, the y-ion series of the sodium adducts contained fragment ions at m/z 818.49 (y7), 707.42 (y6 + H2O), 671.46 (y6 − H2O), 594.35 (y5 + H2O), 481.27 (y4 + H2O), 463.25 (y4), and 364.26 (y3). The b-ion series of the sodium adducts included fragment ions at m/z 917.67 (b7 − CO), 832.50 (b6), 717.49 (b5), and 618.40 (b4). According to these typical CID fragments, the sequence could be deduced as BHA–Glu–Leu–Leu–Val–Asp–Leu–Leu, where BHA represents the shortness of β-hydroxyl fatty acid, and its structure can be identified as surfactin C15 (Fig. 4A4). Similarly, by comparing their mass spectra, another one was identified as surfactin C14. For example, their molecular mass differed by 14 Da (Fig. 4B1), they have the same y-ion series ions, and the b-ion series are different by 14 Da (Fig. 4B2 and B3).
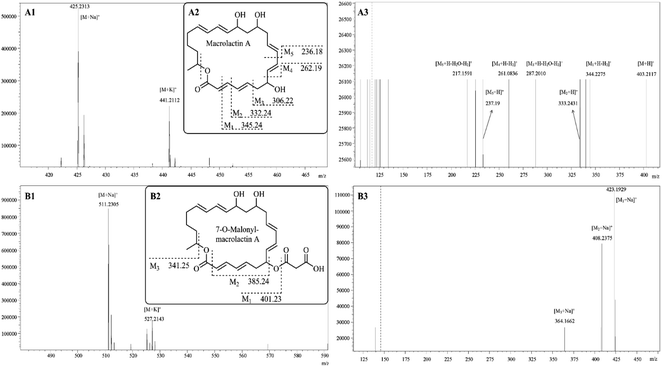
The pair of peaks at m/z 425.23 and m/z 441.21 (Fig. 5A1) corresponded to the sodium adduct [M + Na]+ and potassium adduct [M + K]+, indicating a molecular mass of 402.24 that was in agreement with the molecular mass of macrolactin A (Fig. 5A2).18 The m/z 425.23 was used as the precursor ion for further ESI-MS/MS analyses (Fig. 5A3). The results showed the appearance of protonated fragment ions at m/z 403.21, 344.23, 333.24, 287.20, 261.08, 237.19, and 217.16. The ion at m/z 403.21 corresponded to protonated molecule [M + H]+. The ion at m/z 344.23 corresponded to [M1 + H − H2]+ where M1 (Fig. 5 A2) is derived from the macrolactin A opening from the lactone ring and losing one formic acid residue (COO: 43.99 Da). The ion at m/z 333.24 corresponded to [M2 + H]+ where M2 is derived from M1, losing a methine (CH: 13.01 Da). The ion at m/z 287.20 corresponded to [M3 + H − H2O − H2]+ where M3 is derived from M2, losing a vinylene (CHCH: 26.02 Da). The ion at m/z 261.08 corresponded to [M4 + H − H2]+ where M4 is derived from M3, losing vinyl alcohol (CH2CHOH: 44.03 Da). The ions at m/z 237.19 and 217.16, respectively, corresponded to [M5 + H]+ and [M5 + H − H2O − H2]+ where M5 is derived from M4 losing a vinylene (CHCH: 26.02 Da).
The pair of peaks at m/z 511.23 [M + Na]+ and m/z 527.21 [M + K]+ (Fig. 5B1) indicated a molecular mass of 488.25 that was in agreement with the molecular mass of 7-O-malonyl-macrolactin A (Fig. 5B2).18 The m/z 511.23 was used as the precursor ion for further ESI-MS/MS analyses (Fig. 5B3). The results showed the appearance of product ions at m/z 423.19, 408.24, and 364.17, respectively, corresponded to [M1 + Na]+, [M2 + Na]+ and [M3 + Na]+. As shown in Fig. 5B2, 7-O-malonyl-macrolactin A lost a malonyl group (COCH2COOH: 87.01 Da) to yield M1, and lost a hydrogen malonate ion (COOCH2COOH: 103.00 Da) to produce M2, and further lost one formic acid residue (COO: 43.99 Da) to get M3.
Virtual screening for acaricidal active secondary metabolites
The previous9 and present research resulted that a total of 43 (Table S2†) compounds were identified from the strain W1 through genome mining, CG-MS analysis, LC-MS/MS analysis, MALDI TOF-MS analysis, and bioassay-guided fractionation. These 43 compounds were used to screen for acaricidal activity through BindingDB Web service, in which 16 compounds (Table 5) were selected as candidate compounds for acaricidal activity using the criteria of maximum similarity ≥50%, mainly including four categories of structures, i.e., cyclodipeptides, bacillomycin D, macrolactin A, and surfactins.| No. | Compound | Maximum similarity | Category |
|---|---|---|---|
| 1 | Bacillomycin D C13 | 0.7 | Bacillomycin D |
| 2 | Bacillomycin D C14 | 0.7 | |
| 3 | Bacillomycin D C15 | 0.7 | |
| 4 | Bacillomycin D C16 | 0.7 | |
| 5 | Bacillomycin D C17 | 0.7 | |
| 6 | Cyclo (Gly–Phe) | 0.7 | Cyclodipeptides |
| 7 | Cyclo (Gly–Tyr) | 0.6 | |
| 8 | Cyclo (Phe–Leu) | 0.8 | |
| 9 | Cyclo (Phe–Pro) | 0.7 | |
| 10 | Cyclo (Phe-trans-4-OH–Pro) | 0.7 | |
| 11 | Cyclo (Pro-trans-4-OH–Pro) | 0.5 | |
| 12 | Cyclo (Tyr-trans-4-OH–Pro) | 0.7 | |
| 13 | Macrolactin A | 0.5 | Macrolactin A |
| 14 | Macrolactin A, 7-O-malonyl- | 0.5 | |
| 15 | Surfactin C14 | 0.5 | Surfactin |
| 16 | Surfactin C15 | 0.5 |
Cyclodipeptides, also known as 2,5-diketopiperazines, are the smallest cyclic peptides, and are known to exhibit various biological and pharmaceutical activities, such as antibacterial, antimicrobial, anticancer, cytotoxic, insecticidal, antioxidant, antiviral, and nematicidal.19,20 Our previous research also reported that cyclodipeptides have acaricidal activity.9 Bacillomycins D, a kind of lipopeptides belonging to the iturin family, is well known for its antifungal activity, also showed many biological activities such as antimicrobial, antioxidation, antitumor, and plant growth-promoting activities.14,21 Macrolactin A is a 24-membered polyene macrolide antibiotic with various biological activities including antibacterial, anticancer, antifungal, antiviral, anti-angiogenic, anti-metastatic, and anti-inflammatory.22–24 Surfactin, a cyclic lipopeptide biosurfactant, exhibits a wide range of biocontrol activities, such as antibacterial, antifungal, antiviral, anti-inflammatory, antitumor, and hemolytic action.25–27 Further, we used BindingDB Web service to get potential compounds with acaricidal activity. Candidate compounds present in four categories of structures, i.e., cyclodipeptides, bacillomycin D, macrolactin A, and surfactins capitalize a wide range of biological activities, and have been extensively studied and used in fields of agriculture, environment, food and pharmaceuticals, but there are scarce reports on acaricidal and insecticidal activities. Therefore, our research results open new insights into the four types of compounds. B. velezensis W1 is a biocontrol strain against the phytophagous mite T. urticae, and crude extracts of its secondary metabolites displayed excellent acaricidal activity. Previous studies have obtained information on some metabolites in the form of cyclodipeptides.9
Conclusions
To systematically study W1 metabolites and the acaricidal mechanism, we adopt a comprehensive analysis strategy involving multiple analysis techniques and methods. Initially, genome mining found that 14 BGCs in W1 genome encoded known secondary metabolites are shown in Table 1, in which the bacillomycin D C13–C17 were detected and verified by MALDI-TOF-MS and ESI-MS/MS, and macrolactin A, 7-O-malonyl-macrolactin A, surfactin C14, and surfactin C15 were also identified by ESI-MS/MS. Secondly, the 27 volatile compounds shown in Table 3 were identified by GC-MS, and several cyclodipeptides like cyclo-(Gly–Pro) and cyclo-(Pro–Pro) were consistent with previous research results.9 Finally, through the BindingDB Web service, the acaricidal activities of 43 compounds identified from W1 strain were virtually screened. Among them, 16 compounds had a highly chemical structure similarity to the known acaricides, indicating that they have potential acaricidal activity. These 16 putative active compounds belong to 4 chemical structures, designated as cyclodipeptides, bacillomycins, macrolactins, and surfactins, in which cyclodipeptides have been proven to have acaricidal and insecticidal ability 9, 19, 20, while other compounds have little research report on acaricidal activity. Further, more the emerging research is being conducted to investigate the acaricidal activity and mechanism of action of 16 compounds into a successful interdisciplinary approach.Author contributions
X. Y. L., Y. X., Y. Q. H., and Y. H. W., conceived and designed the study and experiments; X. Y. L., Y. X., and S. M., performed the experiments; analyzed the data; X. Y. L., Y. X., and S. M., wrote the manuscript; and all authors contributed to the final draft of the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the National Natural Science Foundation of China (31660536).References
- O. N. Reva, D. Z. H. Swanevelder, L. A. Mwita, A. D. Mwakilili, D. Muzondiwa, M. Joubert, W. Y. Chan, S. Lutz, C. H. Ahrens, L. V. Avdeeva, M. A. Kharkhota, D. Tibuhwa, S. Lyantagaye, J. Vater, R. Borriss and J. Meijer, Front. Microbiol., 2019, 10, 2610 CrossRef PubMed.
- H. Cawoy, W. Bettiol, P. Fickers and M. Ongena, in Pesticides in the modern world-pesticides use and management, InTech, Rijeka, 2011, pp. 273–302 Search PubMed.
- M. F. Rabbee, M. S. Ali, J. Choi, B. S. Hwang, S. C. Jeong and K. H. Baek, Molecules, 2019, 24(6), 1046 CrossRef CAS PubMed.
- K. Blin, S. Shaw, K. Steinke, R. Villebro, N. Ziemert, S. Y. Lee, M. H. Medema and T. Weber, Nucleic Acids Res., 2019, 47, W81–W87 CrossRef CAS PubMed.
- R. Sigrist, B. S. Paulo, C. F. F. Angolini and L. G. De Oliveira, J. Visualized Exp., 2020,(157), e60825 Search PubMed.
- D. Filimonov, D. Druzhilovskiy, A. Lagunin, T. Gloriozova, A. Rudik, A. Dmitriev, P. Pogodin and V. Poroikov, Biomedical Chemistry: Research and Methods, 2018, 1, e00004 Search PubMed.
- M. K. Gilson, T. Liu, M. Baitaluk, G. Nicola, L. Hwang and J. Chong, Nucleic Acids Res., 2015, 44, D1045–D1053 CrossRef PubMed.
- X.-Y. Li, S. Munir, W.-Y. Cui, P.-J. He, J. Yang, P.-F. He, Y.-X. Wu, Y.-H. Wang and Y.-Q. He, Appl. Ecol. Environ. Res., 2019, 17, 2689–2699 CrossRef.
- X.-Y. Li, Y.-H. Wang, J. Yang, W.-Y. Cui, P.-J. He, S. Munir, P.-F. He, Y.-X. Wu and Y.-Q. He, J. Agric. Food Chem., 2018, 66, 10163–10168 CrossRef CAS PubMed.
- P. Durai, Y.-J. Ko, C.-H. Pan and K. Park, BMC Bioinf., 2020, 21, 309 CrossRef CAS PubMed.
- X.-H. Chen, J. Vater, J. Piel, P. Franke, R. Scholz, K. Schneider, A. Koumoutsi, G. Hitzeroth, N. Grammel and A. W. Strittmatter, J. Bacteriol., 2006, 188, 4024–4036 CrossRef CAS PubMed.
- L. Chen, J. Heng, S. Qin and K. Bian, PLoS One, 2018, 13, e0198560 CrossRef PubMed.
- H. Pan, X. Tian, M. Shao, Y. Xie, H. Huang, J. Hu and J. Ju, Appl. Microbiol. Biotechnol., 2019, 103, 4153–4165 CrossRef CAS PubMed.
- F. Lin, Y. Xue, Z. Huang, M. Jiang, F. Lu, X. Bie, S. Miao and Z. Lu, Appl. Microbiol. Biotechnol., 2019, 103, 7663–7674 CrossRef CAS PubMed.
- N. Thasana, B. Prapagdee, N. Rangkadilok, R. Sallabhan, S. L. Aye, S. Ruchirawat and S. Loprasert, FEBS Lett., 2010, 584, 3209–3214 CrossRef CAS PubMed.
- Q. Gong, C. Zhang, F. Lu, H. Zhao, X. Bie and Z. Lu, Food Control, 2014, 36, 8–14 CrossRef CAS.
- Y. Meng, H.-Z. Gang, S.-Z. Yang, R.-Q. Ye and B.-Z. Mu, Anal. Sci., 2018, 34, 541–545 CrossRef CAS PubMed.
- J. Yuan, B. Li, N. Zhang, R. Waseem, Q. Shen and Q. Huang, J. Agric. Food Chem., 2012, 60, 2976–2981 CrossRef CAS PubMed.
- A. K. Mishra, J. Choi, S.-J. Choi and K.-H. Baek, Molecules, 2017, 22, 1796 CrossRef PubMed.
- X. Wang, Y. Li, X. Zhang, D. Lai and L. Zhou, Molecules, 2017, 22, 2026 CrossRef PubMed.
- T. Wu, M. Chen, L. Zhou, F. Lu, X. Bie and Z. Lu, Appl. Microbiol. Biotechnol., 2020, 1–12 Search PubMed.
- F. Kaspar, P. Neubauer and M. Gimpel, J. Nat. Prod., 2019, 82, 2038–2053 CrossRef CAS PubMed.
- J. P. Marino, M. S. McClure, D. P. Holub, J. V. Comasseto and F. C. Tucci, J. Am. Chem. Soc., 2002, 124, 1664–1668 CrossRef CAS PubMed.
- X. L. Lu, Q. Z. Xu, X. Y. Liu, X. Cao, K. Y. Ni and B. H. Jiao, Chemistry & biodiversity, 2008, 5, 1669–1674 Search PubMed.
- G. Seydlová and J. Svobodová, Open Med., 2008, 3, 123–133 Search PubMed.
- A. W. Zanotto, A. Valério, C. J. de Andrade and G. M. Pastore, Appl. Microbiol. Biotechnol., 2019, 103, 8647–8656 CrossRef CAS PubMed.
- L. Chen, X.-y. Chong, Y.-Y. Zhang, Y.-y. Lv and Y.-S. Hu, Curr. Microbiol., 2020, 77, 71–78 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1ra01326b |
| ‡ Equal contribution of these authors. |
| This journal is © The Royal Society of Chemistry 2021 |