Open Access Article
Open Access ArticleElectromagnetic field-assisted cell-laden 3D printed poloxamer-407 hydrogel for enhanced osteogenesis†
Sayan Deb Dutta‡
a,
Jin Bin‡b,
Keya Gangulya,
Dinesh K. Patela and
Ki-Taek Lim *ac
*ac
aDepartment of Biosystems Engineering, Institute of Forest Science, Kangwon National University, Chuncheon-24341, Republic of Korea. E-mail: ktlim@kangwon.ac.kr
bSchool of Stomatology, Affiliated Hospital of Yanbian University, Yanji-136200, Beijing, China
cBiomechagen Co., Ltd, Chuncheon-24341, Republic of Korea
First published on 7th June 2021
Abstract
3D bioprinted hydrogel has gained enormous attention, especially in tissue engineering, owing to its attractive structure and excellent biocompatibility. In this study, we demonstrated that 3D bioprinted cell-laden ‘thermoresponsive’ poloxamer-407 (P407) gels have the potential to stimulate osteogenic differentiation of apical papilla stem cells (SCAPs) under the influence of low voltage–frequency (5 V–1 Hz, 0.62 mT) electromagnetic fields (EMFs). SCAPs were initially used for cell-laden 3D printing to biomimic the apical papilla of human teeth. The developed hydrogel exhibited higher mechanical strength as well as good printability, showing high-quality micro-architecture. Moreover, the as-printed hydrogels (5 mm × 5 mm) were loaded with plasminogen activator inhibitor-1 (PAI-1) for testing the combined effect of PAI-1 and EMFs on SCAP differentiation. Interestingly, the 3D hydrogels showed improved viability and differentiation of SCAPs under EMFs' influence as examined by live/dead assay and alizarin Red-S staining, respectively. Therefore, our results confirmed that P407 hydrogels are non-toxic for encapsulation of SCAPs, yielding high cell viability and accelerate the cell migration potential. The 3D hydrogels with PAI-1 exhibited high mRNA expression levels for osteogenic/odontogenic gene markers (ALP, Col-1, DSPP, and DMP-1) vis-à-vis control after 14 days of in vitro culture. Our findings suggest that 3D bioprinted P407 hydrogels are biocompatible for SCAP encapsulation, and the applied low voltage–frequency EMFs could effectively improve dental tissue regeneration, particularly for oral applications.
1. Introduction
The effects of electromagnetic fields (EMFs) have been shown to ameliorate bone healing for a long time.1–3 The exogenous EMFs can interfere with various biological processes, including proliferation,4 morphology,5 stemness,1,3 gene expression,6 protein expression,7 apoptosis,8 and cell differentiation.7 Chronic wound response usually generates naturally occurring endogenous EFs that play a crucial role in active cell migration from bone marrow to the wounded site by a process known as electrotaxis (ET). Later on, the role of endogenous EFs has been extensively investigated in both in vivo and in vitro models.9,10 Liu et al. reported the use of pulse-EMFs for osteoporosis treatment.11 A study by Jansen et al. reported that 15 Hz-1 gauss pulse-EMF (5 μs pulse) improved cell proliferation and enhanced the alkaline phosphatase activity, calcium content, and gene expression of adult bone marrow-derived mesenchymal stem cells (hBMSCs).7 Lew et al. also demonstrated that 0.4 tesla static magnetic field (SMFs) enhanced the proliferation and differentiation of dental pulp stem cells (DPSCs) by activating the p38 mitogen-activated protein kinase (MAPK)-family protein.12 Moreover, several reports demonstrated that low voltage–frequency EMFs have the potential to stimulate cell migration and differentiation via inducing the extracellular receptor kinases (ERK), yes-associated proteins (YAP), and transcriptional coactivator PDZ-binding motifs (TAZ) in DPSCs.13,14Mesenchymal stem cells (MSCs) are a promising treatment option compared to the conventional treatment methods in dentistry.1,15 Adult stem cells are multipotent cells capable of differentiation in multiple lineages, such as osteocytes, chondrocytes, adipocytes, and neuron-like cells, respectively.15,16 In addition, stem cell-based tissue regeneration offers several advantages, including the avoidance of complex surgeries, low risk of contamination, high quality of tissue regeneration, and low rate of autoimmune rejection.16 The molar tooth is one of the rich sources of dental stem cells (DSCs). DSCs can be isolated from different parts of dental tissues, such as the dental pulp region (DPSCs), exfoliated primary teeth (SHED), periodontal ligaments (PDLSCs), or form apical papilla (SCAPs).16,17 The human periodontal ligament stem cells (hPDLSCs) are a promising model for dental tissue engineering and regenerative medicine. One of our previous studies showed that stem cells from apical papilla (SCAPs) could differentiate into odontogenic, osteogenic, chondrogenic, and adipogenic lineages.18 In another study, we have reported that hBMSCs encapsulated into three-dimensional (3D) alginate/gelatin/cellulose nanocrystals matrix could potentially trigger osteogenic differentiation in calvaria defect rats.19 Therefore, stem cells combined with suitable scaffolds might be considered ideal candidates for bone tissue engineering applications.
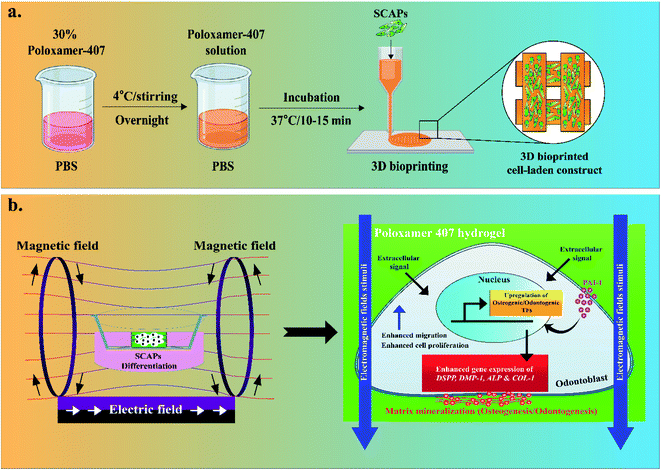
‘3D printing’ or ‘additive manufacturing is a rapidly growing area in tissue engineering and regenerative medicine. 3D printing technique offers a unique micro/nano-structure by using solid or semi-solid structures created by fused deposition (FDM) or direct-ink writing (DIW) methods compared to the conventional hydrogel scaffolds lacking precise control over 3D shape, inner structure, and spatial distribution of different cell types.19,20 3D bioprinting technology has been used in biomedical research for constructing various biomimetic objects, including organs and tissues, customized implants (orthopedic implants), and various surgical implants.21 In particular, the 3D printed smart constructs are widely used pharmaceutical research for studying cellular behavior, drug delivery, toxicological studies, and many others.22
In this study, we have used a DIW-based 3D printing technology to prepare a tissue-engineered construct using poloxamer-407 (P407) with or without plasminogen activator inhibitor (PAI-1) for osteogenic improvement of apical papilla stem cells (SCAPs) for dental tissue regeneration. P407 is a synthetic block copolymer of poly[ethylene glycol] and poly[propylene glycol] and is widely used in tissue engineering for its excellent biocompatibility.23 To accelerate the osteogenic property, an external DC EMF with low voltage–frequency was applied to evaluate EMFs' effects on SCAPs-encapsulated P407 hydrogels, which has not been studied so far. Scheme 1 depicts an overview of EMFs-assisted osteogenic/odontogenic differentiation of SCAPs-laden P407 hydrogels. Our results suggested that the 3D P407 gel was not harmful to SCAPs and improved the proliferation and osteogenic differentiation of SCAPs. Therefore, we hypothesize that EMFs-directed 3D poloxamer hydrogels could be utilized as an ideal candidate for tissue engineering, particularly dental tissue regeneration.
2. Experimental section
2.1. Materials
Thermoresponsive poloxamer 407 (P407; non-ionic, ≤1% water content, pH 7.5) was procured from Sigma-Aldrich, St. Louis, USA. A PASCO® 850 Universal Interface (standard waveforms, frequency range 0.001–100 Hz, amplitude range ± 15 V, and output current range 61 μA at 10 V) with two Helmholtz coils (200 turns; inner radius 10.06 cm, outer radius 10.58 cm. Coil width 1.6 cm, and maximum current change 2 A) was obtained from PASCO Scientific, California, USA. Dulbecco's modified eagle media (DMEM), αMEM, fetal bovine serum (FBS), phosphate-buffered saline (PBS), and 1% P/S were procured from Welgene, Republic of Korea. Mesenchymal stem cell identification kit, live/dead double staining kit, ascorbic acid-2-phosphate, glutamine, alizarin Red-S (ARS), bovine serum albumin (BSA), plasminogen activator inhibitor-1 (PAI-1), and F-actin probe were procured from Sigma-Aldrich, St. Louis, USA. The osteogenic differentiation medium was obtained from Lonza, Maryland, USA. Primary and secondary antibodies against CD34, CD13, CD90, and CD146 were procured from BD Biosciences, San Jose, California, USA. cDNA synthesis kit and SYBR green master mix reagent were procured from Invitrogen, Maryland, USA, and Bio-Rad Laboratories, USA. A fully automated CELLINK® BIO-X 3D printer was procured from CELLINK® Corporation, Sweden.2.2. Isolation of SCAPs
Human third molar teeth were obtained from healthy young males (22–25 years old) and were used to isolate apical papilla stem cells (SCAPs). This protocol was approved by the Institutional Review Board of the Dental Hospital, Seoul National University, Republic of Korea (IRB no. 05004). The written consent was obtained from each patient before surgery. The primary cell culture was carried out, as described previously.24 Briefly, the SCAPs were gradually separated from the teeth by an enzymatic digestion procedure (3 mg mL−1 collagenase type-I and 4 mg mL−1 dispase) at 37 °C for 1 h. After digestion, the cell suspension was procured by passing through a 40 μm strainer. The cells were cultured in αMEM supplemented with 10% FBS, 100 μM L−1 ascorbic acid-2-phosphate, 2 mM glutamine, and 1% P/S at 37 °C in 5% CO2. After 24 h, the media was changed, followed by changes every 2–3 days. At 80–90% confluency, the cells were passaged by 0.25% trypsin–EDTA solution and subcultured accordingly. The primary cells at passage 4 or 5 were used for the proliferation, differentiation, and cell-laden printing experiments.2.3. Characterization and multilineage differentiation of SCAPs
The expression of mesenchymal stem cell-associated immunophenotype markers was assessed by flow cytometry. For this, cells (1 × 106 per mL) were fixed by 3.7% paraformaldehyde (PFA) for 10 min and then resuspended in 1 mL PBS containing 1% BSA for 30 min. After blocking, the cells were incubated with specific primary antibodies against CD34, CD13, CD90, and CD146 at 4 °C for 1 h. Next, the cells were washed with PBS and incubated with mouse anti-human secondary antibodies for another 1 h at 4 °C. Finally, the percentage of CD34−, CD13+, CD90+, and CD146+ cells was measured by a flow cytometer (FACS Caliber, BD Biosciences, California, USA). The data were processed by using Cell-Quest Pro software (BD Biosciences, California, USA).For the multilineage differentiation study, the cells (4 × 104 per mL) were incubated with osteogenic, chondrogenic, adipogenic, and neurogenic media for 21 days (Gibco-BRL, Grand Island, USA). After specific incubation, the cells were stained by a stem cell differentiation kit and staining kit according to the manufacturer's instruction. The stained plates were analyzed by an inverted optical microscope (Zeiss Axio Vert. A1, Germany) to examine the mineralized nodule, proteoglycan, fat deposition, and Nissl body formation as an indicator of osteogenic, chondrogenic, adipogenic, and neurogenic differentiation.
2.4. Preparation of P407 sol–gel
The P407 sol–gel was prepared as described earlier.23 Briefly, 30% (w/v) dissolved in 10 mL ice-cold PBS and stirred for 12 h to obtain a homogenous mixture (sol). The solution was sterile filtered and kept at 4 °C until use. Before 3D printing, the P407 solution was incubated at 37 °C to obtain P407 gel.2.5. 3D printing and cell-laden culture
Before printing, the P407 solution was loaded into the printer cartridge and incubated at 37 °C until the gel was formed. Next, the gels were UV-sterilized for 30 min, and the structure was directed printed onto the sterile 6-well plates placed in the print-bed (pre-heated at 37 °C). The structure of the construct was designed by the Solidworks software (http://www.solidworks.com, Dassault Biosystems, USA) and exported as a ‘stereolithography’ (STL) file. The round-shaped construct (5 mm × 5 mm) was carefully printed using a 27 G (0.2 mm) metal-head needle, and the printing parameters are given in Table 1.| Infill pattern type | Rectilinear |
|---|---|
| Infill density | 20.0% |
| Design type | Cylinder (5 mm × 5 mm) |
| First layer height | 66.0% (0.2 mm) |
| Printing speed | 5 mm s−1 |
| Flow rate | 0.2 mL s−1 |
| Needle size (G) | 27 G |
| Printing pressure | 35 kPa |
| Printing-head temperature | 37 °C |
| Printing-bed temperature | 37 °C |
For cell-laden culture, the 30% P407 (w/v) was dissolved in PBS. The cell-laden construct was prepared, as described in our previous study.19 Briefly, 2 mL of cold poloxamer solution was mixed thoroughly with 2 mL of SCAPs (gel![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) cell = 1
cell = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) to obtain a 2.5 × 104 per mL cell density. Next, the P407 solution was carefully loaded into the plastic cartridges and incubated at 37 °C for 10–15 min. Next, the cell-laden P407 gel was immediately printed onto the sterile 6-well plates at 37 °C. After printing, the cell-laden construct was supplemented by 2 mL of DMEM with 10% FBS and 1% P/S solution and incubated at 37 °C/5% CO2 incubator.
1) to obtain a 2.5 × 104 per mL cell density. Next, the P407 solution was carefully loaded into the plastic cartridges and incubated at 37 °C for 10–15 min. Next, the cell-laden P407 gel was immediately printed onto the sterile 6-well plates at 37 °C. After printing, the cell-laden construct was supplemented by 2 mL of DMEM with 10% FBS and 1% P/S solution and incubated at 37 °C/5% CO2 incubator.
2.6. Printability analysis
The printing quality of the cell-laden constructs was assessed through printing and checking the cell condition immediately after 3D printing. Briefly, 1 or 2 layers were carefully printed and stained with 100 μL of live/dead double staining kit for 5 min and visualized by an inverted fluorescence microscope (Leica DMi8, Germany) to check the strand uniformity and cell printing quality. Images were captured at 10× and 20× magnifications, respectively. Since the P407 gel was stable at 37 °C, we have printed some 3D structures (pyramid, star, cube, and vascular-like structure) to assess the stability of the gel after printing.The printability of the developed hydrogel was evaluated by measuring the uniformity factor (U)19 and printability factor (Pr),25 respectively. For this, 1 or 2 layers of P407 gel was carefully printed with different printing speeds (2 mm s−1, 3 mm s−1, and 5 mm s−1), and the images were captured using an inverted optical microscope (Zeiss, Germany). The printing was repeated thrice (n = 3), and the images were analyzed using ImageJ Fiji plugins (http://www.imagej.nih.gov). The printability was measured by using the following eqn (1) and (2):
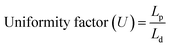 | (1) |
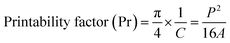 | (2) |
2.7. Rheological characterization
The viscoelastic nature of the prepared hydrogel was assessed by using an ARES-G2 rheometer (TA Instruments, New Castle, USA) with a 25 mm parallel plate (stainless steel) at varying temperatures (25, 30, 35, and 40 °C). The angular frequency (ω) range was set from 0.1 to 100 Rad s−1 with a 1% strain rate. The storage modulus (G′), loss modulus (G′′), complex viscosity (η*), and loss factor (tan![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) δ) values were recorded for 600 s. All the measurements were conducted in three technical replicates (n = 3).
δ) values were recorded for 600 s. All the measurements were conducted in three technical replicates (n = 3).
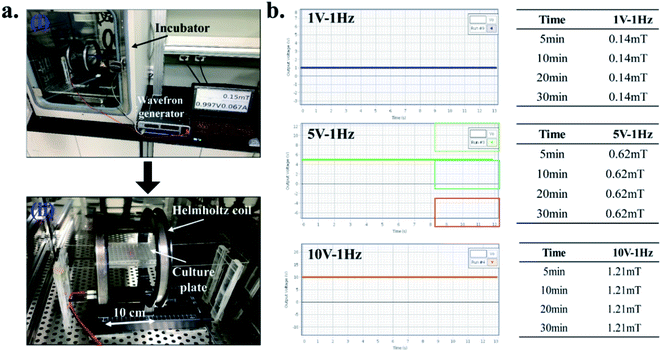
2.8. EMFs stimulation
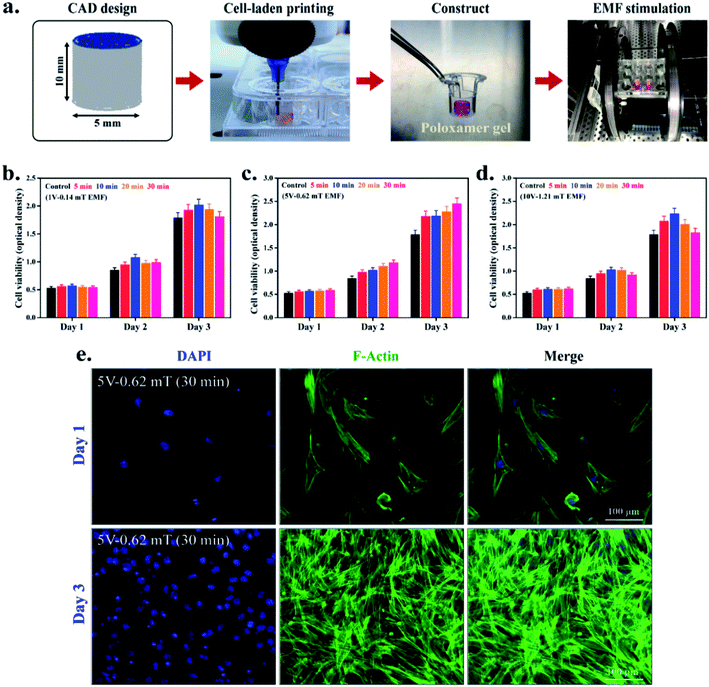
For this, the Helmholtz coils (EM-6722, Pasco Scientific, USA) with a supporting stand were carefully placed inside the BOD incubator. The two coils were placed 10 cm apart and are connected to the external wavefront generator, as shown in Fig. 1(a). During experiments, the 3D construct was placed precisely between the two coils, and DC EMFs was applied in different time interval to achieve the optimum conditions for cell viability Fig. 1(b). During each exposure, the 3D constructs were stimulated at a different time point (5, 10, 20, and 30 min per day) with varying voltage (1 V, 5 V, and 10 V) and magnetic fields (0.14, 0.62, and 1.21 mT), respectively. The Helmholtz coils and the magnetic field (MF) sensor are attached with the wavefront generator (UI-5000, Pasco Scientific, USA). The MF sensor was placed vertically between the two coils. The coils have 200 turns and are supported by pure copper wire (average diameter 0.654 mm) with an inner radius of 10.06 cm, an outer radius of 10.58 cm, and a coil width of 1.6 cm. The distance between the two coils was 10 cm. Ohm's law's resistance (R) of the coils for each EMF setting was calculated as described previously.26 The magnetic flux density (B) was calculated according to eqn (3) (ref. 27) and is given in Table 2:
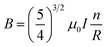 | (3) |
| Voltage (V) | Frequency (Hz) | Magnetic field (mT) | Resistance (Ω) | Magnetic flux density (B) |
|---|---|---|---|---|
| 1 | 1 | 0.14 | 0.5 | 1.2 × 10−7 |
| 5 | 1 | 0.62 | 2.5 | 1.39 × 10−8 |
| 10 | 1 | 1.21 | 5 | 2.79 × 10−8 |
2.9. Cell cytotoxicity assays
The viability of the SCAPs was assessed by the live/dead assay and WST-1 assay. The 3D printed hydrogels were incubated at 37 °C for 5 days. After incubation, the gels were washed by 1× PBS and stained with 100 μL of live/dead double staining kit for 5 min and visualized by an inverted fluorescence microscope (Leica DMi8, Germany). Images were captured at 10× magnification, and the number of cells was quantified by ImageJ software with Fiji plugins using the cell counter filter. For WST-1 assay, the EMF-stimulated gels were washed twice by 1× PBS and replaced by 100 μL of fresh culture media. Next, the cells were incubated with 20 μL of WST-1 dye for 2 h. After incubation, the produced formazan was collected and transferred to a sterile 96-well plate. The absorbance was recorded at 450 nm (625 nm as a reference wavelength) using a spectrophotometer (Infinite® M Nano 200 Pro; TECAN, Switzerland). All the experiments were performed in biological replicates, and data are presented as mean OD ± standard deviations. Statistical significance was considered at *p < 0.05.2.10. Assessment of cell morphology
The morphology of the SCAPs was evaluated by fluorescence microscopy as described earlier.19,28 The gel surface was briefly washed twice by 1× warm PBS, followed by fixation with 3.7% paraformaldehyde (PFA) for 10–15 min. After that, the cells were permeabilized by 100% ice-cold methanol (Sigma-Aldrich, USA) for 5 min and blocked with 1% BSA for 1 h. After blocking, the gels were washed twice by warm PBS and stained with Alexa-Flour 555-conjugated F-actin probe for 30 min at dark. After staining, the gels were washed twice by PBS, and the nucleus was counterstained by DAPI (Sigma-Aldrich, USA). The stained cells were finally visualized by an inverted fluorescence microscope (Leica DMi8, Germany), and the images were captured with appropriate filter channels.2.11. In vitro differentiation study
The mineralization efficiency of the SCAPs was assessed by an ARS staining procedure after 7 and 14 days of incubation in an osteogenic differentiation medium. Briefly, the 3D scaffolds with or without EMF treatment were fixed with 70% ethanol for 15 min and stained by 500 μL of ARS for 5 min. After that, the plates and transwell inserts were washed by distilled water, and the mineralized nodule formation was observed by an inverted optical microscope (Zeiss Axio Vert. A1, Germany). PAI-1 (50 ng mL−1) was taken as a positive control, and a combination of PAI-1 + EMFs stimulation was also taken to observe the synergistic effects on osteogenic differentiation of SCAPs.2.12. RNA isolation and qRT-PCR analysis
The cell-laden scaffolds were incubated in an osteogenic differentiation medium for 7 and 14 days to investigate the gene marker expression in both unstimulated and stimulated cells. The medium was replaced every after 2–3 days. After specific incubation, the total RNA was harvested by RNAzol reagent (Sigma-Aldrich, USA), and the purity was determined by Nanodrop spectrophotometer (Infinite® M Nano 200 Pro; TECAN, Switzerland). 2–4 μg of total RNA was used to synthesize the cDNA using reverse transcriptase and SYBR Green master mix reagent. The relative mRNA expression was recorded by a qRT-PCR (CFX96™ Maestro Real-Time system, Bio-Rad, USA). All the reactions were triplicated and normalized to the housekeeping gene hypoxanthine-guanine phosphoribosyltransferase (HPRT). All the primers (DSPP, DMP-1, ALP, and Col-1) were procured from Bioneer Inc., Daejeon, Republic of Korea, and are listed in Table 3.| Gene ID | GenBank accession no. | Sequences (5′–3′) |
|---|---|---|
| a DSPP; dental sialophosphoprotein, DMP-1; dental matrix protein-1; ALP; alkaline phosphatase; Col-1; collagen type-1. | ||
| DSPP | NC_000004 | (F) CCATGGAGAACGCTGGGG, (R) CAAAGTTGTGATGGATGACC |
| DMP-1 | NM_001079911 | (F) GCAGAGAGTCAGAGCGAGGAA, (R) CCGTGGAGTTGCTATCTTCTTTG |
| ALP | NM_007431 | (F) CCAACTCTTTTGTGCCAGAGA, (R) GGCTACATTGGTGTTGAGCTTTT |
| Col-1 | NM_007742 | (F) GCTCCTCTTAGGGGCCACT, (R) CCACGTCTCACCATTGGGG |
2.13. Statistical analysis
The statistical analysis was carried out using Origin Pro v9.0 software (Origin Lab., USA). One-way ANOVA and Tukey's test post hoc analysis analyzed the statistical significance between control and treatment groups. All the experiments were performed in biological replicates (n = 3), and data are presented as mean OD ± standard deviations. Statistical significance was considered at *p < 0.05.3. Results and discussion
3.1. Characterization and multi-differentiation potential of SCAPs
A schematic presentation of the presence of SCAPs in the human molar tooth is shown in Fig. S1(a).† The cell morphology was captured by an inverted optical microscope after different time intervals and is shown in Fig. S1(b).† Stem cells are mainly characterized by their pluripotentiality and their ability to express various surface markers, and the presence of endogenous growth factors regulates their fate (e.g., TNFs, bFGFs, EGFs, PDGFs), cytokines, and inhibitors.29,30 This property is known as the stemness potential of stem cells.31 We have demonstrated the stemness potential of the extracted SCAPs by FACS analysis, and the results are given in Fig. S1(c).† A variety of stem cells are extracted from dental tissue, including dental pulp stem cells (DPSCs), periodontal ligament stem cells (PDLSCs), dental follicle stem cells (DFSCs). SCAPs can be differentiated into osteoblasts, cementoblasts, adipocytes, chondrocytes, and neuron-like cells.29 Our results indicated the enhanced expression of the stemness positive markers, such as CD13 (∼99%), CD90 (∼99%), and CD146 (∼95%), respectively. However, the presence of negative markers, such as CD34 (∼99%), was also noted, indicating the stemness potential of SCAPs. The multilineage differentiation potential of SCAPs was evaluated after 21 days of culture in osteogenic, chondrogenic, adipogenic, and neurogenic media, and the results are shown in Fig. S1(d).† After specific incubation, the plates were stained by ARS, alcian blue, oil Red-O, and Nissl stain. The presence of mineralized nodule, proteoglycan, oil droplet, and Nissl bodies indicate the ability of SCAPs to differentiate into different lineages, respectively.32,333.2. Characterization of P407 hydrogels
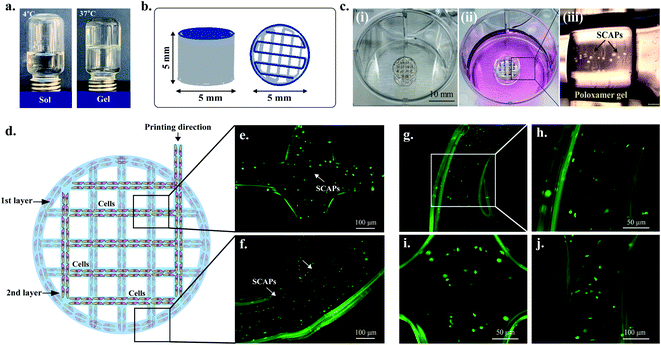
Fig. 2(a) represents the procedure of preparing the sol-gels of P407. The 3D structure (5 mm × 5 mm disc) was designed by Solidworks software and imported as an ‘SLT file’ before printing. The SLT file was imported into the bioprinter and sliced by the Bio-X in-built software to achieve a uniform slicing of the 3D structure (Fig. 2(b)). The infill density was set up to 20% so that the pores were visible clearly during the printing procedure. The printing was carried out on the 6-well plates in a single printing setting, and the printer-bed temperature was set up to 37 °C to allow a uniform temperature distribution during the printing process. The gels were immediately fridge-dried and kept at an ambient temperature until use. The resulting structure was approximately 5 ± 0.2 mm in height with 4.8–4.9 mm in diameter. The average weight of the structure was 0.06 g. Interestingly, when the 3D structure was immersed into 2 mL warm PBS (37 °C), the structure absorbed the buffer and turns into a gel-like structure. Moreover, when cold PBS (4 °C) was added into the printed structure, the structure absorbed the buffer and turned into a liquid structure after 30 min. This is due to the amphiphilic nature of the copolymers of poly(ethylene glycol) and poly(propylene glycol) present in P407 gel, which makes it an ideal ‘thermoreversible gel’.343.3. Printability and rheological properties
Next, we have evaluated the printing quality of the cell-laden P407 gels. Fig. 2(c) represents the cell-laden 3D printed construct of P407 gel with corresponding optical microscopy images. The printed strand indicates the presence of SCAPs, as shown in Fig. 2(c)(iii). For cell-laden printing, the P407 was dissolved in PBS and mixed with SCAPs at a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio to achieve a 2.5 × 104 per mL cell density. The mixed solution was carefully loaded into the printer cartridges and incubated at 37 °C for about 10–15 min. The schematic illustration of cell printing is given in Fig. 2(d). Initially, the first and second layers were printed and stained by a live/dead staining kit to check the gel's cell homogeneity. As shown in Fig. 2(e–j), the cells were homogeneously distributed inside the gel, and the printed structure was nearly uniform as predicted in the original design. Both the cross-connecting junctions and strands have superior printing quality. These results indicate that P407 gels exhibited superior cell-based printing and could be used as an ideal biomaterial for tissue engineering and drug delivery.23,34
1 ratio to achieve a 2.5 × 104 per mL cell density. The mixed solution was carefully loaded into the printer cartridges and incubated at 37 °C for about 10–15 min. The schematic illustration of cell printing is given in Fig. 2(d). Initially, the first and second layers were printed and stained by a live/dead staining kit to check the gel's cell homogeneity. As shown in Fig. 2(e–j), the cells were homogeneously distributed inside the gel, and the printed structure was nearly uniform as predicted in the original design. Both the cross-connecting junctions and strands have superior printing quality. These results indicate that P407 gels exhibited superior cell-based printing and could be used as an ideal biomaterial for tissue engineering and drug delivery.23,34
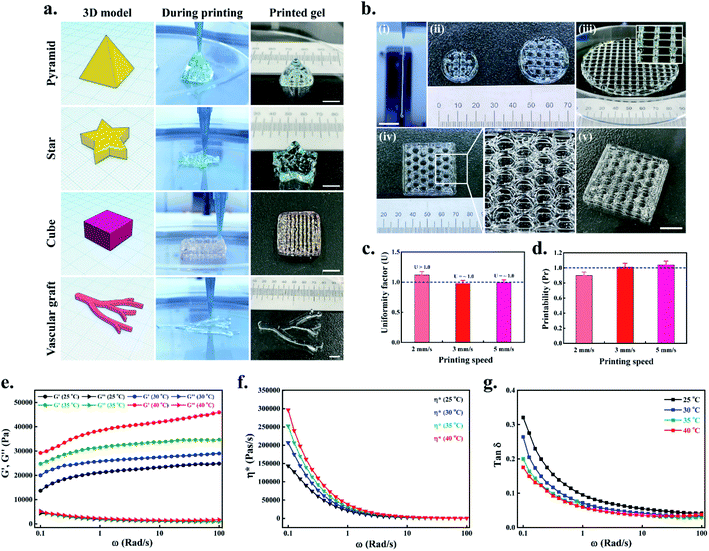
Next, we have evaluated the printing quality of complex structures, such as pyramid (5 × 5 × 5 mm, 70 layers), star (5 × 5 × 5 mm, 20 layers), cube (5 × 5 × 5 mm, 20 layers), and vascular-like structure (6 × 5 × 2 mm, 10 layers). All the structures were printed layer-by-layer up to desired height, and the final architecture was shown in Fig. 3(a). The results indicate that all the structures were printed uniquely, meaning that the theoretical design is retained in the actual printed structure. Moreover, the filament formation test indicated that after incubating P407 gel for 10–15 min at 37 °C, the extruded filament was relatively uniform without any crack or rupture with a length of >15 mm (Fig. 3(b)(i)). Since P407 is an amphipathic copolymer, the critical micellization temperature (CMC) decreases with increasing temperature, and the poly(ethylene oxide)–poly(propylene oxide) (PEO–PPO) units self-assembled to form hydrogel network rather than producing micelle-like structures.35 Therefore, 3D printing of P407 hydrogel with varying resolution sustained the mechanical integrity, and the printed filaments appeared to be nearly perfect with any patterns or infill densities, as shown in Fig. 3(b)(ii–v).
The printability of the 3D printed scaffolds was evaluated by uniformity factor (U) and printability factor (Pr). For this, 1 or 2 layers were printed with different printing speeds (2 mm s−1, 3 mm s−1, and 5 mm s−1) at 37 °C and photographed using an inverted optical microscope. As shown in Fig. 3(c), the uniformity (U) of the printed filament was slightly increased (U > 1) when printing speed was 2 mm s−1, while at 3 mm s−1 and 5 mm s−1, the filament was nearly uniform (U = ∼1). Similarly, the Pr, which is inversely proportional to the pore geometry, demonstrated that higher printing speed (3 mm s−1 and 5 mm s−1) exhibited perfect rectangular pore geometry (Pr = ∼1) (Fig. S2†), while the low printing speed (2 mm s−1) exhibited a slight drop of Pr value (Fig. 3(d)). These results demonstrate that the P407 hydrogel could print perfectly when the printing speed was between 3–5 mm s−1.
The viscoelastic nature of the P407 hydrogel was evaluated using a rotational rheometer with an angular frequency (ω) ranging from 0.1 to 100 Rad s−1 at different temperature (25, 30, 35, and 40 °C), and the changes in the storage modulus (G′) and loss modulus (G′′) is shown in Fig. 3(e). Notably, an enhancement of G′ was observed with the gradual increase in the temperature of the P407 hydrogel. A two-fold increase in the G′ value was observed when the temperature was increased to 40 °C compared to the 25 °C. Similarly, the G′′ value was also enhanced when the temperature was increased to 40 °C. A typical complex viscosity (η*) vs. shear rate curves of the P407 hydrogel at different temperatures are given in Fig. 3(f). An enhancement of η* was observed in the P407 hydrogel when the temperature was slowly increased from 25 °C to 40 °C. The loss factor (tan![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) δ) in various temperatures is also given in Fig. 3(g). When the P407 solution was kept at 20–25 °C, the polymer chains exist as extended coils, and the molecular arrangement is more or less regular. At 25–30 °C, the P407 solution slightly forms a gel-like structure owing to intermolecular rearrangement of the PEO–PPO units. However, at 37–40 °C, the extended coils become random due to the unstable hydrogen bonding between PPO and water, leading to increased solution entropy.36,37 As a result, the polymer chains of P407 move closer, and more interconnected networks were formed, increasing G′ values. This was also reflected by an enhancement of η* as shown in Fig. 3(f). At 37–40 °C, the loss factor of the P407 hydrogel (tan
δ) in various temperatures is also given in Fig. 3(g). When the P407 solution was kept at 20–25 °C, the polymer chains exist as extended coils, and the molecular arrangement is more or less regular. At 25–30 °C, the P407 solution slightly forms a gel-like structure owing to intermolecular rearrangement of the PEO–PPO units. However, at 37–40 °C, the extended coils become random due to the unstable hydrogen bonding between PPO and water, leading to increased solution entropy.36,37 As a result, the polymer chains of P407 move closer, and more interconnected networks were formed, increasing G′ values. This was also reflected by an enhancement of η* as shown in Fig. 3(f). At 37–40 °C, the loss factor of the P407 hydrogel (tan![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) δ = 0.03) showed typical elastic nature (shear-thickening), which indicates the proper gelation of the developed gel. These properties are crucial for 3D printing applications.38 Taken together, our results suggested that the developed hydrogel has good printability and could be used as an ideal material for future bioink development.
δ = 0.03) showed typical elastic nature (shear-thickening), which indicates the proper gelation of the developed gel. These properties are crucial for 3D printing applications.38 Taken together, our results suggested that the developed hydrogel has good printability and could be used as an ideal material for future bioink development.
3.4. P407 hydrogel sustains SCAPs viability
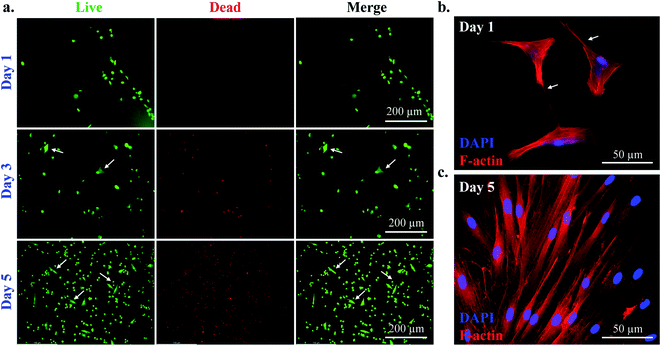
The cytotoxicity of the cell-laden P407 hydrogel was evaluated by live/dead staining kit after five days of incubation, and the results are shown in Fig. 4(a). After cell-laden printing, the 6-well plates were supplemented by 1 mL DMEM media containing 10% FBS and 1% P/S. The media was replaced every two days. The as-prepared hydrogel showed no adverse effect on the SCAPs after five days of culture. Interestingly, at the 3 day point, the cells started protruding inside the gel, and fibroblast-like morphology appeared. At the 5 day point, most of the cells appeared proliferating inside the hydrogel, suggesting the biocompatibility of P407. The quantification data for live dead staining is given in Fig. S3.†To investigate cell morphology, we have also performed immunocytochemical staining to observe the F-actin arrangement. Adhesion of cells to their native ECM is one of the pivotal properties required for proliferation and differentiation.19,39 Therefore, various biomimetic scaffolds could control the ECM structure to increase stem cells' proliferation.40 The morphology of the SCAPs at the desired time interval is given in Fig. 4(b and c). At day 1 samples, the cells produced sufficient numbers of filopodia. However, at the 3 day point, the gel was entirely filled by the SCAPs, and the F-actin filament appeared in regular shape and distribution as indicated in Fig. 3(c). These results suggest that P407 gels are highly biocompatible for cell-laden printing and could be used as a candidate biomaterial for tissue engineering.
3.5. EMF stimulation improves the proliferation and migration of SCAPs
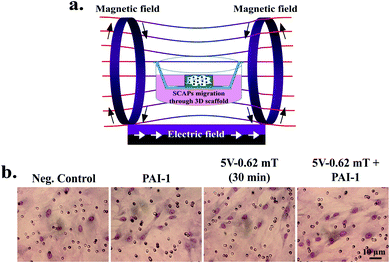
The cell-laden constructs (10 mm × 5 mm) were directly utilized for DC EMFs stimulation experiments. Fig. 5(a) represents the schematic procedure of cell-printing in transwell chamber and EMF stimulation, respectively. The PASCO EMF device has various waveforms, including triangular, sinusoidal, square, and ramp (DC offset), with a frequency range of 0.001 Hz to 100 kHz. The EMF stimulation may be beneficial clinically for bone fracture treatment because it induces the proliferation and migration of stem cells, such as hBMSCs, PDLSCs, and SCAPs.7,41,42 We have chosen the low voltage–frequency DC EMFs, such as 1 V–1 Hz (0.14 mT), 5 V–1 Hz (0.62 mT), and 10 V–1 Hz (1.21 mT). Their exposure time was 5 min per day, 10 min per day, 20 min per day, and 30 min per day for each hydrogel group, respectively. The cell-laden constructs were printed directly onto the 8 μm transwell inserts and supplemented by 200 μL of serum-free media. 10% serum-containing media supplemented the outer chamber as a chemoattractant. The culture plates were kept at 37 °C. Plates without any stimulation were taken as control. After the desired time interval, the cell viability was measured by WST-1 dye, and the results are given in Fig. 5(b–d). The results indicate that none of the treatment conditions were harmful to the culture cells. Notably, an enhancement of cell viability was found in 5 V–1 Hz (0.62 mT) EMFs exposure at a dose of 30 min per day. Moreover, an increasing trend of viability was observed in 5 V-treated groups, which was significantly higher than the control groups after 3 days of culture. A similar kind of observation was reported by Samiei et al., where they have demonstrated the higher proliferation of hDPSCs in the presence of sinusoidal EMFs.1Furthermore, we have also evaluated the cell morphology in 5 V EMF-treated groups after 3 days of culture. The cell morphology of the SCAPs was examined by fluorescence microscopy, and the results are given in Fig. 5(e). Interestingly, the cell morphology appeared to be usual with regular shape and filopodia distribution. At the 3 day point, the entire 3D matrix was covered by cells, suggesting that 5 V–1 Hz (0.62 mT) EMFs exposure is biocompatible for SCAPs and could be used as a stimulating agent for dental tissue regeneration.1,43 The migration potential of SCAPs was also assessed by culturing the cell-hydrogel directly onto the transwell migration chamber, and the results are given in Fig. 6. A schematic representation for the EMF-induced migration is shown in Fig. 6(a). The cell-laden constructs were printed onto the 8 μm inserts and stimulated by 5 V–1 Hz (0.62 mT) exposure at 30 min per day. Next, the cells were allowed to migrate up to 12 h. After 12 h, the transwell inserts were stained by Giemsa stain, photographed by an inverted optical microscope, and the number of migrated cells was quantified by ImageJ software. PAI-1-treated plates were considered as the positive control. To investigate the synergistic effect, the cell-laden construct was further supplemented by PAI-1 along with EMFs exposure, and the results are shown in Fig. 6(b). Notably, the number of migrated cells was significantly increased in both EMFs-treated and PAI-1 + EMFs-treated samples, comparable to the negative control groups, as shown in Fig. S4.† According to previously published reports, a few papers are relevant to EMFs' effects on dental stem cell proliferation and migration. The biophysical stimuli, mainly the effect of the electromagnetic field, are ill-explored for dental tissue regeneration.1 Hence, we have investigated the effects of low voltage–frequency DC EMFs with relatively short exposure time for SCAPs proliferation and migration. We anticipate that the 5 V–1 Hz (0.62 mT) EMFs (30 min per day) treatment could be used as an ideal condition for stem cell migration and proliferation during cell-based printing for dental tissue regeneration.
3.6. In vitro differentiation and gene expression study
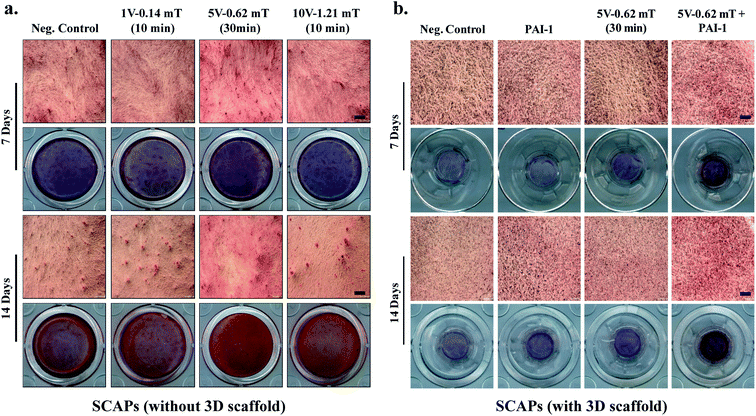
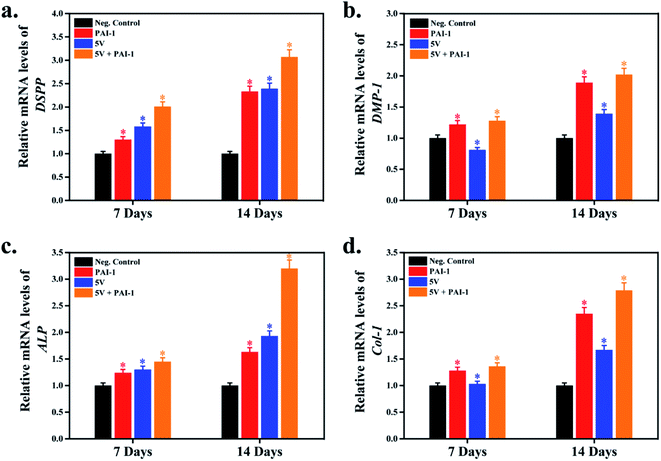
The in vitro differentiation potential of cell-laden hydrogels was evaluated by the ARS staining procedure. For this, the printed constructs were cultured in 200 μL of DMEM media for 2–3 days. After that, the printed hydrogels were washed by PBS buffer and supplemented by 200 μL of osteogenic differentiation media and cultured for 7 and 14 days. After the desired incubation period, the cells were stained by ARS to study the mineralization efficiency, and the results are shown in Fig. 7. Plates without scaffolds were compared side-by-side with the printed constructs (Fig. 7(a)). Moreover, PAI-1-treated plates were taken as the positive control. Notably, in both cases, the 5 V–1 Hz (0.62 mT) EMFs stimulation exhibited higher mineralization potential. However, a more intense mineralized nodule formation was observed in the presence of PAI-1-loaded 3D scaffold (Fig. 7(b)), suggesting that a combination of growth factors with EMFs stimulation could synergistically trigger the osteoblastic differentiation of SCAPs.The expression of osteogenesis and odontogenesis-related genes (ALP, Col-1, DSPP, and DMP-1) were evaluated by qRT-PCR, and the results are shown in Fig. 8. PAI-1-treated samples were taken as the positive control, and plates without any scaffold were taken as the negative control. Moreover, to study the synergistic effect of SCAPs gene expression, we have used PAI-1 + 5 V EMFs as an inducer of osteogenesis. Osteogenesis is a complex biological process, and several osteogenic/odontogenic genes are associated with this process.44 A large number of studies reported that SCAPs are found to express several gene markers related to osteogenesis/odontogenesis, such as alkaline phosphatase (ALP), runt-related transcription factor-2 (Runx-2), osteocalcin (OCN), dental sialophosphoprotein (DSPP), dental matrix protein (DMP), collagen type-1 (Col-1), and many others.45,46 ALP is an early gene marker and acts as an indicator of early osteogenesis. Similarly, DSPP, DMP-1, and Col-1 indicate the presence of late osteogenesis/odontogenesis.29 Our results demonstrated that DSPP and DMP-1 markers' expression significantly increased in 5 V (0.62 mT) EMFs treatment; however, in synergistic treatment, the expression profile was more significant than the positive control, suggesting that a combination of PAI-1 with EMFs treatment could trigger osteogenic/odontogenic differentiation (Fig. 8(a and b)). A similar fashion was also observed in ALP and Col-1, comparable to the control groups (Fig. 8(c and d)). Therefore, the higher expression of theses gene markers (DSPP, DMP-1, ALP, and Col-1) indicated their better osteogenic efficiency of P407-encapsulated SCAPs in the presence of EMFs.
Cell proliferation and differentiation are crucial factors for growth and development. Cellular differentiation, in particular, osteogenesis, is profoundly affected by nature, concentration, and shape (2D/3D structure) of the biomaterials present in the osteogenic differentiation media.47 Liu et al. reported that simvastatin/poloxamer 407 hydrogels improved the osteo–angiogenic properties of stem cells.48 In another study, poloxamer-encapsulated hDPSCs showed improved proliferation and osteogenic differentiation and higher expression of OCN and Runx2 transcription factors.15 Based on these previous reports, we have conducted a synergistic exposure system, where various DC EMFs directly stimulated the cell-laden P407 gel. We anticipate that SCAPs-encapsulated 3D poloxamer hydrogels with EMFs stimulation (5 V–1 Hz, 0.62 mT) could be used as a promising candidate for bone tissue engineering, particularly dental tissue regeneration.
4. Conclusions
In this study, apical papilla stem cells (SCAPs) were encapsulated in a ‘thermoresponsive’ hydrogel and 3D bioprinted to biomimic the apical papilla region of human teeth. The as-prepared P407 hydrogel was found biodegradable, non-toxic, and enhanced the viability of SCAPs in vitro. Moreover, the cell-laden printed construct was ultimately tested via low voltage–frequency electromagnetic fields (EMFs) stimulation. Our results suggested that the applied DC EMFs (5 V–1 Hz, 0.62 mT) enhanced the viability and raised the osteogenic/odontogenic potential of SCAPs by increasing DSPP, DMP-1, ALP, and Col-1 activity. However, the effective molecular mechanism of this process is still unknown. Therefore, in detail molecular mechanism behind this process would be the future direction of our present study. This study's findings revealed that cell-laden pluronic hydrogels are incredibly biocompatible, and the approach of EMFs-based cell stimulation provides a promising strategy for dental tissue engineering, mostly for oral applications.Conflicts of interest
The authors declare no competing financial interests.Acknowledgements
This work was supported by the Starting growth Technological R&D program (S2840309, Development of a novel bioreactor system with stimuli for stem cell culture) funded by the Small and Medium Business Administration (SMBA) and the ‘Basic Science Research Program’ through ‘National Research Foundation of Korea’ (NRF - 2018R1A6A1A03025582 & 2019R1D1A3A03103828), Republic of Korea. We would also like to thank the Postdoctoral International Exchange Program (postdoctoral management committee office of china 2018-115).References
- M. Samiei, Z. Aghazadeh, E. D. Abdolahinia, A. Vahdati, S. Daneshvar and A. Noghani, Acta Odontol. Scand., 2020, 1–7 Search PubMed.
- J. Borrelli, W. D. Prickett and W. M. Ricci, Clin. Orthop. Relat. Res., 2003, 411, 245–254 CrossRef PubMed.
- P. Hannemann, K. Göttgens, B. van Wely, K. Kolkman, A. Werre, M. Poeze and P. Brink, J. Bone Jt. Surg., Br. Vol., 2012, 94, 1403–1408 CrossRef CAS PubMed.
- M. T. Tsai, W. J. Li, R. S. Tuan and W. H. Chang, J. Orthop. Res., 2009, 27, 1169–1174 CrossRef PubMed.
- B. Noriega-Luna, M. Sabanero, M. Sosa and M. Avila-Rodriguez, Micron, 2011, 42, 600–607 CrossRef CAS PubMed.
- C. Grassi, M. D'Ascenzo, A. Torsello, G. Martinotti, F. Wolf, A. Cittadini and G. B. Azzena, Cell Calcium, 2004, 35, 307–315 CrossRef CAS PubMed.
- J. H. Jansen, O. P. van der Jagt, B. J. Punt, J. A. Verhaar, J. P. van Leeuwen, H. Weinans and H. Jahr, BMC Musculoskeletal Disord., 2010, 11, 188 CrossRef PubMed.
- S. Mayer-Wagner, A. Passberger, B. Sievers, J. Aigner, B. Summer, T. S. Schiergens, V. Jansson and P. E. Müller, Bioelectromagnetics, 2011, 32, 283–290 CrossRef CAS PubMed.
- M. Zhao, B. Song, J. Pu, T. Wada, B. Reid, G. Tai, F. Wang, A. Guo, P. Walczysko and Y. Gu, Nature, 2006, 442, 457–460 CrossRef CAS PubMed.
- K. E. Hammerick, M. T. Longaker and F. B. Prinz, Biochem. Biophys. Res. Commun., 2010, 397, 12–17 CrossRef CAS PubMed.
- H. F. Liu, L. Yang, H. C. He, J. Zhou, Y. Liu, C. Y. Wang, Y. C. Wu and C. Q. He, Bioelectromagnetics, 2013, 34, 323–332 CrossRef CAS PubMed.
- W. Z. Lew, Y. C. Huang, K. Y. Huang, C. T. Lin, M. T. Tsai and H. M. Huang, J. Tissue Eng. Regener. Med., 2018, 12, 19–29 CrossRef CAS PubMed.
- L. Zheng, L. Zhang, L. Chen, J. Jiang, X. Zhou, M. Wang and Y. Fan, J. Tissue Eng. Regener. Med., 2018, 12, 2029–2040 CrossRef CAS PubMed.
- S.-h. Hsu and J.-C. Chang, Cytotechnology, 2010, 62, 143–155 CrossRef PubMed.
- I. M. Diniz, C. Chen, X. Xu, S. Ansari, H. H. Zadeh, M. M. Marques, S. Shi and A. Moshaverinia, J. Mater. Sci.: Mater. Med., 2015, 26, 153 CrossRef PubMed.
- N. Sultan, L. E. Amin, A. R. Zaher, B. A. Scheven and M. E. Grawish, World J. Stomatol., 2019, 7, 1–19 CrossRef.
- L. Ma, Y. Makino, H. Yamaza, K. Akiyama, Y. Hoshino, G. Song, T. Kukita, K. Nonaka, S. Shi and T. Yamaza, PLoS One, 2012, 7, e51777 CrossRef CAS PubMed.
- B. Jin and P.-H. Choung, Tissue Eng., Part A, 2016, 22, 721–732 CrossRef CAS PubMed.
- S. D. Dutta, J. Hexiu, D. K. Patel, K. Ganguly and K.-T. Lim, Int. J. Biol. Macromol., 2021, 167, 644–658 CrossRef CAS PubMed.
- T. A. Mir and M. Nakamura, Tissue Eng., Part B, 2017, 23, 245–256 CrossRef PubMed.
- K. C. Kolan, J. A. Semon, B. Bromet, D. E. Day and M. C. Leu, Int. J. Bioprint., 2019, 5, 204 Search PubMed.
- M. Ojansivu, A. Rashad, A. Ahlinder, J. Massera, A. Mishra, K. Syverud, A. Finne-Wistrand, S. Miettinen and K. Mustafa, Biofabrication, 2019, 11, 035010 CrossRef CAS PubMed.
- H. Cho, U. Jammalamadaka, K. Tappa, C. Egbulefu, J. Prior, R. Tang and S. Achilefu, Mol. Pharmaceutics, 2019, 16, 552–560 CrossRef CAS PubMed.
- H. Jin, H.-W. Choung, K.-T. Lim, B. Jin, C. Jin, J.-H. Chung and P.-H. Choung, Tissue Eng., Part A, 2015, 21, 2817–2828 CrossRef CAS PubMed.
- T. Distler, C. Polley, F. Shi, D. Schneidereit, M. D. Ashton, O. Friedrich, J. F. Kolb, J. G. Hardy, R. Detsch and H. Seitz, Adv. Healthcare Mater., 2021, 2001876 CrossRef CAS PubMed.
- A. I. Aldebs, F. T. Zohora, N. Nosoudi, S. P. Singh and J. E. Ramirez-Vick, Bioelectromagnetics, 2020, 41, 175–187 CrossRef CAS PubMed.
- T. Gupta, V. Jain and P. Tandon, Med. Biol. Eng. Comput., 1991, 29, 113–120 CrossRef CAS PubMed.
- D. K. Patel, S. D. Dutta, J. Hexiu, K. Ganguly and K.-T. Lim, Int. J. Biol. Macromol., 2020, 162, 1429–1441 CrossRef CAS PubMed.
- J. Kang, W. Fan, Q. Deng, H. He and F. Huang, BioMed Res. Int., 2019, 2019, 1–8 Search PubMed.
- M. Al-Habib and G. T.-J. Huang, in Odontogenesis, Springer, 2019, pp. 59–76 Search PubMed.
- P. M. Aponte and A. Caicedo, Stem Cells Int., 2017, 2017, 1–17 CrossRef PubMed.
- Y. Cao, D. Xia, S. Qi, J. Du, P. Ma, S. Wang and Z. Fan, Cell Proliferation, 2013, 46, 447–456 CrossRef CAS PubMed.
- R. Karamzadeh and M. B. Eslaminejad, in Regenerative Medicine and Tissue Engineering, IntechOpen, 2013 Search PubMed.
- E. Russo and C. Villa, Pharmaceutics, 2019, 11, 671 CrossRef CAS PubMed.
- P. Alexandridis, J. F. Holzwarth and T. A. Hatton, Macromolecules, 1994, 27, 2414–2425 CrossRef CAS.
- I.-C. Tung, Int. J. Pharm, 1994, 107, 85–90 CrossRef CAS.
- J.-J. Xuan, P. Balakrishnan, D. H. Oh, W. H. Yeo, S. M. Park, C. S. Yong and H.-G. Choi, Int. J. Pharm, 2010, 395, 317–323 CrossRef CAS PubMed.
- M. Müller, J. Becher, M. Schnabelrauch and M. Zenobi-Wong, Biofabrication, 2015, 7, 035006 CrossRef PubMed.
- S. D. Dutta, D. K. Patel, Y.-R. Seo, C.-W. Park, S.-H. Lee, J.-W. Kim, J. Kim, H. Seonwoo and K.-T. Lim, J. Nanomater., 2019, 2019, 1–11 CrossRef.
- J. H. Lee, J.-H. Park, Y.-R. Yun, J.-H. Jang, E.-J. Lee, W. Chrzanowski, I. B. Wall and H.-W. Kim, J. Mater. Chem. B, 2013, 1, 2731–2741 RSC.
- B. Hiemer, M. Krogull, T. Bender, J. Ziebart, S. Krueger, R. Bader and A. Jonitz-Heincke, Mol. Med. Rep., 2018, 18, 2133–2141 CAS.
- A. Lacy-hulbert, J. C. Metcalfe and R. Hesketh, FASEB J., 1998, 12, 395–420 CrossRef CAS PubMed.
- E. M. El-Maghraby, D. H. El-Rouby and A. M. Saafan, Arch. Oral Biol., 2013, 58, 796–805 CrossRef PubMed.
- H.-B. Kim, B. Jin, D. K. Patel, J.-W. Kim, J. Kim, H. Seonwoo and K.-T. Lim, IEEE Trans. NanoBioscience, 2019, 18, 463–468 Search PubMed.
- S. Tetè, E. Nargi, F. Mastrangelo, V. Zizzari, G. D'Apolito, T. Traini, G. Costanzo, V. Dadorante, I. D'Alimonte and S. Caputi, Int. J. Immunopathol. Pharmacol., 2008, 21, 309–318 CrossRef PubMed.
- R. Patil, B. M. Kumar, W.-J. Lee, R.-H. Jeon, S.-J. Jang, Y.-M. Lee, B.-W. Park, J.-H. Byun, C.-S. Ahn and J.-W. Kim, Exp. Cell Res., 2014, 320, 92–107 CrossRef CAS PubMed.
- N. Kawazoe and G. Chen, Biomaterials, 2015, 54, 226–236 CrossRef PubMed.
- H. Liu, W. Li, C. Liu, J. Tan, H. Wang, B. Hai, H. Cai, H.-J. Leng, Z.-J. Liu and C.-L. Song, Biofabrication, 2016, 8, 045012 CrossRef PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1ra01143j |
| ‡ These two authors have contributed equally to this manuscript. |
| This journal is © The Royal Society of Chemistry 2021 |