Open Access Article
Open Access ArticleConformational stability as a quality attribute for the cell therapy raw material human serum albumin†
Evelien Wynendaelea,
Gamaliel Junren Mab,
Xiaolong Xua,
Nam-Joon Cho b and
Bart De Spiegeleer
b and
Bart De Spiegeleer *a
*a
aFaculty of Pharmaceutical Sciences, Ghent University, Ottergemsesteenweg 460, Ghent 9000, Belgium. E-mail: Bart.DeSpiegeleer@UGent.be; Tel: +32 9 264 81 00
bSchool of Materials Science and Engineering, Nanyang Technological University, 50 Nanyang Avenue, Nanyang 639798, Singapore
First published on 23rd April 2021
Abstract
Although human serum albumin (HSA) has been used for many decades, there is still a lack of suitable quality control (QC) attributes. Its current use as a raw material in gene-, cell- and tissue-therapies requires more appropriate functionally-relevant quality attributes and methods. This study investigated the conformational stability of serum albumin using circular dichroism (CD) spectroscopy and dynamic light scattering (DLS) for evaluating the thermal sensitivity, and quartz crystal microbalance-dissipation (QCM-D) and localized surface plasmon resonance (LSPR) for assessing the adsorption behavior. Different serum albumin samples were used, encompassing plasma-derived HSA (pHSA), recombinant octanoate-stabilized HSA (rHSA) and bovine serum albumin (BSA). The melting temperature (Tm) as well as the onset temperature (Tonset) were obtained from the derivative curves of the temperature gradient CD data at 222 nm. The results from DLS, as well as from real-time QCM-D and LSPR silica-adsorption kinetic profiles confirmed the relatively higher conformational stability of the octanoate (fatty acid) containing rHSA, while the additional negative charge resulted in a lower amount adsorbed to the silica surface compared to the non-stabilized HSA and BSA. Adsorption studies further revealed that BSA has a lower conformational stability and undergoes more extensive adsorption-induced spreading compared to the non-stabilized HSA. Collectively, the temperature-based (CD and DLS) as well as adsorption-based biosensor (QCM-D and LSPR) approaches gave congruent and discriminatory information about the conformational stability of different serum albumins, indicating that these techniques provide information on valuable QC attributes.
1. Introduction
Human serum albumin (HSA), the most abundant protein in human plasma, is a heart-shaped protein with a molecular weight of about 66 kDa that consists of 585 amino acids in a single polypeptide, forming three domains with specific structure and function.1–3 Crystal structures of several albumin types, including possible complexes such as with fatty acids, have already been elucidated, indicating different structural compositions.4–7 In vivo, it can not only help maintain osmotic blood pressure but also transport endogenous (e.g. hormones, fatty acids, metals and toxic metabolites) and exogenous compounds (e.g. drug molecules).8–10 Besides its widespread use in clinical applications,11 serum albumin is often used as a supplement in mammalian cell cultures, replacing the traditionally used serum supplements like fetal bovine serum (FBS) which are complex mixtures, difficult to pharmaceutically keep consistent.12,13 The beneficial effects of serum albumin on cells in culture can be divided into several functional groups: antioxidant and radical scavenging activity,14,15 attachment of cells,16 regulation of apoptosis,17 mediating cell uptake,12 providing amino acids and its synergistic effect on other substances in the cell medium.16,18,19 As such, it is also an important raw material used in the production of advanced therapy medicinal products (ATMPs),20 promoting growth and proliferation of the ATMP-cells. In the spirit of the 3Rs (reduction, refinement and replacement) and considering the need for well-known, chemically defined raw materials in cell cultures used in the manufacturing of medicinal products for therapeutic purposes as well as reducing the risk of blood-derived pathogen contaminations, animal (mostly bovine) and human plasma-derived serum albumins (pBSA and pHSA) are often replaced by recombinant HSA (rHSA).12,21Although albumin has been used for many decades, a simple set of formal, biorelevant, robust and discriminatory attributes and methods to assure quality consistency, as is required in recent guidelines,22 do not yet exist. This situation is very similar to the former heparin quality situation, also a well-established and abundantly used medicine, until the 2007–2008 crisis which upgraded the quality attributes.23 Current Ph. Eur. and USP monographs of plasma-derived HSA (pHSA) in the form of aqueous solutions are available; however, the quality attributes described in these monographs are limited, and do not include conformational sensitivity, as measured by thermostability and adsorption behaviour. While its relation to its cell-culture functionality is not yet elucidated, it is considered a discriminatory overall quality control attribute for biopharmaceuticals.24
CD spectroscopy is an absorption spectroscopy method based on the differential absorption of left and right circularly polarized light, which is used to determine the secondary structural composition of proteins.25 As such, this technique is also used for the investigation of the (un)folding behavior of proteins as a function of temperature, determining the thermodynamics of protein unfolding and binding interactions.26 However, up till now, the application of CD for quality control purposes of biopharmaceuticals is rarely reported, despite its presence as a general pharmaceutical technique in different pharmacopoeia. The same is also true for DLS, which provides particle size information (hydrodynamic diameter) in the nanometer range, based on Brownian motion and using the Stokes–Einstein equation. QCM-D and LSPR, on the contrary, are biosensor devices currently not yet described in any pharmacopoeia, utilizing bulk acoustic and optical waves, respectively, to quantitatively measure in real time the mass and refractive index changes occurring on the sensing surface upon protein adsorption. Moreover, QCM-D provides additional information on the viscoelastic properties of the protein adlayer. This makes them highly useful to investigate the conformational stability of proteins through their adsorption behavior as a measure of quality and they have been extensively used to study how modulating the conformational stability of both BSA and HSA through various experimental conditions can influence their adsorption behavior.27–30 Other techniques can also be used, such as DSC,31,32 MALDI-TOF-MS or AUC (analytical ultracentrifuge) which evaluate more directly a selected quality attribute (the degree of oligomerization), or surface hydrophobicity.33 However, the techniques used here are not yet well integrated in pharmaceutical quality control environments, while they are well suited for adequately controlling overall quality consistency.
This study aims to investigate the conformational stability as a pharmaceutical QC quality attribute of three different albumin types (human serum albumin, derived from plasma and recombinant, and pBSA), using different complementary techniques.
2. Experimental
2.1. Materials and equipment
The HSA samples investigated in this study were donated by Oxyrane Belgium NV company with following information: rHSA (Novozymes, lot# PDP150055), recombinant HSA expressed in Saccharomyces cerevisiae, was received as a 100 mg ml−1 solution in 20 mM phosphate buffer with 8 mM octanoate; pHSA (Sigma-Aldrich, product code A3782, lot# SLBN5035V) was a lyophilized powder made from human serum as fatty acid-free material. The bovine serum albumin (BSA) was obtained from Merck and Sigma-Aldrich (product code 1.12018.0100 and A7030, lot# K44575418 and SLBK3074V, respectively). The other chemicals used in this study are all of analytical grade and were purchased from VWR Chemicals (Leuven, Belgium).Aqueous serum albumin solutions used in the analytics were prepared by dissolving the powder or diluting the corresponding solutions with 10 mM phosphate buffer pH 7.4 to obtain the stated concentrations.
2.2. Circular dichroism (CD) analysis
CD analysis was performed on a nitrogen-flushed Jasco J-815 CD spectrometer (Tokyo, Japan). The quartz cuvette (path length of 1 mm) was obtained from Hellma Analytics (Müllheim, Germany). At a scan speed of 50 nm min−1, 3 continuous scans were recorded. The data pitch was set at 0.2 nm and the bandwidth at 1.00 nm. The wavelength region of 190–260 nm was recorded for all the samples. The concentration of all serum albumin samples was 0.1 mg ml−1, corresponding to approximately 1–2 μM. All measurements were done in independent triplicates, using the obtained mean value for data analysis. Quantitative secondary structure analysis was performed by CONTIN algorithm (protein reference set 4) from Dichroweb (London, UK).34,35For the temperature gradient test, the gradient was set at 80 °C h−1 in the range of 20 °C to 95 °C (with temperature intervals of 5 °C). Although 1 °C intervals would give more datapoints and hence will result in a more precise Tm value, i.e. with a decreased uncertainty around its estimated value, the interval of 5 °C was adequate for our purpose. Tm is the temperature at which the protein possesses 50% of folded fraction, determined by evaluating the CD signal at a wavelength of 222 nm in function of temperature.36,37 The first derivative of the CD data was obtained and further fitted to a cubic function y = ax3 + bx2 + cx + d from the five maximum points using Origin software (Northampton, USA). The x value at apex y point of the fitted cubic function corresponds to Tm. The wavelength 222 nm corresponds to the negative band signal typical of α-helix, and is a most-often used wavelength to evaluate Tm of α-helix dominated proteins.38–40 Besides Tm, the onset point (Tonset), where the thermal unfolding transition starts, was also investigated in this study. The second derivative of the CD data was obtained using Origin software (Northampton, USA). Tonset is the x value at apex y point of the polynomial curve, fitted from the three or four maximum points of the second derivative curve.
2.3. Dynamic light scattering (DLS) measurement
DLS was performed using a particle size analyzer (ZetaPALS, Brookhaven Instruments, Holtsville, NY, USA) that is equipped with a 658.0 nm monochromatic laser. Measurements were taken at a 90° scattering angle and the BIC Particle Sizing software (v5.27; Brookhaven Instruments) was used to analyze the intensity autocorrelation function in order to obtain the intensity-weighted Gaussian size distribution. The temperature in the measurement chamber was controlled with a feedback loop and measurements of each 150 μM serum albumin sample were first recorded at 25 °C, followed by heating to 50 °C, then increasing the temperature to 75 °C in 5 °C increments. After each temperature step increase, the measurement chamber was equilibrated for 5 min before the DLS measurement was performed. All reported values were obtained from 5 technical replicates. The standard deviation (s.d.), which is related to the size distribution of the protein samples, was obtained by dividing the full-width-at-half-maximum (FWHM) of the Gaussian size distribution from DLS measurements by 2√(2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ln(2)) ≈ 2.355.
ln(2)) ≈ 2.355.
2.4. QCM-D monitoring
The quartz crystal microbalance-dissipation (QCM-D) experiments were conducted using the QSense E4 instrument (Biolin Scientific AB, Sweden) with silica-coated quartz crystal sensor chips (QSX 303, Biolin Scientific). Before each experiment, the sensor chips were washed by sequentially rinsing with 1% (m/v) aqueous SDS solution, water and ethanol. After drying under a gentle stream of nitrogen gas and oxygen plasma treatment (PDC-002, Harrick Plasma, USA) for 3 min, the chips were assembled into the measurement chamber. For experiments, liquid samples were injected into the measurement chamber at a flow rate of 100 μl min−1. A stable baseline was first established with buffer solution, then 10 μM protein samples were injected for 10 min, followed by a buffer washing step. All measurements were conducted at 25 °C. The corresponding resonance frequency (ΔF) and energy dissipation (ΔD) shifts were recorded in real-time at multiple odd overtones, as previously described.41 Data collected at the fifth overtone are reported. Measurement operations were controlled by the QSoft 401 (Biolin Scientific) software package.2.5. Localized surface plasmon resonance (LSPR) characterization
Ensemble-averaged localized surface plasmon resonance (LSPR) experiments in transmission mode were performed with silica-coated sensor chips containing gold nanodisk arrays using an Insplorion XNano instrument (Insplorion AB, Sweden), under analogous conditions as the QCM-D experiments (10 μM solutions, flow: 100 μl min−1 for 10 min, followed by a buffer washing step).42 Before each experiment, the sensor chips were washed and surface-treated in the same way as in QCM-D experiments (sequentially rinsing with 1% (m/v) aqueous SDS solution, water and ethanol followed by oxygen plasma treatment). The Insplorer software package (Insplorion AB) was used to record shifts in the LSPR centroid (peak) position (Δλ) from the extinction spectrum.3. Results and discussion
3.1. CD thermal stability
The plasma HSA remained a relatively stable conformation up to a temperature of 60 °C, while unfolding became significant when further increasing the temperature (Fig. 1A). Deconvolution of the CD spectrum, using a reference database of well-characterized proteins, was performed to quantify the secondary structural fractions. At 20 °C, the α-helix content was 60.8% (RSD = 0.5%, n = 3), β-sheet 2.3% (RSD = 2.0%, n = 3), β-turn 12.7% (RSD = 1.7%, n = 3) and the unordered 24.2% (RSD = 2.2%, n = 3), indicating the high level of secondary, not denatured structure. When the temperature increased, the α-helix content rapidly decreased and β-sheet content increased from 60 °C onwards, indicating temperature-induced unfolding. A smaller increase in β-turns and unordered secondary structure was also observed, which is frequently observed for proteins during a heating procedure (Fig. 1B).43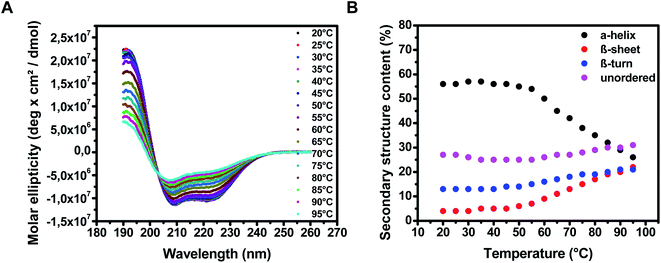 | ||
| Fig. 1 CD spectra of pHSA in the temperature range of 20–95 °C. (A) Spectra of pHSA at different temperatures; (B) the calculated relative secondary structure contents (Dichroweb). | ||
The same data processing procedure was applied for the other serum albumin samples (Fig. S1 and S2 in ESI†). Similar behaviors following the increase of temperature were observed for these serum albumin samples. As expected, secondary structure differences among these serum albumin samples were observed at the beginning of the heating process, while differences were not observed at the end temperature, i.e. 95 °C, where complete unfolding and oligomerization was present.
After smoothing the CD signal data by averaging consecutive three points, the first derivative of the CD data was obtained using Origin software (Northampton, USA). The plots of the CD signal and their first derivative plots at 222 nm as a function of temperature are given in Fig. 2. Tm is the temperature at the apex of the cubic function (where the first derivative of the fitted function equals zero) fitted from the five maximum points of the first derivative curve. The calculated Tm values of the albumin samples are presented in Table 1.
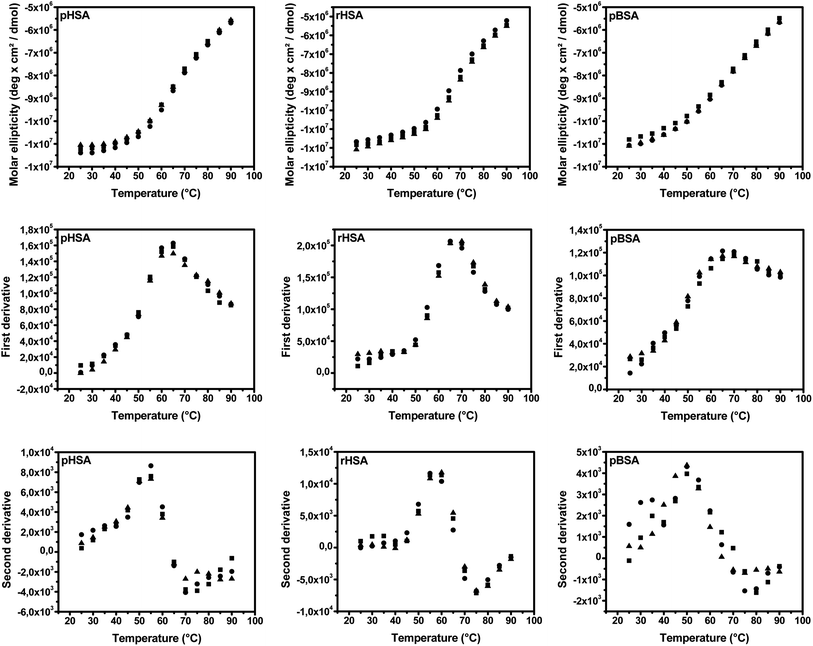 | ||
| Fig. 2 CD signals (up), their first (middle) and second derivative plots (down) of pHSA, rHSA and pBSA at 222 nm in function of temperature (n = 3). Symbols represent individual data points. | ||
| Sample | Tm (°C) (RSD%, n = 3) | Tonset (°C) (RSD%, n = 3) |
|---|---|---|
| pHSA | 63.0 (0.4%) | 53.5 (1.3%) |
| rHSA | 67.9 (1.1%) | 57.1 (0.9%) |
| pBSA | 67.7 (2.5%) | 50.3 (1.7%) |
Besides Tm, the onset temperature (Tonset) where the thermal unfolding transition starts, was also determined using the second-derivative method. The second derivative of the smoothed CD data was obtained using Origin software (Northampton, USA). Tonset was defined as the temperature corresponding to the apex of the polynomial curve, fitted from the three or four maximum points of the second-derivative curve. The second-derivative plots are also given in Fig. 2, while the calculated Tonset values are presented in Table 1.
The combination of Tm and Tonset differentiates clearly the 3 different types of serum albumin.
It is well-known that changes in temperature can cause alterations in the conformation of the protein, due to folding/unfolding, aggregation and denaturation. Thermal stability, as an indication of the protein conformational folding characteristic, is also related to several protein biological properties, such as the biological half-life.44–47 Thus, thermal stability is a possible quality attribute that can be used to reflect the functional consistency of proteins. While thermal stability using CD was mostly used in the fundamental characterization of binding properties of selected compounds with serum albumin, its use as a quality control attribute was not yet reported. In our previous work, the thermal stability of L-ASNase was evaluated by CD only using two different parameters, Tm and slope.24 The slope was determined by fitting the CD data in function of temperature using a sigmoidal function, which required the CD signal reaching a plateau. However, this is not applicable for proteins like serum albumin, which cannot reach a signal maximum within the practical temperature range (Fig. 2). Thus, a new parameter Tonset, the temperature where the thermal unfolding transition starts, was used in this study. Different methods are available to estimate this Tonset value. Firstly, it can be regarded as the lowest experimental temperature where the conformation change starts; thus, Tonset is then similar to a common toxicological parameter, i.e. the lowest observed effect level (LOEL), determined by probabilistic approaches, which is widely used in dose–response assessments.48 Secondly, the Tonset value can be determined using a cross-line method, which is derived from a previously reported method of using the bending point in dissolution profiles.49 The bending region, including the onset point, can be located in three or four data points by visual verification, e.g. 50–65 °C for pHSA (Fig. 2). The three or four consecutive points on both sides of the bending region were found to be linear. Thus, two lines were fitted from these data points next to the estimated bending point and the crossing point of these two lines can be used as Tonset. Thirdly, the onset point can be calculated from the second-derivative plot as the x-coordinate of the apex. In this study, the second-derivative method was used due to its easier calculation and the calculated Tonset values were close to the values obtained using the cross-line method with a minor system shift of 0.7 ± 0.5 °C (±SD, n = 3) on average (see ESI Table S1†).
The Tm of pHSA and pBSA was found to be 63.0 °C (RSD 0.4%, n = 3), and 67.7 °C (RSD 2.5%, n = 3), respectively, which is consistent with the literature findings.50,51 The significant difference of Tm between rHSA and pHSA can be explained by the presence of 8 μM octanoate in the 2 μM rHSA sample, which was used as a stabilizer according to the information from the supplier. This observation was confirmed by the literature: the presence of fatty acid can stabilize HSA, leading to an increase in Tm (from ∼62 °C to ∼64–72 °C).50
Only far-UV CD was investigated in our study. The backbone amide chromophores do show absorbance in this region and relatively low concentrations of 0.1 mg ml−1 in a 1 mm cuvette yield acceptable spectra. The evaluation of structural characteristics in near-UV CD would require significantly more sample in the path (higher concentrations and/or increased path lengths). Moreover, these far-UV CD investigations would require more time (more scans) to increase the signal/noise ratio.
3.2. DLS thermal stability
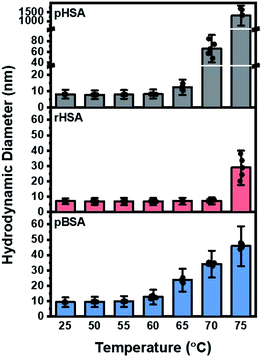
At room temperature, and up to 55 °C, rHSA showed the lowest hydrodynamic diameter (∼6.5 nm), followed by pHSA (∼8.5 nm) and pBSA (∼10 nm), which is in agreement with the literature reported albumin monomers.52 BSA gradually increased in size from 60 °C onwards up to ∼45 nm at 75 °C. This markedly contrasted both human serum albumins: pHSA rapidly and significantly increased in size from 65 °C, reaching up to more than 1000 nm at 75 °C, while rHSA remained much more stable, and only increased at 75 °C to approximately 30 nm (Fig. 3). The observed oligomerization behavior is closely related to the temperature-induced unfolding of the albumin proteins, which exposes hydrophobic regions leading to intermolecular hydrophobic interactions between the albumin molecules. The DLS data support that rHSA is more stable against temperature-induced oligomerization than pHSA and pBSA due to its fatty acid stabilizer. At 75 °C, pHSA oligomerized to micron-scale sizes, i.e. 200–500 times larger than pBSA and rHSA protein aggregates.The DLS and CD spectroscopy data indicate that pBSA had a lower onset temperature of oligomerization compared to pHSA, a similar Tm (where ΔG for the unfolding equals zero) and smaller protein aggregates when increasing the temperature above Tm which supports that pBSA has a lower conformational stability but greater colloidal stability than pHSA. That is, pHSA has a higher onset temperature of unfolding, but undergoes more extensive oligomerization when unfolded to form larger aggregates than pBSA. Taken together, our CD and DLS analysis support that the Tm and Tonset from CD as well as the onset temperature of oligomerization and extent of oligomerization from DLS are useful parameters for characterizing the thermal stability as a quality attribute of albumin proteins.
3.3. QCM-D
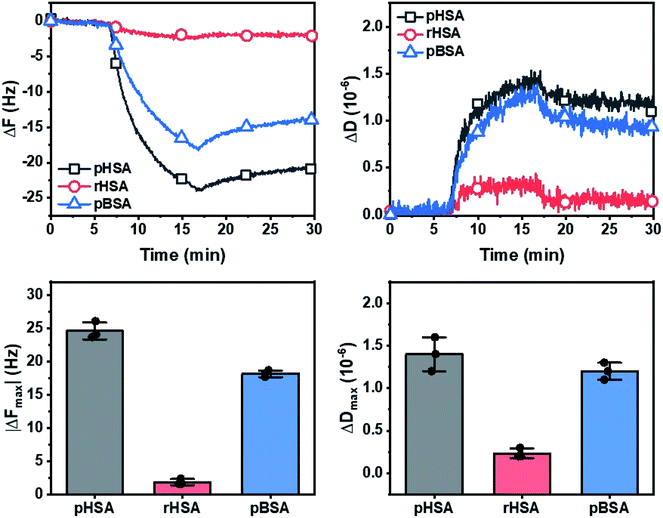
At adsorption saturation, the adsorption uptake of pHSA, rHSA and pBSA in terms of mean absolute final frequency shift (|ΔFmax|) was 25.0, 2.0, and 18.0 Hz, respectively (Fig. 4, left). The corresponding mean dissipation shifts (ΔDmax) were 1.4, 0.2, and 1.2 × 10−6, respectively (Fig. 4, right).The resonance frequency shifts (ΔF) observed showed a significant and almost irreversible adsorption to the silica surface for pHSA and to a lesser extent for pBSA, compared to a minimal adsorption for rHSA, indicating a weak interaction of rHSA with the silica surface. This is due to the additional octanoate (fatty acid) stabilizer in rHSA which increased its conformational stability while additionally adding a negative charge, resulting in a lower amount adsorbed and less extensive adsorption-related deformation on the silica surface compared to the non-stabilized HSA and BSA. While this was previously demonstrated for the adsorption of BSA onto a silica surface,53,54 a similar effect is observed here for HSA. The corresponding energy dissipation factor shift (ΔD) indicated a minor degree of viscoelastic character within the protein adlayers for pHSA and pBSA, and which was almost negligible for rHSA.
3.4. LSPR
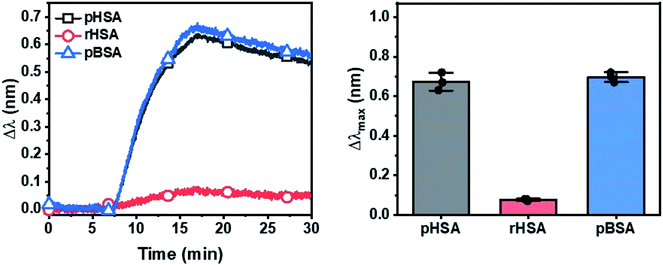
Compared to the traditional SPR, which, similar to QCM-D, shows a bulk effect extending to ∼200 nm, the localized surface plasmon resonance (LSPR) sensing technique, which consists of silica-coated gold nanodisk arrays, has a sensing depth of only ∼20 nm distance. Therefore, the LSPR technique is not only sensitive to protein adsorption uptake but is also sensitive to the extent of adsorption-related deformation of the protein molecules on the sensor surface.55–57 pBSA shows a slightly higher optical extinction peak shift (Δλ) than pHSA, i.e. the reverse of the ΔF shifts observed in QCM-D (Fig. 5). This indicates that pBSA underwent more extensive deformation and spreading and occupied a larger surface area per molecule on the surface compared to pHSA, resulting in pBSA having a lower adsorption amount observed using QCM-D, but a higher LSPR signal response.52 At the same time, the low LSPR signal response from the adsorption of rHSA agrees with that observed from QCM-D experiments.Therefore, multiple lines of experimental evidence from the solution-phase characterization by CD and DLS techniques as well as the adsorption studies characterized by both QCM-D and LSPR techniques are consistent. Together, they support that pBSA has a lower conformational stability than pHSA and more easily undergoes conformational changes upon exposure to external stresses such as high temperatures and solid–liquid interfaces. By contrast, the fatty acid-stabilized rHSA has the greatest conformational stability among the three tested albumin samples and not only underwent the least extensive temperature-induced unfolding and oligomerization, but also the least amount of adsorption and adsorption-related deformation.
4. Conclusions
In this study, the potential of CD and DLS for assessing the thermal stability and QCM-D and LSPR for assessing the adsorption behavior of different serum albumins as functionally relevant quality attributes, measuring aspects of conformational stability, was investigated. Analysis of the Tm and Tonset parameters from CD data together with the temperature-induced oligomerization behavior from DLS data revealed that pHSA had a greater solution-phase conformational stability but a lower colloidal stability than pBSA. Moreover, pHSA yielded higher QCM-D and lower LSPR signal responses from adsorption experiments onto silica surfaces compared to pBSA, indicating that pHSA underwent less adsorption-related deformation than pBSA agreeing with pHSA's greater solution-phase conformational stability. At the same time, rHSA contained an additional octanoate stabilizer and consistently exhibited the greatest solution-phase conformational stability and least extensive adsorption-related deformation compared to the non-stabilized, fatty acid-free pHSA and pBSA. Taken together, the data indicate that all four methods gave consistent and discriminatory results, with overlapping information while also providing more nuanced aspects in the context of resolving protein conformational and colloidal stability. The different techniques and analysis methods described here form the basis for setting acceptance specifications for human albumin used as a raw material during the ATMP development, based on information from statistical process analysis and leading to a more consistent ATMP-product. Moreover, extensive quality-by-design functional studies can be performed to support the risk-based performance approach towards specification settings as currently recommended in the pharmaceutical industry.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors gratefully acknowledge the financial support granted to X. X. by the PhD grant of the China Scholarship Council (CSC). The authors would like to thank Oxyrane Belgium NV company for the donation of the human serum albumin samples. The authors thank Bo Kang and Tijl De Bie (Department of Electronics and Information Systems, Faculty of Engineering and Architecture, Ghent University) for stimulating discussions.References
- P. Akbarzadehlaleh, M. Mirzaei, M. Mashahdi-Keshtiban, K. Shamsasenjan and H. Heydari, Adv. Pharm. Bull., 2016, 6, 309 CrossRef CAS PubMed.
- M. Dockal, D. C. Carter and F. Rüker, J. Biol. Chem., 2000, 275, 3042 CrossRef CAS PubMed.
- T. Chatterjee, A. Pal, S. Dey, B. K. Chatterjee and P. Chakrabarti, PLoS One, 2012, 7, e37468 CrossRef CAS PubMed.
- X. M. He and D. C. Carter, Nature, 1992, 358, 209 CrossRef CAS PubMed.
- S. Curry, H. Mandelkow, P. Brick and N. Franks, Nat. Struct. Biol., 1998, 5, 827 CrossRef CAS PubMed.
- A. Bujacz, Acta Crystallogr., 2012, D68, 1278 CrossRef PubMed.
- A. Kawaa, V. T. G. Chuang, Y. Kouno, K. Yamasaki, S. Miyamoto, M. Anraku and M. Otagiri, Biochim. Biophys. Acta, Proteins Proteomics, 2017, 1865, 979 CrossRef PubMed.
- K. Yamasaki, V. T. G. Chuang, T. Maruyama and M. Otagiri, Biochim. Biophys. Acta, Gen. Subj., 2013, 1830, 5435 CrossRef CAS PubMed.
- T. Peters, All about albumin: biochemistry, genetics, and medical applications, Academic Press, 1995 Search PubMed.
- J. Simard, P. Zunszain, C.-E. Ha, J. Yang, N. Bhagavan, I. Petitpas, S. Curry and J. Hamilton, Proc. Natl. Acad. Sci., 2005, 102, 17958 CrossRef CAS PubMed.
- G. Fanali, A. Di Masi, V. Trezza, M. Marino, M. Fasano and P. Ascenzi, Mol. Aspects Med., 2012, 33, 209 CrossRef CAS PubMed.
- G. L. Francis, Cytotechnology, 2010, 62, 1 CrossRef CAS PubMed.
- C. Tekkatte, G. P. Gunasingh, K. Cherian and K. Sankaranarayanan, Stem Cells Int., 2011, 2011, 504723 Search PubMed.
- M. Roche, P. Rondeau, N. R. Singh, E. Tarnus and E. Bourdon, FEBS Lett., 2008, 582, 1783 CrossRef CAS PubMed.
- M. De Castro, G. Orive, A. Gascon, R. Hernandez and J. Pedraz, Int. J. Pharm., 2006, 310, 8 CrossRef CAS PubMed.
- D. B. Horváthy, M. Simon, C. M. Schwarz, M. Masteling, G. Vácz, I. Hornyák and Z. Lacza, BioFactors, 2017, 43, 315 CrossRef PubMed.
- H. Zoellner, M. Hofler, R. Beckmann, P. Hufnagl, E. Vanyek, E. Bielek, J. Wojta, A. Fabry, S. Lockie and B. R. Binder, J. Cell Sci., 1996, 109, 2571 CrossRef CAS.
- M. T. Larsen, M. Kuhlmann, M. L. Hvam and K. A. Howard, Mol Cell Ther., 2016, 4, 3 CrossRef PubMed.
- A. M. Merlot, D. S. Kalinowski and D. R. Richardson, Front. Physiol., 2014, 5, 299 Search PubMed.
- European Medicines Agency, EPAR summary: Provenge: autologous peripheral blood mononuclear cells activated with PAP-GM-CSF (sipuleucel-T), EMA/410890/2013, 2013 Search PubMed.
- J. Keenan, D. Pearson and M. Clynes, Cytotechnology, 2006, 50, 49 CrossRef CAS PubMed.
- Committee for Advanced Therapies (CAT), European Medicines Agency, Guideline on quality, non-clinical and clinical requirements for investigational advanced therapy medicinal products in clinical trials, EMA/CAT/852602/2018, 31 January 2019 Search PubMed.
- M. Guerrini, D. Beccati, Z. Shriver, A. Naggi, K. Viswanathan, A. Bisio, I. Capila, J. C. Lansing, S. Guglieri and B. Fraser, Nat. Biotechnol., 2008, 26, 669 CrossRef CAS PubMed.
- H. Yao, E. Wynendaele and B. De Spiegeleer, Drug Test. Anal., 2019, 12, 67 CrossRef PubMed.
- H. Yao, E. Wynendaele, X. Xu, A. Kosgei and B. De Spiegeleer, J. Pharm. Biomed. Anal., 2018, 147, 50 CrossRef CAS PubMed.
- N. J. Greenfield, Nat. Protoc., 2006, 1, 2527 CrossRef CAS PubMed.
- J. H. Park, T. N. Sut, J. A. Jackman, A. R. Ferhan, B. K. Yoon and N.-J. Cho, Phys. Chem. Chem. Phys., 2017, 19, 8854 RSC.
- J. H. Park, J. A. Jackman, A. R. Ferhan, G. J. Ma, B. K. Yoon and N.-J. Cho, ACS Appl. Mater. Interfaces, 2018, 10, 32047 CrossRef CAS PubMed.
- J. H. Park, A. R. Ferhan, J. A. Jackman and N.-J. Cho, Colloids Surf., B, 2019, 180, 306 CrossRef CAS PubMed.
- J. Y. B. Tan, B. K. Yoon, G. J. Ma, T. N. Sut, N.-J. Cho and J. A. Jackman, Langmuir, 2020, 36, 9215 CrossRef CAS PubMed.
- C. B. Bleustein, M. Sennett, R. T. V. Kung, D. Felsen, D. P. Poppas and R. B. Stewart, Lasers Surg. Med., 2000, 27, 465 CrossRef CAS PubMed.
- M. G. Gorobets, L. A. Wasserman, A. V. Bychkova and M. A. Rosenfeld, Chem. Phys., 2019, 523, 34 CrossRef CAS.
- L. Jiang, Z. Wang, Y. Li, X. Meng, X. Sui, B. Qi and L. Zhou, Int. J. Food Prop., 2015, 18, 1059 CrossRef CAS.
- L. Whitmore and B. A. Wallace, Biopolymers, 2008, 89, 392 CrossRef CAS PubMed.
- L. Whitmore and B. A. Wallace, Nucleic Acids Res., 2004, 32, W668 CrossRef CAS PubMed.
- N. J. Greenfield, Nat. Protoc., 2007, 1, 2876 CrossRef PubMed.
- N. J. Greenfield, Nat. Protoc., 2006, 1, 2527 CrossRef CAS PubMed.
- S. Verma, R. K. Mehta, P. Maiti, K. H. Rohm and A. Sonawane, Biochim. Biophys. Acta, 2014, 1844, 1219 CrossRef CAS PubMed.
- S. Benjwal, S. Verma, K. H. Rohm and O. Gursky, Protein Sci., 2006, 15, 635 CrossRef CAS PubMed.
- L. Facchinetti de Castro Girao, S. L. Goncalves da Rocha, R. S. Sobral, A. P. Dinis Ano Bom, A. L. Franco Sampaio, J. Godinho da Silva Jr, M. A. Ferrara, E. Pinto da Silva Bon and J. Perales, Protein Expression Purif., 2016, 120, 118 CrossRef CAS PubMed.
- N.-J. Cho, C. W. Frank, B. Kasemo and F. Höök, Nat. Protoc., 2010, 5, 1096 CrossRef CAS PubMed.
- J. A. Jackman, V. P. Zhdanov and N.-J. Cho, Langmuir, 2014, 30, 9494 CrossRef CAS PubMed.
- J. Ioannou, A. M. Donald and R. Tromp, Food Hydrocolloids, 2015, 46, 216 CrossRef CAS.
- Y. Liu, J. Lee, K. M. Mansfield, J. H. Ko, S. Sallam, C. Wesdemiotis and H. D. Maynard, Bioconjugate Chem., 2017, 28, 836 CrossRef CAS PubMed.
- M. J. Borrok, N. Mody, X. Lu, M. L. Kuhn, H. Wu, W. F. Dall'Acqua and P. Tsui, J. Pharm. Sci., 2017, 106, 1008 CrossRef CAS PubMed.
- Y. Xie, J. An, G. Yang, G. Wu, Y. Zhang, L. Cui and Y. Feng, J. Biol. Chem., 2014, 289, 7994 CrossRef CAS PubMed.
- X. t. Mai, J. Huang, J. Tan, Y. Huang and Y. Chen, J. Pept. Sci., 2015, 21, 561 CrossRef CAS PubMed.
- W. A. Chiu, D. A. Axelrad, C. Dalaijamts, C. Dockins, K. Shao, A. J. Shapiro and G. Paoli, Environ. Health Perspect., 2018, 126, 067009 CrossRef PubMed.
- L. Van Vooren, G. Krikilion, J. Rosier and B. De Spiegeleer, Drug Dev. Ind. Pharm., 2001, 27, 885 CrossRef CAS PubMed.
- B. E. Lang and K. D. Cole, Biotechnol. Prog., 2015, 31, 62 CrossRef CAS PubMed.
- A. Michnik and J. Therm, Anal. Calorim., 2003, 71, 509 CrossRef CAS.
- G. J. Ma, A. R. Ferhan, T. N. Sut, J. A. Jackman and N.-J. Cho, Colloids Surf., B, 2020, 194, 111194 CrossRef CAS PubMed.
- G. J. Ma, A. R. Ferhan, J. A. Jackman and N.-J. Cho, Communications Materials, 2020, 1, 1 CrossRef.
- G. J. Ma, A. R. Ferhan, J. A. Jackman and N.-J. Cho, Langmuir, 2020, 36, 10606 CrossRef CAS PubMed.
- J. A. Jackman, A. R. Ferhan, B. K. Yoon, J. H. Park, V. P. Zhdanov and N.-J. Cho, Anal. Chem., 2017, 89, 12976 CrossRef CAS PubMed.
- A. R. Ferhan, J. A. Jackman, T. N. Sut and N.-J. Cho, Sensors, 2018, 18, 1283 CrossRef PubMed.
- H. Park, G. J. Ma, B. K. Yoon, N.-J. Cho and J. A. Jackman, Langmuir, 2021, 37, 1306 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1ra01064f |
| This journal is © The Royal Society of Chemistry 2021 |